Application of Causality Modelling for Prediction of Molecular Properties for Textile Dyes Degradation by LPMO
Abstract
:1. Introduction
2. Experimental
2.1. Enzyme
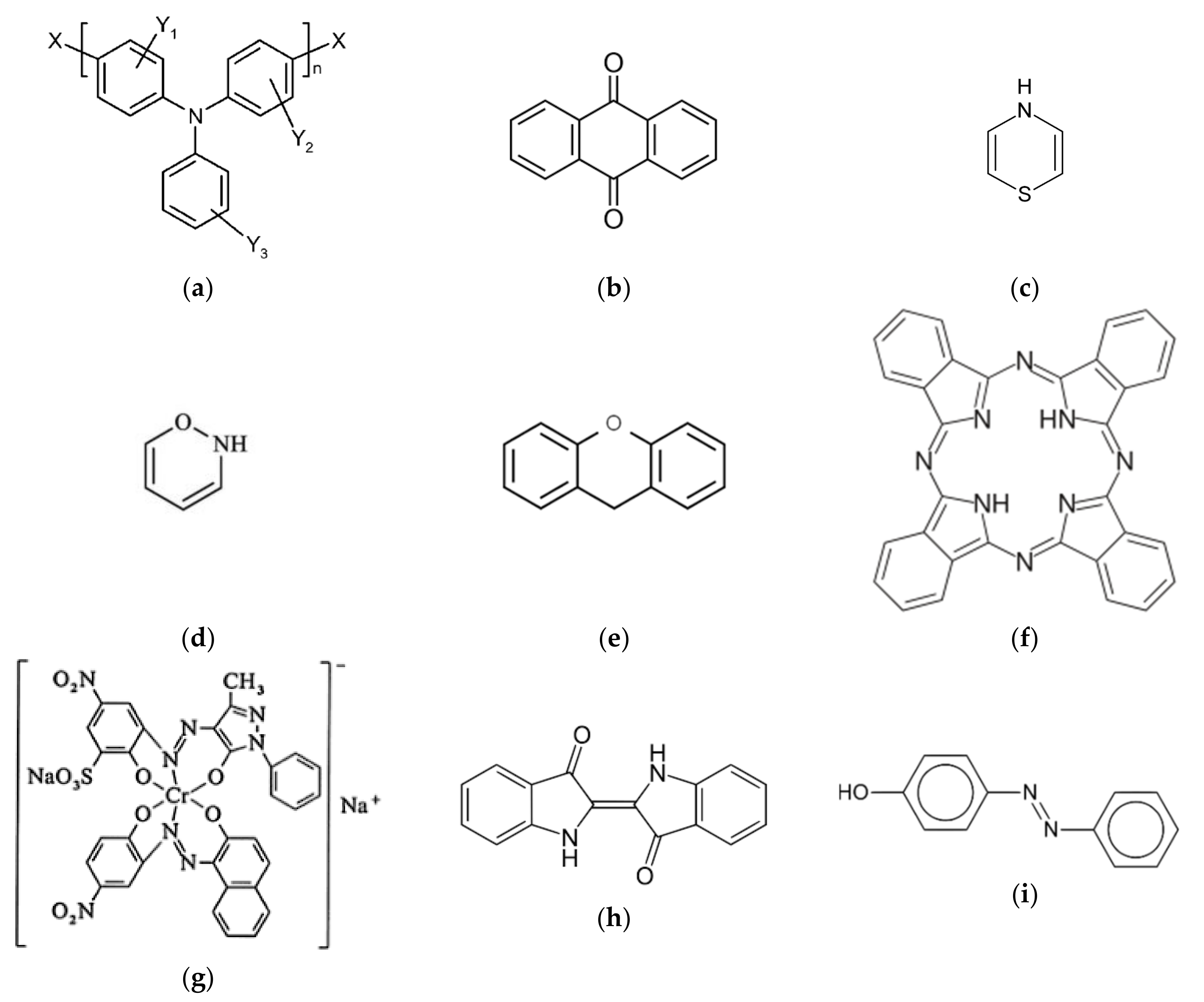
2.2. Dyes
2.3. Sample Preparation and Screening
2.4. Calculation of Molecular Descriptors
3. Results and Discussion
3.1. Establishment and Validation of SCM Models for the Prediction of the Molecular Properties Important for the Enzymatic Degradation of Days
3.2. Utilisation of SCM and DTM Models for the Prediction of the Molecular Properties Important for the LPMO Degradation of Days
3.3. Effect of Textile Dyes Molecular Descriptors on LPMO Activity
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Slama, H.B.; Bouket, A.C.; Pourhassan, Z.; Alenezi, F.N.; Silini, A.; Cherif-Silini, H.; Oszako, T.; Luptakova, L.; Golińska, L.; Belbahri, P. Diversity of Synthetic Dyes from Textile Industries, Discharge Impacts and Treatment Methods. Appl. Sci. 2021, 11, 6255. [Google Scholar] [CrossRef]
- Majumdar, R.; Shaikh, W.A.; Chakraborty, S.; Chowdhury, S. Chapter 12—A review on microbial potential of toxic azo dyes bioremediation in aquatic system. In Microbial Biodegradation and Bioremediation; Das, S., Dash, H.R., Eds.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 241–261. [Google Scholar]
- Robinson, P.K. Enzymes: Principles and biotechnological applications. Essays Biochem. 2015, 59, 1–41. [Google Scholar] [CrossRef] [PubMed]
- Bahtiyari, M.I.; Körlü, A.E.; Bilisik, K. Bioprocessing of natural textile fibres and clothes. In Fundamentals of Natural Fibres and Textiles; Woodhead Publishing: Sawston, UK, 2021; pp. 221–262. [Google Scholar]
- El-Gendi, H.; Saleh, A.K.; Badierah, R.; Redwan, E.M.; El-Maradny, Y.A.; El-Fakharany, E.M. A Comprehensive Insight into Fungal Enzymes: Structure, Classification, and Their Role in Mankind’s Challenges. J. Fungi 2022, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Kabir, S.M.M.; Koh, J. Sustainable Textile Processing by Enzyme Applications. In Biodegradation Technology of Organic and Inorganic Pollutants; Mendes, P.K.F., de Sousa, R.N., Mielke, K.C., Eds.; IntechOpen: London, UK, 2021. [Google Scholar]
- Wilson, D.B. Cellulases, Encyclopedia of Microbiology, 3rd ed.; Academic Press: Cambridge, MA, USA, 2009; pp. 252–258. [Google Scholar]
- Kumar, D.; Bhardwaj, R.; Jassal, S.; Goyal, T.; Khullar, A.; Gupta, N. Application of enzymes for an eco-friendly approach to textile processing. Environ. Sci. Pollut. Res. Int. 2021, 14, 11. [Google Scholar] [CrossRef]
