Toxicity Assessment of the Binary Mixtures of Aquatic Organisms Based on Different Hypothetical Descriptors
Abstract
1. Introduction
2. Results
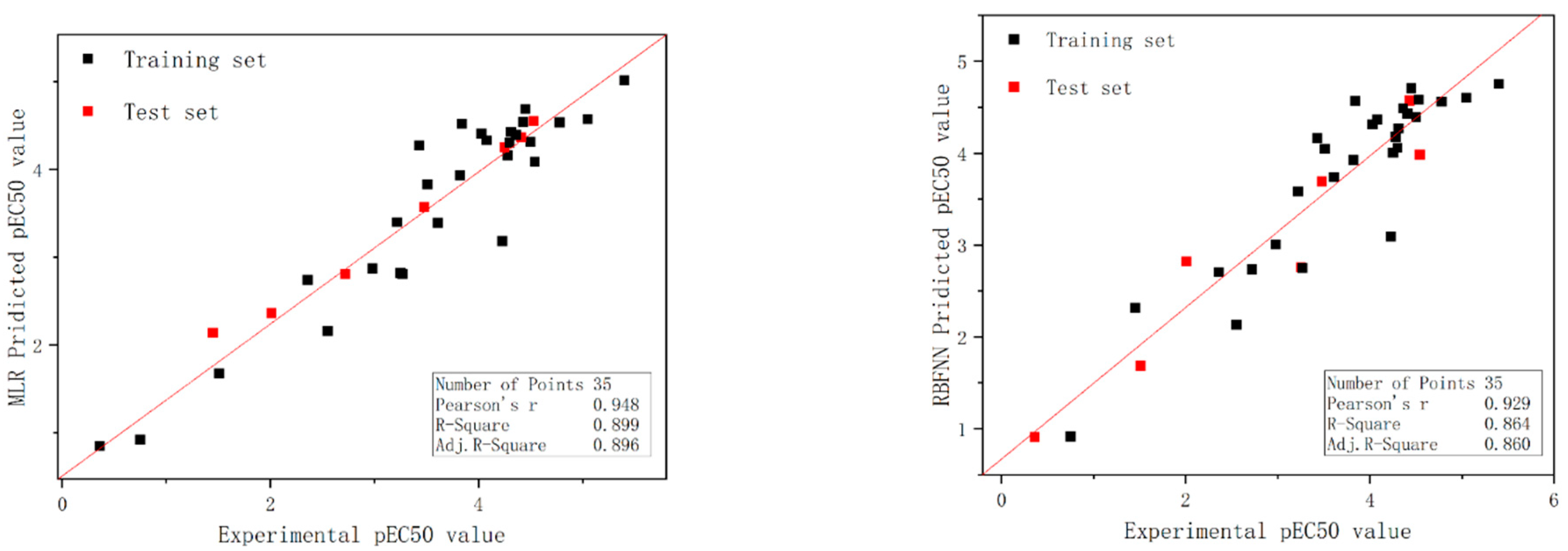
2.1. Model Development for Individual Compounds
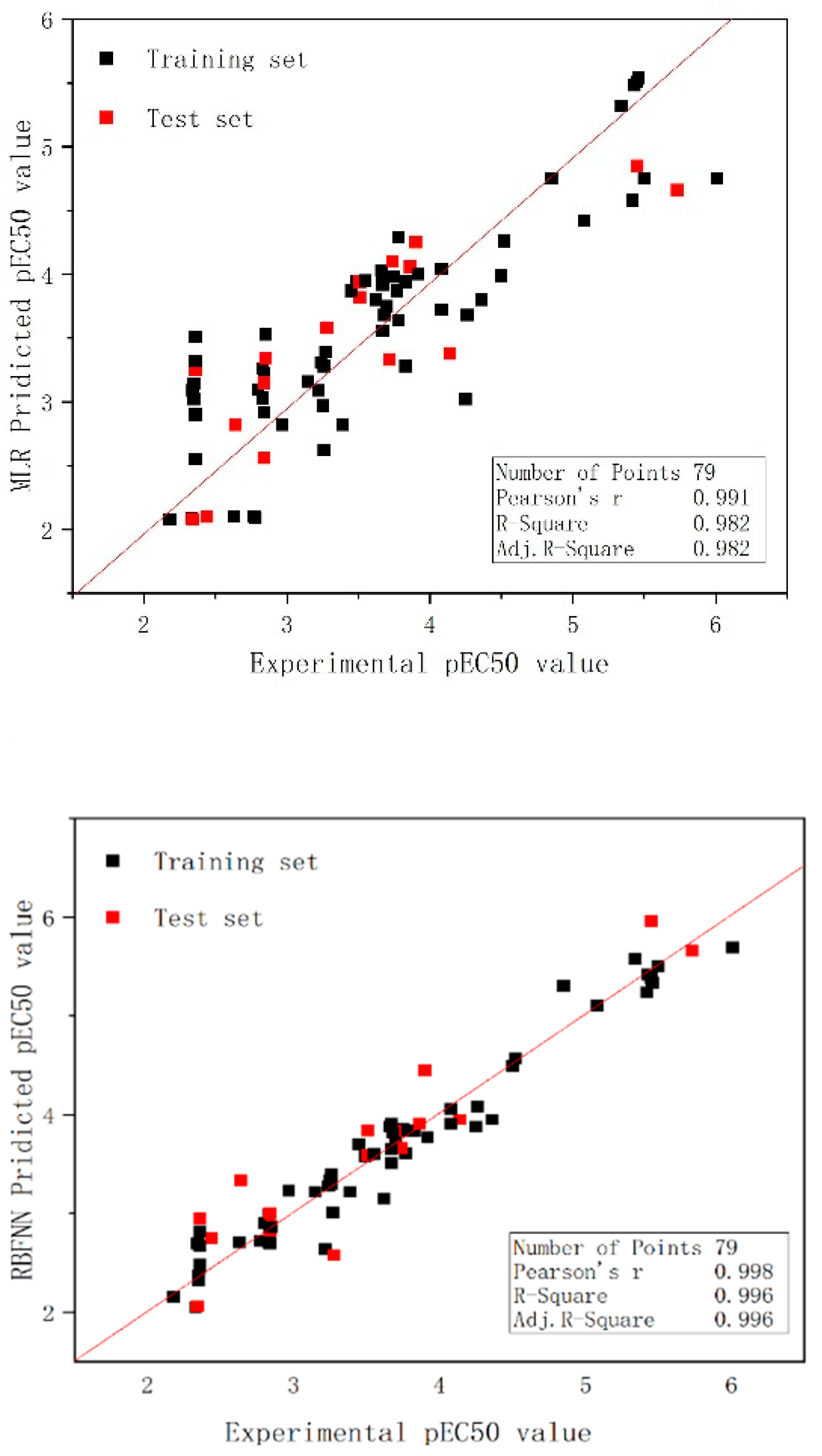
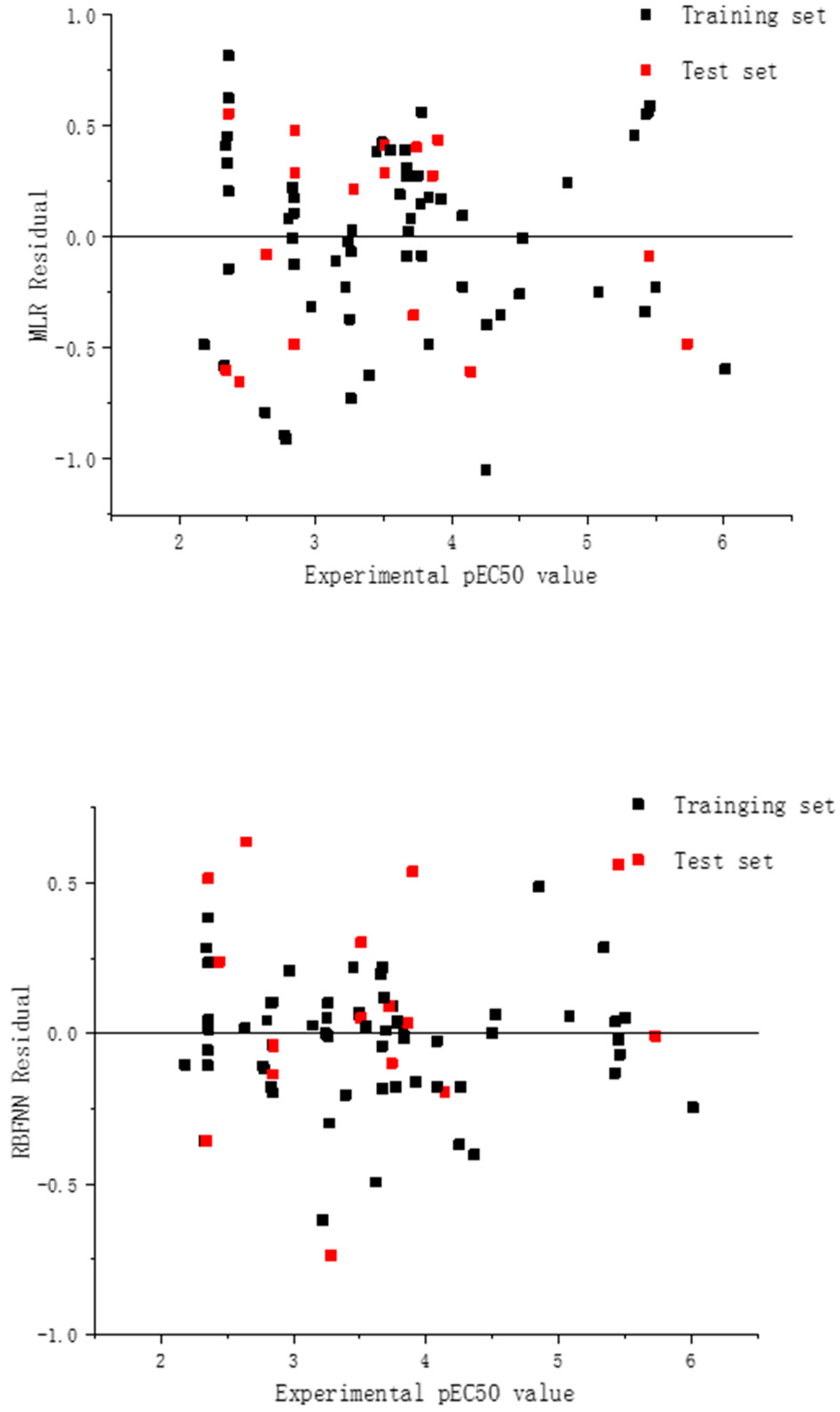
2.2. Model Development for Binary Mixture Compounds
2.3. RBFNN Results Analysis
2.4. Validation of the Models
2.5. Model Applicability Domain Analysis
2.6. Discussion of Selected Descriptors
3. Materials and Methods
3.1. Datasets
3.2. Molecular Descriptors Generation and Selection
3.3. Model Building Technique
3.3.1. Multiple Linear Regressions
3.3.2. Radial Basis Function Neural Networks (RBFNN)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Wang, T.; Zhou, X.; Wang, D.; Yin, D.; Lin, Z. Using molecular docking between organic chemicals and lipid membrane to revise the well known octanol–water partition coefficient of the mixture. Environ. Toxicol. Pharmacol. 2012, 34, 59–66. [Google Scholar] [CrossRef]
- Escher, B.I.; Baumer, A.; Bittermann, K.; Henneberger, L.; König, M.; Kühnert, C.; Klüver, N. General baseline toxicity QSAR for nonpolar, polar and ionisable chemicals and their mixtures in the bioluminescence inhibition assay with Aliivibrio fischeri. Environ. Sci. Processes Impacts 2017, 19, 414–428. [Google Scholar] [CrossRef]
- Schwarzenbach, R.P.; Escher, B.I.; Fenner, K.; Hofstetter, T.B.; Johnson, C.A.; von Gunten, U.; Wehrli, B. The challenge of micropollutants in aquatic systems. Science 2006, 313, 1072–1077. [Google Scholar] [CrossRef]
- Rand, G.M.; Wells, P.G.; McCarty, L.S. Introduction to Aquatic Toxicology//Fundamentals of Aquatic Toxicology; CRC Press: Boca Raton, FL, USA, 2020; pp. 3–67. [Google Scholar] [CrossRef]
- Yang, R.S.; Thomas, R.S.; Gustafson, D.L.; Campain, J.; A Benjamin, S.; Verhaar, H.J.; Mumtaz, M.M. Approaches to developing alternative and predictive toxicology based on PBPK/PD and QSAR modeling. Environ. Health Perspect. 1998, 106 (Suppl. 6), 1385–1393. [Google Scholar] [CrossRef]
- Heys, K.A.; Shore, R.F.; Pereira, M.G.; Jones, K.C.; Martin, F.L. Risk assessment of environmental mixture effects. RSC Adv. 2016, 6, 47844–47857. [Google Scholar] [CrossRef]
- Usepa, U. Guidelines for the health risk assessment of chemical mixtures. Fed. Regist. 1986, 51, 34014–34025. [Google Scholar]
