In Vitro Anticancer Screening, Molecular Docking and Antimicrobial Studies of Triazole-Based Nickel(II) Metal Complexes
Abstract
:1. Introduction
2. Experimental
2.1. Materials and Methods
2.2. Synthesis
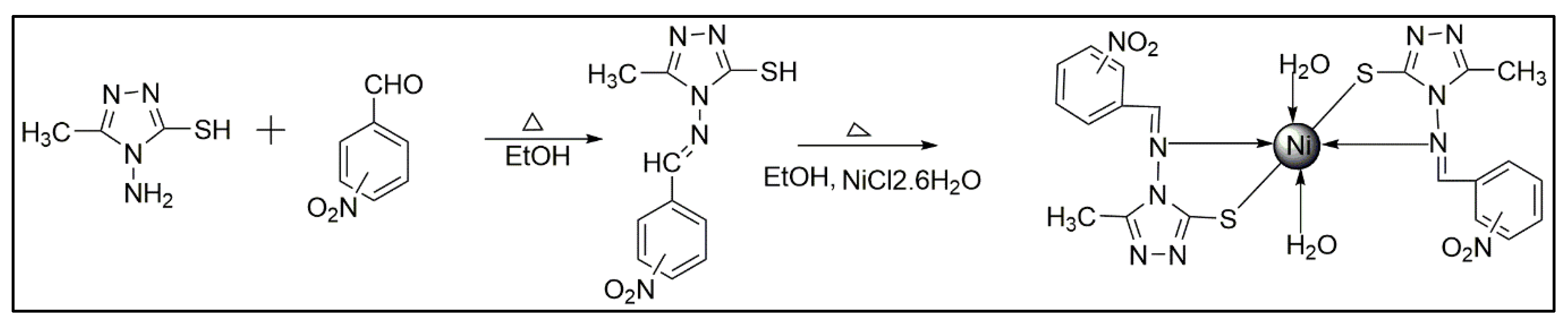
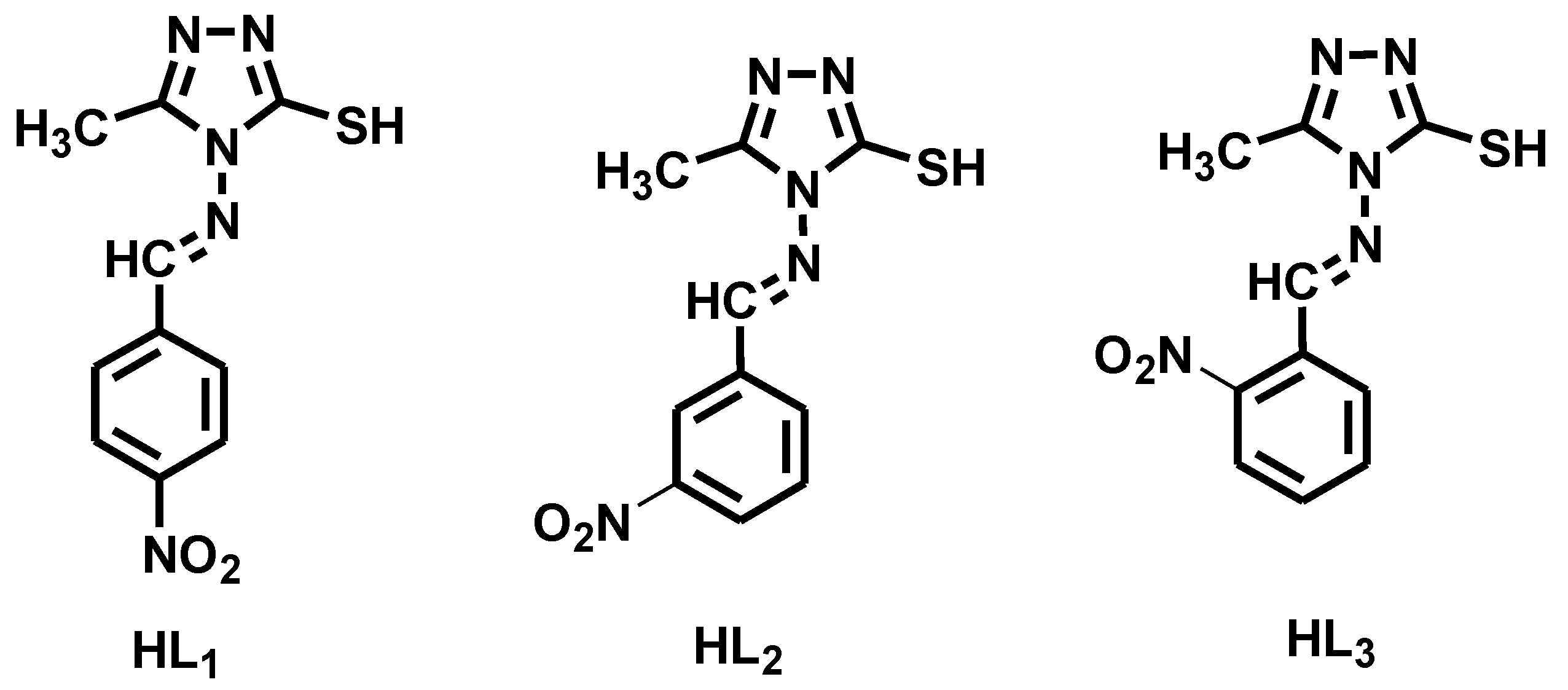
2.2.1. Synthesis of Ligands (HL1–HL3)
2.2.2. Synthesis of Metal Complexes (C1–C3)
2.3. Biological Studies
2.3.1. In Vitro Antimicrobial Studies
2.3.2. Molecular Docking Studies
2.3.3. Antiproliferative Activities against OVCAR-3 Cell Line
2.4. Conceptual DFT Studies
3. Results and Discussion
3.1. H-NMR and Mass Spectral Analysis
3.2. FT-IR Spectral Studies
3.3. Electronic and Magnetic Moment Studies
3.4. Thermal Analysis
3.5. X-ray Diffraction Analysis
3.6. Conceptual DFT Report
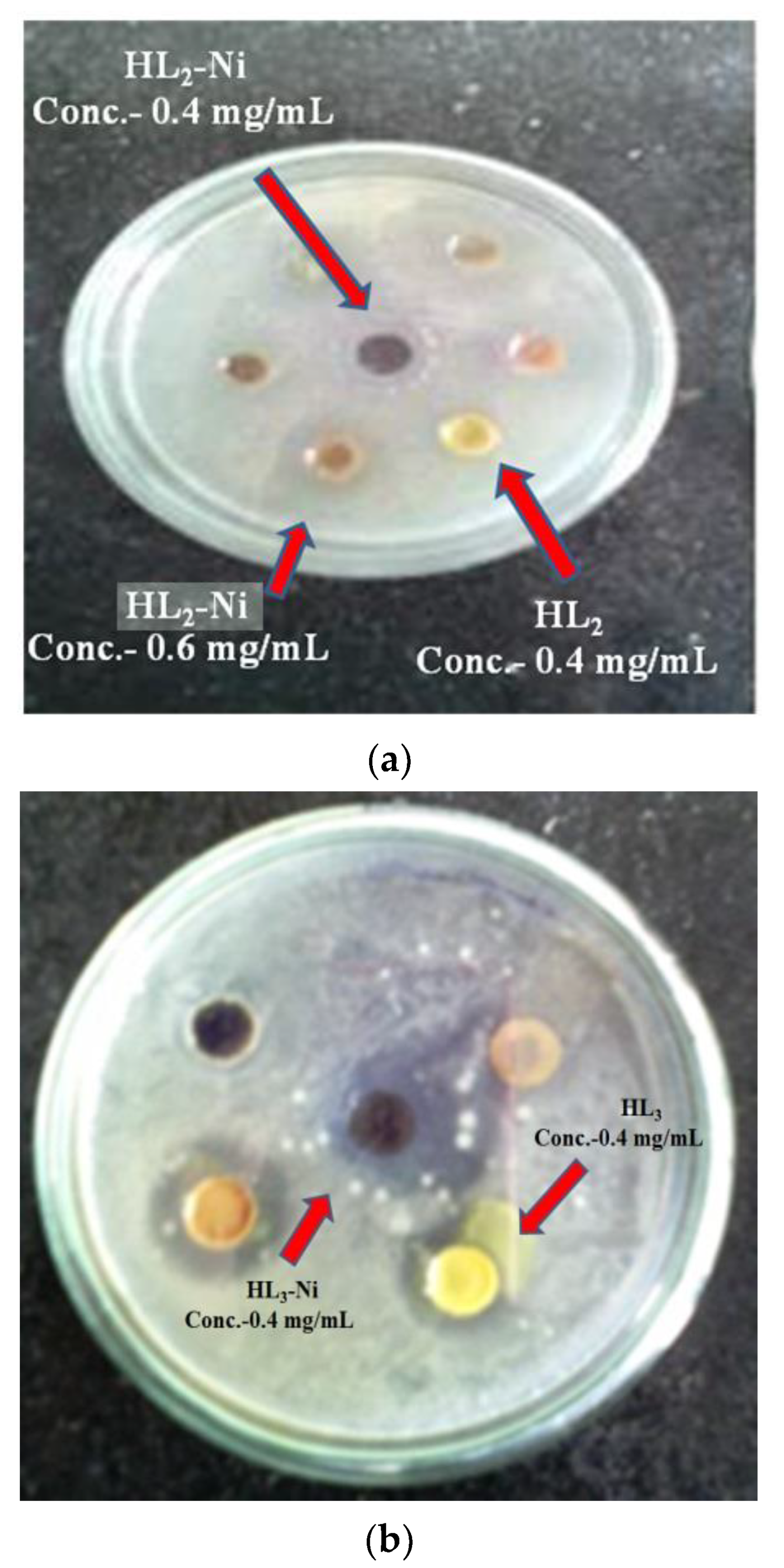
3.7. In Vitro Antimicrobial and Antifungal Activities
3.8. Molecular Docking Studies
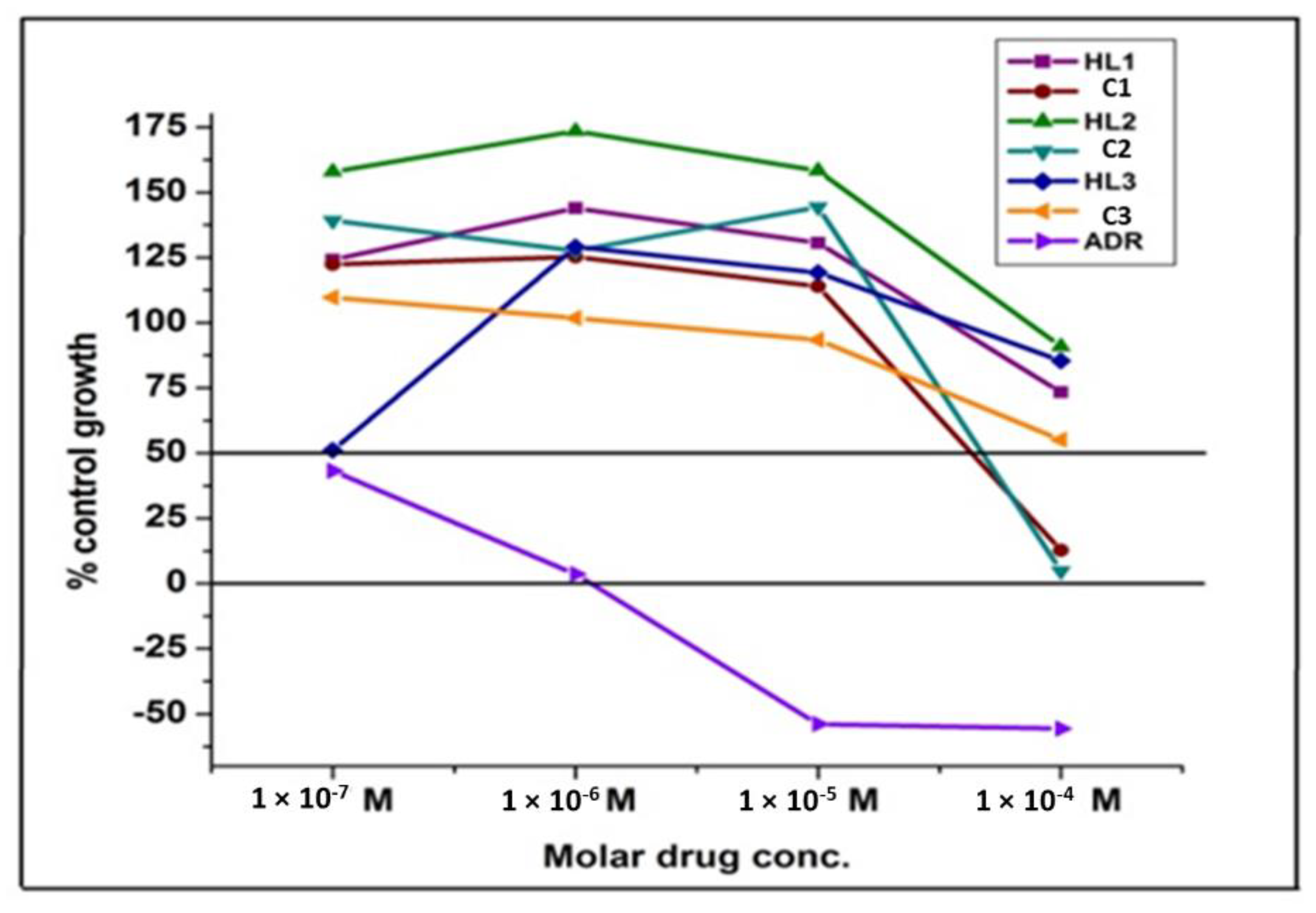
3.9. Effect of ligands (HL1–HL3) and its complexes (C1–C3) on antiproliferative activity