- Vaaje-Kolstad, G.; Westereng, B.; Horn, S.J.; Liu, Z.; Zhai, H.; Sorlie, M.; Eijsink, V.G. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 2010, 330, 219–222. [Google Scholar] [CrossRef]
- Singhania, R.R.; Dixit, P.; Patel, A.K.; Giri, B.S.; Kuo, C.H.; Chen, C.W.; Dong, C.D. Role and significance of lytic polysaccharide monooxygenases (LPMOs) in lignocellulose deconstruction. Bioresour. Technol. 2021, 335, 125261. [Google Scholar] [CrossRef]
- Li, F.; Zhao, H.; Shao, R.; Zhang, X.; Yu, H. Enhanced Fenton Reaction for Xenobiotic Compounds and Lignin Degradation Fueled by Quinone Redox Cycling by Lytic Polysaccharide Monooxygenases. J. Agric. Food Chem. 2021, 69, 7104–7114. [Google Scholar] [CrossRef]
- Ikram, M.; Naeem, M.; Zahoor, M.; Rahim, A.; Hanafiah, M.M.; Oyekanmi, A.A.; Shah, A.B.; Mahnashi, M.H.; Al Ali, A.; Jalal, N.A.; et al. Biodegradation of Azo Dye Methyl Red by Pseudomonas aeruginosa: Optimization of Process Conditions. Int. J. Environ. Res. Public Health 2022, 19, 9962. [Google Scholar] [CrossRef]
- Ikram, M.; Naeem, M.; Zahoor, M.; Hanafiah, M.M.; Oyekanmi, A.A.; Ullah, R.; Farraj, D.A.A.; Elshikh, M.S.; Zekker, I.; Gulfam, N. Biological Degradation of the Azo Dye Basic Orange 2 by Escherichia coli: A Sustainable and Ecofriendly Approach for the Treatment of Textile Wastewater. Water 2022, 14, 2063. [Google Scholar] [CrossRef]
- Ikram, M.; Naeem, M.; Zahoor, M.; Hanafiah, M.M.; Oyekanmi, A.A.; Ullah, R.; Farraj, D.A.A.; Elshikh, M.S.; Zekker, I.; Gulfam, N. Correction: Ikram et al. Biological Degradation of the Azo Dye Basic Orange 2 by Escherichia coli: A Sustainable and Ecofriendly Approach for the Treatment of Textile Wastewater. Water 2022, 14, 2063. Water 2022, 14, 2969. [Google Scholar] [CrossRef]
- Ikram, M.; Zahoor, M.; Gaber, S.B. Biodegradation and decolorization of textile dyes by bacterial strains: A biological approach for wastewater treatment. Z. Phys. Chem. 2021, 235, 10. [Google Scholar] [CrossRef]
- Khan, A.U.; Zahoor, M.; Rehman, M.U.; Shah, A.B.; Zekker, I.; Khan, F.A.; Ullah, R.; Albadrani, G.M.; Bayram, R.; Mohamed, H.R.H. Biological Mineralization of Methyl Orange by Pseudomonas aeruginosa. Water 2022, 14, 1551. [Google Scholar] [CrossRef]
- Khan, A.U.; Rehman, M.U.; Zahoor, M.; Shah, A.B.; Zekker, I. Biodegradation of Brown 706 Dye by Bacterial Strain Pseudomonas aeruginosa. Water 2021, 13, 2959. [Google Scholar] [CrossRef]