- Kortenkamp, A.; Backhaus, T.; Faust, M. State of the art report on mixture toxicity. Contract 2009, 70307, 94–103. [Google Scholar]
- Altenburger, R.; Backhaus, T.; Boedeker, W.; Faust, M.; Scholze, M.; Grimme, L.H. Predictability of the toxicity of multiple chemical mixtures to Vibrio fischeri: Mixtures composed of similarly acting chemicals. Environ. Toxicol. Chem. Int. J. 2000, 19, 2341–2347. [Google Scholar] [CrossRef]
- Bliss, C.I. The toxicity of poisons jointly applied. Ann. Appl. Biol. 1939, 26, 585–615. [Google Scholar] [CrossRef]
- Bureš, M.S.; Ukić, Š.; Cvetnić, M.; Prevarić, V.; Markić, M.; Rogošić, M.; Kušić, H.; Bolanča, T. Toxicity of binary mixtures of pesticides and pharmaceuticals toward Vibrio fischeri: Assessment by quantitative structure-activity relationships. Environ. Pollut. 2021, 275, 115885. [Google Scholar] [CrossRef] [PubMed]
- Luan, F.; Xu, X.; Liu, H.; Cordeiro, M.N.D.S. Prediction of the baseline toxicity of non-polar narcotic chemical mixtures by QSAR approach. Chemosphere 2013, 90, 1980–1986. [Google Scholar] [CrossRef] [PubMed]
- Roy, K.; Kar, S.; Das, R.N. A Primer on QSAR/QSPR Modeling: Fundamental Concepts; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar] [CrossRef]
- Chatterjee, M.; Roy, K. Prediction of aquatic toxicity of chemical mixtures by the QSAR approach using 2D structural descriptors. J. Hazard. Mater. 2021, 408, 124936. [Google Scholar] [CrossRef] [PubMed]
- Khan, P.M.; Rasulev, B.; Roy, K. QSPR modeling of the refractive index for diverse polymers using 2D descriptors. ACS Omega 2018, 3, 13374–13386. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, H.; Yang, X.; Wang, L. Aquatic toxicity and aquatic ecological risk assessment of wastewater-derived halogenated phenolic disinfection byproducts. Sci. Total Environ. 2022, 809, 151089. [Google Scholar] [CrossRef] [PubMed]
- Cassani, S.; Kovarich, S.; Papa, E.; Roy, P.P.; Rahmberg, M.; Nilsson, S.; Sahlin, U.; Jeliazkova, N.; Kochev, N.; Pukalov, O.; et al. Evaluation of CADASTER QSAR models for the aquatic toxicity of (benzo) triazoles and prioritisation by consensus prediction. Altern. Lab. Anim. 2013, 41, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.; Benfenati, E.; Roy, K. Consensus QSAR modeling of toxicity of pharmaceuticals to different aquatic organisms: Ranking and prioritization of the DrugBank database compounds. Ecotoxicol. Environ. Saf. 2019, 168, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Topliss, J.G.; Edwards, R.P. Chance factors in studies of quantitative structure-activity relationships. J. Med. Chem. 1979, 22, 1238–1244. [Google Scholar] [CrossRef]