3.10. Structure Activity Relationship (SAR Studies)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Bakale, R.P.; Naik, G.N.; Mangannavar, C.V.; Muchchandi, I.S.; Shcherbakov, I.; Frampton, C.; Gudasi, K.B. Mixed ligand complex via zinc (II)-mediated in situ oxidative heterocyclization of hydrochloride salt of 2-chlorobenzaldehyde hydralazine hydrazone as potential of antihypertensive agent. Eur. J. Med. Chem. 2014, 73, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Júnior, W.B.; Alexandre-Moreira, M.S.; Alves, M.A.; Perez-Rebolledo, A.; Parrilha, G.L.; Castellano, E.E.; Piro, O.E.; Barreiro, E.J.; Lima, L.M.; Beraldo, H. Analgesic and anti-inflammatory activities of salicylaldehyde 2-chlorobenzoyl hydrazone (H(2)LASSBio-466), salicylaldehyde-4-chlorobenzoyl hydrazone (H(2)LASSBio-1064) and their zinc (II)complexes. Molecules 2011, 16, 6902. [Google Scholar] [CrossRef] [PubMed]
- Inam, A.; Siddiqui, S.M.; Macedo, T.S.; Moreira, D.R.M.; Leite, A.C.L.; Soares, M.B.P.; Azam, A. Design, synthesis and biological evaluation of 3-[4-(7-chloro-quinolin-4-yl)-piperazin-1-yl]-propionic acid hydrazones as antiprotozoal agents. Eur. J. Med. Chem. 2014, 75, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, G.G.; Zayed, E.M.; Hindy, A.M. Coordination behavior of new bis Schiff base ligand derived from 2-furan carboxaldehyde and propane-1,3-diamine. Spectroscopic, thermal, anticancer and antibacterial activity studies. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 145, 76–84. [Google Scholar] [CrossRef]
- de Almeida, A.; Oliveira, B.L.; Correia, J.D.; Soveral, G.; Casini, A. Emerging protein targets for metal-based pharmaceutical agents: An update. Co-Ord. Chem. Rev. 2013, 257, 2689–2704. [Google Scholar] [CrossRef]
- Dheer, D.; Singh, V.; Shankar, R. Medicinal attributes of 1,2,3-triazoles: Current developments. Bioorg. Chem. 2017, 71, 30–54. [Google Scholar] [CrossRef]
- Yan, W.; Wang, X.; Li, K.; Li, T.-X.; Wang, J.-J.; Yao, K.-C.; Cao, L.-L.; Zhao, S.-S.; Ye, Y.-H. Design, synthesis, and antifungal activity of carboxamide derivatives possessing 1,2,3-triazole as potential succinate dehydrogenase inhibitors. Pestic. Biochem. Physiol. 2019, 156, 160–169. [Google Scholar] [CrossRef]
- Schulze, B.; Schubert, U.S. Beyond click chemistry-supramolecular interactions of 1,2,3-triazoles. Chem. Soc. Rev. 2014, 43, 2522–2571. [Google Scholar] [CrossRef]
- Kirsch, C.; Pulst, M.; Samiullah, M.H.; Ruda, P.; Hasan, N.; Kressler, J. 1,2,3-Triazole mediated Li+-ion conductivity in poly(ethylene oxide) based electrolytes. Solid State Ion. 2017, 309, 163–169. [Google Scholar] [CrossRef]
- Xu, Z.; Zhao, S.-J.; Liu, Y. 1,2,3-Triazole-containing hybrids as potential anticancer agents: Current developments, action mechanisms and structure-activity relationships. Eur. J. Med. Chem. 2019, 183, 111700. [Google Scholar] [CrossRef]
- Singh, H.; Kumar, M.; Nepali, K.; Gupta, M.K.; Saxena, A.K.; Sharma, S.; Bedi, P.M.S. Triazole tethered C5-curcuminoid-coumarin based molecular hybrids as novel antitubulin agents: Design, synthesis, biological investigation and docking studies. Eur. J. Med. Chem. 2016, 116, 102–115. [Google Scholar] [CrossRef]
- Kumar, K.; Pradines, B.; Madamet, M.; Amalvict, R.; Benoit, N.; Kumar, V. 1H-1,2,3-triazole tethered isatin-ferrocene conjugates: Synthesis and in vitro antimalarial evaluation. Eur. J. Med. Chem. 2014, 87, 801–804. [Google Scholar] [CrossRef]