- Sharma, A.; Kiciman, E. DoWhy: A Python Library for Causal Inference. 2020. Available online: https://github.com/py-why/dowhy (accessed on 29 November 2021).
- Scott, S.L. 2021. Available online: https://cran.r-project.org/web/packages/BoomSpikeSlab/index.html (accessed on 29 November 2021).
- Grubinger, T.; Zeileis, A.; Pfeiffer, K.P. evtree: Evolutionary Learning of Globally Optimal Classification and Regression Trees in R. J. Stat. Softw. 2014, 64, 1–29. [Google Scholar] [CrossRef]
- Rezić, T.; Vrsalović, A.; Kurtanjek, Ž. New approach to the evaluation of lignocellulose-derived by-products impact on lytic-polysaccharide monooxygenase activity by using molecular descriptor structural causality model. Bioresour. Technol. 2021, 342, 125990. [Google Scholar] [CrossRef] [PubMed]
- Sygmund, C.; Kracher, D.; Scheiblbrandner, S.; Zahma, K.; Felice, A.K.G.; Harreither, W.; Kittl, R.; Ludwig, R. Characterization of the Two Neurospora crassa Cellobiose Dehydrogenases and Their Connection to Oxidative Cellulose Degradation. Appl. Environ. Microbiol. 2012, 78, 6161–6171. [Google Scholar] [CrossRef] [PubMed]
- Kittl, R.; Kracher, D.; Burgstaller, D.; Haltrich, D.; Ludwig, R. Production of four Neurospora crassa lytic polysaccharide monooxygenases in Pichia pastoris monitored by a fluorimetric assay. Biotechnol. Biofuels 2012, 5, 79. [Google Scholar] [CrossRef] [PubMed]
- Yap, C.W. PaDEL-descriptor: An open-source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 2011, 32, 1466–1474. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Soft. 2010, 33, 1–22. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- R Core Team. R: A Language and Environment for Foundation for Statistical Computing; R Core Team: Vienna, Austria, 2017. [Google Scholar]
- Chavan, S.; Gaikwad, A. Extraction and recovery of lignin-derived phenolic inhibitors to enhance enzymatic glucose production. Biomass Bioenergy 2021, 144, 105897. [Google Scholar] [CrossRef]
- Breslmayr, E.; Daly, S.; Požgajčić, A.; Chang, H.C.; Rezić, T.; Oostenbrink, C.; Ludwig, R. Improved spectrophotometric assay for lytic polysaccharide monooxygenase. Biotechnol. Biofuels 2019, 12, 283. [Google Scholar] [CrossRef] [PubMed]
- Hedison, T.M.; Breslmayr, E.; Shanmugam, M.; Karnpakdee, K.; Heyes, D.J.; Green, A.P.; Ludwig, R.; Scrutton, N.S.; Kracher, D. Insight into the H2O2-driven catalytic mechanism of fungal lytic polysaccharide monooxygenases. FEBS J. 2021, 288, 4115–4128. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Tang, J.; Chen, J.; Deng, J.; Wang, J.; Zhang, Q. Evaluation of inhibition of lignocellulose-derived by-products on bioethanol production by using the QSAR method and mechanism study. Biochem. Eng. J. 2019, 147, 153–162. [Google Scholar] [CrossRef]