- Yao, Z.; Lin, Z.; Wang, T.; Tian, D.; Zou, X.; Gao, Y.; Yin, D. Using molecular docking-based binding energy to predict toxicity of binary mixture with different binding sites. Chemosphere 2013, 92, 1169–1176. [Google Scholar] [CrossRef]
- Kaneko, H. Estimation of predictive performance for test data in applicability domains using y-randomization. J. Chemom. 2019, 33, e3171. [Google Scholar] [CrossRef]
- Roy, K.; Kar, S.; Das, R.N. Understanding the Basics of QSAR for Applications in Pharmaceutical Sciences and Risk Assessment; Academic Press: Cambridge, MA, USA, 2015. [Google Scholar] [CrossRef]
- ChemDraw Professional 20.0.0.41; PerkinElmer Informatics, Inc.: Massachusetts, MA, USA, 2011.
- HyperChem 6.01; Hypercube, Inc.: Waterloo, ON, Canada, 2000.
- Stewart, J.P.P. MOPAC6.0, Quantum Chemistry Program Exchange, No.455; Indiana University: Bloomington, IN, USA, 1989. [Google Scholar]
- Katritzky, A.R.; Lobanov, V.S.; Karelson, M. CODESSA 2.63: T Raining Manual; University of Florida: Gainesville, FL, USA, 1995. [Google Scholar]
- Muratov, E.N.; Varlamova, E.V.; Artemenko, A.G.; Polishchuk, P.G.; Kuz’Min, V.E. Existing and developing approaches for QSAR analysis of mixtures. Mol. Inform. 2012, 31, 202–221. [Google Scholar] [CrossRef]
- Ajmani, S.; Rogers, S.C.; Barley, M.H.; Livingstone, D.J. Application of QSPR to mixtures. J. Chem. Inf. Modeling 2006, 46, 2043–2055. [Google Scholar] [CrossRef] [PubMed]
- Gaudin, T.; Rotureau, P.; Fayet, G. Mixture descriptors toward the development of quantitative structure–property relationship models for the flash points of organic mixtures. Ind. Eng. Chem. Res. 2015, 54, 6596–6604. [Google Scholar] [CrossRef]
- Qin, L.T.; Chen, Y.H.; Zhang, X.; Mo, L.Y.; Zeng, H.H.; Liang, Y.P. QSAR prediction of additive and non-additive mixture toxicities of antibiotics and pesticide. Chemosphere 2018, 198, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Sobati, M.A.; Abooali, D.; Maghbooli, B.; Najafi, H. A new structure-based model for estimation of true critical volume of multi-component mixtures. Chemom. Intell. Lab. Syst. 2016, 155, 109–119. [Google Scholar] [CrossRef]