- Kracher, D.; Scheiblbrandner, S.; Felice, A.K.G.; Breslmayr, E.; Preims, M.; Ludwicka, K.; Haltrich, D.; Eijsink, V.G.H.; Ludwig, R. Extracellular electron transfer systems fuel cellulose oxidative degradation. Science 2016, 352, 1098–1101. [Google Scholar] [CrossRef]
- Bhatia, S.K.; Jagtap, S.S.; Bedekar, A.A.; Bhatia, R.K.; Patel, A.K.; Pant, D.; Rajesh Banu, J.; Rao, C.V.; Kim, Y.G.; Yang, Y.H. Recent developments in pretreatment technologies on lignocellulosic biomass: Effect of key parameters, technological improvements, and challenges. Bioresour. Technol. 2020, 300, 122724. [Google Scholar] [CrossRef] [PubMed]
- Kalisch, M.; Mächler, M.; Colombo, D.; Maathuis, M.H.; Bühlmann, P. Causal inference using graphical models with the R package pcalg. J. Stat. Softw. 2012, 47, 1–26. [Google Scholar] [CrossRef]
- Chylenski, P.; Bissaro, B.; Sørlie, M.; Røhr, A.K.; Várnai, A.; Horn, S.J.; Eijsink, V.G.H. Lytic polysaccharide monooxygenases in enzymatic processing of lignocellulosic biomass. ACS Catal. 2019, 9, 4970–4991. [Google Scholar] [CrossRef]




| Molecule | GATS6c | ATSC8e | AATSC3v | AATSC7i | IC4 | ATSC6e | I/Io | DTM Decision |
|---|---|---|---|---|---|---|---|---|
| Methyl Orange | 0.361 | −0.171 | −10.697 | 0.231 | 4.4 | 1.97 | 0.79 | ATSC8e |
| Neolan gruen E-B 400% | 0.691 | 4.06 | 9.176 | −0.244 | 5.223 | 1.182 | 0.65 | GATS6c |
| Basic Blue 1 | 0.666 | 0.635 | −5.089 | −0.309 | 4.703 | 0.045 | 0.58 | GATS6c |
| Malachite Green | 0.647 | −0.159 | −6.006 | −0.298 | 4.663 | 0.194 | 0.54 | GATS6c |
| Malachite oxalate Green | 0.647 | −0.159 | −6.006 | −0.298 | 4.663 | 0.194 | 0.5 | GATS6c |
| Indigocarmin | 0.767 | 1.13 | −9.065 | 0.067 | 4.559 | 2.736 | 0.45 | ATSC8e |
| Basic Blue 5 | 0.659 | 0.185 | 5.578 | −0.177 | 5.326 | 0.029 | 0.43 | GATS6c |
| Potassium Indigotrisulfonate | 0.675 | 0.64 | −9.937 | 0.113 | 4.875 | 6.033 | 0.43 | GATS6c |
| Orbantin crveno 4BL | 1.163 | 11.912 | −8.834 | 0.033 | 5.295 | −5.012 | 0.27 | AATSC7i |
| Direct Blue 71 | 1.094 | −1.669 | −3.376 | 0.453 | 5.822 | −0.867 | 0.25 | ATSC8e |
| CibacetRot 3B | 1.473 | 0.317 | −0.213 | −0.179 | 4.385 | −1.895 | 0.2 | AATSC7i |
| Fuchsin | 0.531 | −0.382 | −7.857 | −0.571 | 4.737 | 0.111 | 0.19 | ATSC8e |
| Crystal Violet | 0.591 | −0.313 | −7.137 | −0.308 | 4.159 | 0.36 | 0.18 | ATSC8e |
| Thionin | 1.199 | 0.229 | −7.186 | −0.698 | 4.393 | −0.228 | 0.18 | AATSC7i |
| Basic Blue 3 (60%) | 1.227 | −0.617 | 12.15 | 0.119 | 4.543 | −0.077 | 0.14 | ATSC8e |
| Reactive Black 5 | 0.861 | −4.2 | −2.996 | −0.148 | 5.254 | 3.782 | 0.14 | ATSC8e |
| Gallocyanine | 1.34 | −0.616 | −8.258 | −0.002 | 4.684 | −0.944 | 0.14 | ATSC8e |
| Meldola’s Blue | 1.456 | −0.021 | −5.727 | 0.212 | 4.682 | −0.368 | 0.14 | AATSC7i |
| Congo red | 0.696 | −0.945 | −2.662 | 0.252 | 5.146 | 1.637 | 0.13 | ATSC8e |
| Cuprophenyl grey 2BL | 0.847 | 0.41 | −4.807 | 0.268 | 6.236 | 3.328 | 0.12 | AATSC7i |
| Diphenylechtblau 4GL | 1.07 | −2.134 | −3.741 | 0.277 | 5.744 | 1.682 | 0.12 | ATSC8e |
| beta-naphthol orange | 0.928 | 0.335 | −4.667 | 0.238 | 4.852 | 0.579 | 0.12 | AATSC7i |
| Basic Blue 41 | 0.917 | 0.055 | 4.46 | 0.122 | 5.061 | 0.244 | 0.12 | AATSC7i |
| Metilenblau | 0.857 | 0.08 | −7.51 | 0.227 | 3.905 | −0.108 | 0.11 | AATSC7i |
| Bromocresol Purple | 0.756 | 0.357 | −6.449 | −0.631 | 4.836 | −0.91 | 0.11 | AATSC7i |
| Bromophenol blue | 0.739 | 0.531 | −6.513 | −0.307 | 4.516 | −0.601 | 0.11 | AATSC7i |
| Victoria Blue B | 0.615 | −0.177 | −3.964 | −0.259 | 5.337 | 0.209 | 0.1 | ATSC8e |
| Acid Blue 45 | 1.219 | −0.548 | −3.501 | −0.195 | 4.564 | −3.876 | 0.1 | ATSC8e |
| Remazol Brilliant Blue R | 0.861 | −4.2 | −2.996 | −0.148 | 5.254 | 3.782 | 0.09 | ATSC8e |
| Acid green 3 | 0.653 | −1.767 | −0.957 | −0.148 | 5.681 | 1.259 | 0.09 | ATSC8e |
| Lanaynrein? rot 2BL | 1.113 | −0.361 | −3.896 | −0.256 | 5.176 | −0.603 | 0.08 | ATSC8e |
| Brilliant cresyl blue | 1.024 | −0.056 | 1.547 | 0.008 | 4.635 | −0.282 | 0.08 | AATSC7i |
| Solarrot B | 0.897 | −0.924 | −5.142 | 0.078 | 5.628 | −0.038 | 0.08 | ATSC8e |
| Azure A | 0.963 | 0.151 | −7.67 | −0.093 | 4.39 | −0.162 | 0.08 | AATSC7i |
| Reactive Blue 2 | 1.04 | −5.485 | −3.569 | −0.246 | 5.236 | −0.989 | 0.07 | ATSC8e |
| Bromocrezol-green | 0.764 | 1.565 | −2.266 | −0.184 | 4.836 | −0.18 | 0.07 | AATSC7i |
| Orbacid R | 0.761 | 2.2 | −5.174 | −0.049 | 5.423 | 0.228 | 0.07 | AATSC7i |
| Toluidine Blue | 0.982 | 0.14 | −8.79 | −0.18 | 4.436 | −0.253 | 0.07 | AATSC7i |
| Acid Violet 43 | 1.176 | 0.792 | −3.758 | −0.116 | 5.083 | −4.591 | 0.06 | AATSC7i |
| Tris (2) | 0.729 | 0.071 | 0.801 | −0.155 | 3.322 | −0.079 | 0.06 | AATSC7i |
| Cuprophenylreinblau 2BL | 0.973 | −2.945 | −4.35 | 0.091 | 5.77 | −2.56 | 0.06 | ATSC8e |