- Zeng, X.; Zhen, Z.; He, J.; Han, L. A feature selection approach based on sensitivity of RBFNNs. Neurocomputing 2018, 275, 2200–2208. [Google Scholar] [CrossRef]
- Derks, E.; Pastor, M.S.S.; Buydens, L.M.C. Robustness analysis of radial base function and multi-layered feed-forward neural network models. Chemom. Intell. Lab. Syst. 1995, 28, 49–60. [Google Scholar] [CrossRef]
- Wang, T.; Tang, L.; Luan, F.; Cordeiro, M.N.D.S. Prediction of the toxicity of binary mixtures by QSAR approach using the hypothetical descriptors. Int. J. Mol. Sci. 2018, 19, 3423. [Google Scholar] [CrossRef]


| NO. | Equation | R2 | R2adj | F | Q2LOO |
|---|---|---|---|---|---|
| 1 | 0.730 | 0.711 | 39.395 | 0.731 | |
| 2 | 0.754 | 0.737 | 44.663 | 0.756 | |
| 3 | 0.735 | 0.716 | 40.419 | 0.805 | |
| 4 | 0.525 | 0.493 | 16.145 | 0.498 | |
| 5 | 0.569 | 0.539 | 19.257 | 0.546 | |
| 6 | 0.581 | 0.552 | 20.225 | 0.557 | |
| 7 | 0.561 | 0.531 | 18.681 | 0.541 | |
| 8 | 0.734 | 0.716 | 40.304 | 0.737 | |
| 9 | 0.726 | 0.707 | 38.665 | 0.727 | |
| 10 | 0.700 | 0.679 | 34.016 | 0.704 | |
| 11 | 0.715 | 0.696 | 36.689 | 0.719 |
| Nos. | Individual Compounds | MLR | RBFNN | ||||
|---|---|---|---|---|---|---|---|
| Toxicity (pEC50 (mol/L)) | Toxicity (pEC50 (mol/L)) | ||||||
| Experimental | Predicted | Residual | Experimental | Predicted | Residual | ||
| 1 | Acetaldehyde | 2.36 | 2.74 | −0.38 | 2.36 | 2.7 | −0.34 |
| 2 | Propionaldehyde* | 2.72 | 2.81 | −0.09 | 2.72 | 2.74 | −0.02 |
| 3 | Butyraldehyde | 3.25 | 2.82 | 0.43 | 3.25 | 2.76 | 0.49 |
| 4 | Valeraldehyde | 3.27 | 2.81 | 0.46 | 3.27 | 2.75 | 0.52 |
| 5 | Benzaldehyde | 3.43 | 4.27 | −0.84 | 3.43 | 4.17 | −0.74 |
| 6 | p-Nitrobenzaldehyde | 4.28 | 4.17 | 0.11 | 4.28 | 4.17 | 0.11 |
| 7 | p-Terephthaldehyde | 4.31 | 4.43 | −0.12 | 4.31 | 4.27 | 0.04 |
| 8 | p-Chlorobenzaldehyde* | 4.25 | 4.25 | 0 | 4.25 | 4 | 0.25 |
| 9 | p-Bromobenzaldehyde | 4.3 | 4.31 | −0.01 | 4.3 | 4.06 | 0.24 |
| 10 | p-Hydrobenzaldehyde | 4.54 | 4.09 | 0.45 | 4.54 | 3.98 | 0.56 |
| 11 | p-Methylbenzaldehyde | 3.82 | 3.93 | −0.11 | 3.82 | 3.93 | −0.11 |
| 12 | p-Methoxybenzaldehyde | 4.03 | 4.41 | −0.38 | 4.03 | 4.31 | −0.28 |
| 13 | p-Dimethylaminobenzaldehyde | 5.4 | 5.02 | 0.38 | 5.4 | 4.75 | 0.65 |
| 14 | Malononitrile | 2.55 | 2.16 | 0.39 | 2.55 | 2.13 | 0.42 |
| 15 | Glycolonitrile | 2.98 | 2.87 | 0.11 | 2.98 | 3.01 | −0.03 |
| 16 | α-Hydroxyisobutyronitrile | 3.61 | 3.39 | 0.22 | 3.61 | 3.74 | −0.13 |
| 17 | Allyl cyanide* | 1.45 | 2.14 | −0.69 | 1.45 | 2.32 | −0.87 |
| 18 | Benzonitrile* | 3.48 | 3.57 | −0.09 | 3.48 | 3.69 | −0.21 |
| 19 | Benzyl cyanide | 4.23 | 3.18 | 1.05 | 4.23 | 3.09 | 1.14 |
| 20 | Acetonitrile | 0.75 | 0.92 | −0.17 | 0.75 | 0.92 | −0.17 |
| 21 | Acrylonitrile | 1.51 | 1.68 | −0.17 | 1.51 | 1.69 | −0.18 |
| 22 | Succinonitrile | 0.36 | 0.85 | −0.49 | 0.36 | 0.91 | −0.55 |
| 23 | Phthalonitrile | 3.51 | 3.83 | −0.32 | 3.51 | 4.05 | −0.54 |
| 24 | Lactonitrile* | 2.01 | 2.36 | −0.35 | 2.01 | 2.82 | −0.81 |
| 25 | Sulfamethazine | 4.08 | 4.33 | −0.25 | 4.08 | 4.37 | −0.29 |
| 26 | Sulfapyridine | 3.84 | 4.52 | −0.68 | 3.84 | 4.57 | −0.73 |
| 27 | Sulfamethoxazole | 4.45 | 4.69 | −0.24 | 4.45 | 4.7 | −0.25 |
| 28 | Sulfadiazine | 4.5 | 4.32 | 0.18 | 4.5 | 4.39 | 0.11 |
| 29 | Sulfisoxazole | 4.43 | 4.54 | −0.11 | 4.43 | 4.57 | −0.14 |
| 30 | Sulfamonomethoxine | 5.05 | 4.58 | 0.47 | 5.05 | 4.6 | 0.45 |
| 31 | Sulfachloropyridazine | 4.78 | 4.54 | 0.24 | 4.78 | 4.56 | 0.22 |
| 32 | Sulfachinoxalin* | 4.53 | 4.56 | −0.03 | 4.53 | 4.58 | −0.05 |
| 33 | Sulfamethoxydiazine* | 4.41 | 4.37 | 0.04 | 4.41 | 4.43 | −0.02 |
| 34 | Sulfamethoxypyridazine | 4.36 | 4.4 | −0.04 | 4.36 | 4.49 | −0.13 |
| 35 | Trimethoprim | 3.22 | 3.4 | −0.18 | 3.22 | 3.58 | −0.36 |
| Rn-N | HOMO | Tot-pc | Min-C | Max-C-H | |
|---|---|---|---|---|---|
| Rn-N | +1.000 | ||||
| HOMO | −0.297 | +1.000 | |||
| Tot-pc | +0.217 | +0.630 | +1.000 | ||
| Min-C | −0.049 | −0.622 | −0.696 | +1.000 | |
| Max-C-H | −0.423 | +0.311 | +0.197 | −0.145 | +1.000 |
| MLR | RBFNN | |
|---|---|---|
| R2 | 0.721 | 0.880 |
| F | 38.773 | 110.980 |
| K | 0.999 | 1.030 |
| RMS q2ext | 0.508 0.720 | 0.367 0.853 |
| Mixture No. | Chemicals in the Mixture | The Ratio of Toxic Unit | Experimental pEC50 Mix (mol/L) | MLR | RBFNN | ||
|---|---|---|---|---|---|---|---|
| Toxicity (pEC50 (mol/L)) | Toxicity (pEC50 (mol/L)) | ||||||
| Predicted | Residual | Predicted | Residual | ||||
| 1 * | 1\14 | 1\1 | 2.44 | 2.1 | 0.34 | 2.75 | −0.31 |
| 2 | 2\14 | 1\1 | 2.63 | 2.1 | 0.53 | 2.71 | −0.08 |
| 3 | 3\14 | 1\1 | 2.77 | 2.1 | 0.67 | 2.72 | 0.05 |