| Lanasynrein gr BL | 1.071 | −0.118 | −5.891 | −0.091 | 4.736 | −7.915 | 0.06 | AATSC7i |
| Neolan Blue 3R | 1.071 | −0.118 | −5.891 | −0.091 | 4.736 | −7.915 | 0.06 | AATSC7i |
| Lanaset Gr B | 0.907 | −3.633 | −0.471 | −0.198 | 5.195 | −2.521 | 0.05 | ATSC8e |
| Lanaset Braun B | 1.342 | −0.093 | −1.984 | −0.215 | 5.089 | −0.2 | 0.05 | AATSC7i |
| Acid Violet 9 | 0.944 | −2.707 | −3.534 | −0.152 | 5.889 | −0.198 | 0.05 | ATSC8e |
| Cuprophenylbraun 2RL | 0.927 | −0.016 | −3.093 | −0.255 | 5.547 | −0.007 | 0.04 | AATSC7i |
| Neolan Black WA extra N | 1.185 | −2.163 | −0.679 | 0.029 | 5.037 | −2.737 | 0.04 | ATSC8e |
| Reactive Blue 7 | 0.972 | 0.112 | −5.99 | −0.467 | 4.054 | −0.167 | 0.04 | AATSC7i |
| Rhodamine B | 1.176 | 0.792 | −3.758 | −0.116 | 5.083 | −4.591 | 0.04 | AATSC7i |
| Neolan Yellow RE | 0.959 | 0.004 | −4.26 | −0.253 | 5.053 | 2.604 | 0.03 | AATSC7i |
| Orbodisperz marine S-BL | 1.198 | 3.051 | 5.66 | −0.045 | 5.157 | 0.95 | 0.03 | AATSC7i |
| Cuprophenyl marine BL | 0.731 | −0.611 | −7.352 | 0.1 | 6.016 | 0.313 | 0.03 | ATSC8e |
| Irgalanbraun 2GL | 0.859 | 5.513 | −7.265 | −0.286 | 5.395 | 1.342 | 0.02 | AATSC7i |
| LanasetGelb 4 GN | 0.859 | 5.513 | −7.265 | −0.286 | 5.395 | 1.342 | 0.01 | AATSC7i |
| Cuprophenylrot BL | 0.762 | −0.966 | −2.273 | 0.209 | 5.529 | 2.026 | 0.01 | ATSC8e |
| Alizarin S | 1.157 | −1.655 | −2.011 | −0.216 | 4.418 | 0.1 | 0.01 | ATSC8e |
| Lanasynobraun GRL | 0.643 | −4.319 | −5.771 | −0.393 | 5.337 | 7.44 | 0.01 | ATSC8e |
| Sulforhodamine B | 1.413 | −0.736 | 8.627 | 0.105 | 4.824 | 6.331 | 0.01 | ATSC8e |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rezić, I.; Kracher, D.; Oros, D.; Mujadžić, S.; Anđelini, M.; Kurtanjek, Ž.; Ludwig, R.; Rezić, T. Application of Causality Modelling for Prediction of Molecular Properties for Textile Dyes Degradation by LPMO. Molecules 2022, 27, 6390. https://doi.org/10.3390/molecules27196390
Rezić I, Kracher D, Oros D, Mujadžić S, Anđelini M, Kurtanjek Ž, Ludwig R, Rezić T. Application of Causality Modelling for Prediction of Molecular Properties for Textile Dyes Degradation by LPMO. Molecules. 2022; 27(19):6390. https://doi.org/10.3390/molecules27196390
Chicago/Turabian StyleRezić, Iva, Daniel Kracher, Damir Oros, Sven Mujadžić, Magdalena Anđelini, Želimir Kurtanjek, Roland Ludwig, and Tonči Rezić. 2022. "Application of Causality Modelling for Prediction of Molecular Properties for Textile Dyes Degradation by LPMO" Molecules 27, no. 19: 6390. https://doi.org/10.3390/molecules27196390