| 4 | 4\14 | 1\1 | 2.78 | 2.09 | 0.69 | 2.72 | 0.06 |
| 5 | 5\14 | 1\1 | 2.8 | 3.1 | −0.3 | 2.9 | −0.1 |
| 6 * | 6\14 | 1\1 | 2.84 | 2.56 | 0.28 | 2.76 | 0.08 |
| 7 | 7\14 | 1\1 | 2.84 | 2.92 | −0.08 | 2.86 | −0.02 |
| 8 | 8\14 | 1\1 | 2.83 | 3.26 | −0.43 | 2.71 | 0.12 |
| 9 | 9\14 | 1\1 | 2.84 | 3.22 | −0.38 | 2.7 | 0.14 |
| 10 * | 10\14 | 1\1 | 2.85 | 3.34 | −0.49 | 2.87 | −0.02 |
| 11 | 11\14 | 1\1 | 2.83 | 3.03 | −0.2 | 2.99 | −0.16 |
| 12 * | 12\14 | 1\1 | 2.84 | 3.15 | −0.31 | 3 | −0.16 |
| 13 | 13\14 | 1\1 | 2.85 | 3.53 | −0.68 | 2.86 | −0.01 |
| 14 | 5\15 | 1\1 | 3.15 | 3.16 | −0.01 | 3.22 | −0.07 |
| 15 | 6\15 | 1\1 | 3.26 | 2.62 | 0.64 | 3.4 | −0.14 |
| 16 | 7\15 | 1\1 | 3.25 | 2.97 | 0.28 | 3.34 | −0.09 |
| 17 | 8\15 | 1\1 | 3.24 | 3.31 | −0.07 | 3.28 | −0.04 |
| 18 | 9\15 | 1\1 | 3.26 | 3.28 | −0.02 | 3.29 | −0.03 |
| 19 | 10\15 | 1\1 | 3.27 | 3.39 | −0.12 | 3.01 | 0.26 |
| 20 | 11\15 | 1\1 | 3.22 | 3.09 | 0.13 | 2.64 | 0.58 |
| 21 * | 13\15 | 1\1 | 3.28 | 3.58 | −0.3 | 2.58 | 0.7 |
| 22 * | 1\16 | 1\1 | 2.64 | 2.82 | −0.18 | 3.34 | −0.7 |
| 23 | 2\16 | 1\1 | 2.97 | 2.82 | 0.15 | 3.23 | −0.26 |
| 24 | 3\16 | 1\1 | 3.39 | 2.82 | 0.57 | 3.22 | 0.17 |
| 25 * | 5\16 | 1\1 | 3.51 | 3.82 | −0.31 | 3.84 | −0.33 |
| 26 | 6\16 | 1\1 | 3.83 | 3.28 | 0.55 | 3.84 | −0.01 |
| 27 | 7\16 | 1\1 | 3.78 | 3.64 | 0.14 | 3.83 | −0.05 |
| 28 | 8\16 | 1\1 | 3.75 | 3.98 | −0.23 | 3.86 | −0.11 |
| 29 | 9\16 | 1\1 | 3.83 | 3.94 | −0.11 | 3.83 | 0 |
| 30 * | 10\16 | 1\1 | 3.86 | 4.06 | −0.2 | 3.91 | −0.05 |
| 31 | 11\16 | 1\1 | 3.7 | 3.75 | −0.05 | 3.73 | −0.03 |
| 32 | 12\16 | 1\1 | 3.77 | 3.87 | −0.1 | 3.61 | 0.16 |
| 33* | 13\16 | 1\1 | 3.9 | 4.25 | −0.35 | 4.45 | −0.55 |
| 34 | 1\17 | 1\1 | 2.18 | 2.08 | 0.1 | 2.16 | 0.02 |
| 35 | 3\17 | 1\1 | 2.33 | 2.09 | 0.24 | 2.05 | 0.28 |
| 36 * | 4\17 | 1\1 | 2.34 | 2.08 | 0.26 | 2.06 | 0.28 |
| 37 | 5\17 | 1\1 | 2.34 | 3.09 | −0.75 | 2.7 | −0.36 |
| 38 | 6\17 | 1\1 | 2.36 | 2.55 | −0.19 | 2.48 | −0.12 |
| 39 | 7\17 | 1\1 | 2.36 | 2.9 | −0.54 | 2.67 | −0.31 |
| 40 * | 8\17 | 1\1 | 2.36 | 3.25 | −0.89 | 2.95 | −0.59 |
| 41 | 10\17 | 1\1 | 2.36 | 3.32 | −0.96 | 2.82 | −0.46 |
| 42 | 11\17 | 1\1 | 2.35 | 3.02 | −0.67 | 2.32 | 0.03 |
| 43 | 12\17 | 1\1 | 2.35 | 3.14 | −0.79 | 2.37 | −0.02 |
| 44 | 13\17 | 1\1 | 2.36 | 3.51 | −1.15 | 2.45 | −0.09 |
| 45 | 5\18 | 1\1 | 3.45 | 3.87 | −0.42 | 3.7 | −0.25 |
| 46 * | 6\18 | 1\1 | 3.72 | 3.33 | 0.39 | 3.83 | −0.11 |
| 47 | 7\18 | 1\1 | 3.68 | 3.68 | 0 | 3.82 | −0.14 |
| 48 | 8\18 | 1\1 | 3.66 | 4.03 | −0.37 | 3.88 | −0.22 |
| 49 * | 10\18 | 1\1 | 3.74 | 4.1 | −0.36 | 3.66 | 0.08 |
| 50 | 11\18 | 1\1 | 3.62 | 3.8 | −0.18 | 3.15 | 0.47 |
| 51 | 12\18 | 1\1 | 3.67 | 3.92 | −0.25 | 3.51 | 0.16 |
| 52 | 13\18 | 1\1 | 3.78 | 4.29 | −0.51 | 3.84 | −0.06 |
| 53 | 5\19 | 1\1 | 3.67 | 3.56 | 0.11 | 3.91 | −0.24 |
| 54 | 6\19 | 1\1 | 4.25 | 3.02 | 1.23 | 3.88 | 0.37 |
| 55 * | 7\19 | 1\1 | 4.14 | 3.38 | 0.76 | 3.95 | 0.19 |
| 56 | 8\19 | 1\1 | 4.08 | 3.72 | 0.36 | 4.06 | 0.02 |
| 57 | 9\19 | 1\1 | 4.26 | 3.68 | 0.58 | 4.08 | 0.18 |
| 58 | 10\19 | 1\1 | 4.36 | 3.8 | 0.56 | 3.95 | 0.41 |
| 59 | 13\19 | 1\1 | 4.5 | 3.99 | 0.51 | 4.49 | 0.01 |
| 60 | 25\35 | 1\1 | 5.08 | 4.42 | 0.66 | 5.1 | −0.02 |
| 61 | 26\35 | 1\1 | 4.85 | 4.75 | 0.1 | 5.31 | −0.46 |
| 62 | 27\35 | 1\1 | 5.5 | 4.75 | 0.75 | 5.5 | 0 |
| 63 | 28\35 | 1\1 | 5.42 | 4.58 | 0.84 | 5.24 | 0.18 |
| 64 * | 29\35 | 1\1 | 5.45 | 4.85 | 0.6 | 5.96 | −0.51 |
| 65 | 30\35 | 1\1 | 6.01 | 4.75 | 1.26 | 5.69 | 0.32 |
| 66 * | 31\35 | 1\1 | 5.73 | 4.66 | 1.07 | 5.66 | 0.07 |
| 67 | 27\35 | 13396\1 | 3.49 | 3.94 | −0.45 | 3.58 | −0.09 |
| 68 | 27\35 | 8587\1 | 3.49 | 3.94 | −0.45 | 3.58 | −0.09 |
| 69 | 27\35 | 2747\1 | 3.49 | 3.94 | −0.45 | 3.59 | −0.1 |
| 70 * | 27\35 | 858\1 | 3.51 | 3.94 | −0.43 | 3.59 | −0.08 |
| 71 | 27\35 | 274\1 | 3.55 | 3.95 | −0.4 | 3.6 | −0.05 |
| 72 | 27\35 | 85\1 | 3.67 | 3.96 | −0.29 | 3.65 | 0.02 |
| 73 | 27\35 | 27\1 | 3.92 | 4 | −0.08 | 3.77 | 0.15 |
| 74 | 27\35 | 15\1 | 4.08 | 4.04 | 0.04 | 3.91 | 0.17 |
| 75 | 27\35 | 4\1 | 4.52 | 4.26 | 0.26 | 4.57 | −0.05 |
| 76 | 27\35 | 1\6 | 5.34 | 5.32 | 0.02 | 5.58 | −0.24 |
| 77 | 27\35 | 1\21 | 5.43 | 5.48 | −0.05 | 5.42 | 0.01 |
| 78 | 27\35 | 1\37 | 5.45 | 5.51 | −0.06 | 5.38 | 0.07 |
| 79 | 27\35 | 1\116 | 5.46 | 5.54 | −0.08 | 5.34 | 0.12 |
| MLR | RBFNN | ||||
|---|---|---|---|---|---|
| R2 | RMS | MAE | R2 | RMS | MAE |
| 0.027 | 1.347 | 1.130 | 0.139 | 1.549 | 1.258 |
| 0.013 | 1.318 | 1.130 | 0.117 | 1.531 | 1.226 |
| 0.077 | 1.410 | 1.212 | 0.120 | 1.533 | 1.226 |
| 0.089 | 1.421 | 1.153 | 0.149 | 1.556 | 1.282 |
| 0.071 | 1.404 | 1.155 | 0.141 | 1.550 | 1.258 |
| 0.001 | 1.263 | 1.101 | 0.139 | 1.549 | 1.247 |
| 0.057 | 1.388 | 1.136 | 0.174 | 1.573 | 1.325 |
| 0.084 | 1.416 | 1.187 | 0.170 | 1.571 | 1.267 |
| 0.061 | 1.392 | 1.146 | 0.145 | 1.553 | 1.258 |
| 0.040 | 1.367 | 1.156 | 0.167 | 1.569 | 1.280 |
| Training Set | R2 | F | RMS | Test Set | R2 | F | RMS |
|---|---|---|---|---|---|---|---|
| B + C + D + T | 0.711 | 150.419 | 0.506 | A | 0.790 | 52.605 | 0.458 |
| A + C + D + T | 0.712 | 150.482 | 0.513 | B | 0.799 | 55.744 | 0.426 |
| A + B + D + T | 0.723 | 158.899 | 0.506 | C | 0.752 | 42.370 | 0.456 |
| A + B + C + T | 0.743 | 176.638 | 0.478 | D | 0.674 | 28.902 | 0.563 |
| A + B + C + D | 0.744 | 180.233 | 0.479 | T | 0.674 | 26.113 | 0.566 |
| Average | 0.727 | 163.334 | 0.496 | 0.737 | 41.147 | 0.494 |
| Training Set | R2 | F | RMS | Test Set | R2 | F | RMS |
|---|---|---|---|---|---|---|---|
| B + C + D + T | 0.937 | 912.852 | 0.237 | A | 0.931 | 187.779 | 0.277 |
| A + C + D + T | 0.932 | 837.216 | 0.212 | B | 0.956 | 306.656 | 0.216 |
| A + B + D + T | 0.933 | 845.700 | 0.252 | C | 0.944 | 235.582 | 0.218 |
| A + B+ C + T | 0.929 | 799.833 | 0.254 | D | 0.958 | 317.511 | 0.207 |
| A + B+ C + D | 0.942 | 1014.217 | 0.231 | T | 0.904 | 122.236 | 0.298 |
| Average | 0.935 | 881.963 | 0.237 | 0.939 | 233.952 | 0.243 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, M.; Zhang, L.; Zhuang, X.; Tian, C.; Luan, F.; Cordeiro, M.N.D.S. Toxicity Assessment of the Binary Mixtures of Aquatic Organisms Based on Different Hypothetical Descriptors. Molecules 2022, 27, 6389. https://doi.org/10.3390/molecules27196389
Ji M, Zhang L, Zhuang X, Tian C, Luan F, Cordeiro MNDS. Toxicity Assessment of the Binary Mixtures of Aquatic Organisms Based on Different Hypothetical Descriptors. Molecules. 2022; 27(19):6389. https://doi.org/10.3390/molecules27196389
Chicago/Turabian StyleJi, Meng, Lihong Zhang, Xuming Zhuang, Chunyuan Tian, Feng Luan, and Maria Natália D. S. Cordeiro. 2022. "Toxicity Assessment of the Binary Mixtures of Aquatic Organisms Based on Different Hypothetical Descriptors" Molecules 27, no. 19: 6389. https://doi.org/10.3390/molecules27196389
APA StyleJi, M., Zhang, L., Zhuang, X., Tian, C., Luan, F., & Cordeiro, M. N. D. S. (2022). Toxicity Assessment of the Binary Mixtures of Aquatic Organisms Based on Different Hypothetical Descriptors. Molecules, 27(19), 6389. https://doi.org/10.3390/molecules27196389