- González-Calderón, D.; Mejía-Dionicio, M.G.; Morales-Reza, M.A.; Ramírez-Villalva, A.; Morales-Rodríguez, M.; Jauregui-Rodríguez, B.; Díaz-Torres, E.; González-Romero, C.; Fuentes-Benítes, A. Azide-enolate 1,3-dipolar cycloaddition in the synthesis of novel triazole-based miconazole analogues as promising antifungal agents. Eur. J. Med. Chem. 2016, 112, 60–65. [Google Scholar] [CrossRef]
- Zhang, B. Comprehensive review on the anti-bacterial activity of 1,2,3-triazole hybrids. Eur. J. Med. Chem. 2019, 168, 357–372. [Google Scholar] [CrossRef]
- Kant, R.; Kumar, D.; Agarwal, D.; Gupta, R.D.; Tilak, R.; Awasthi, S.K.; Agarwal, A. Synthesis of newer 1,2,3-triazole linked chalcone and flavone hybrid compounds and evaluation of their antimicrobial and cytotoxic activities. Eur. J. Med. Chem. 2016, 113, 34–49. [Google Scholar] [CrossRef]
- Chinthala, Y.; Thakur, S.; Tirunagari, S.; Chinde, S.; Domatti, A.K.; Arigari, N.K.; Srinivas, K.V.N.S.; Alam, S.; Jonnala, K.K.; Khan, F.; et al. Synthesis, docking and ADMET studies of novel chalcone triazoles for anti-cancer and anti-diabetic activity. Eur. J. Med. Chem. 2015, 93, 564–573. [Google Scholar] [CrossRef]
- Kuang, G.-C.; Michaels, H.A.; Simmons, J.T.; Clark, R.J.; Zhu, L. Chelation-assisted, copper (II)-acetate-accelerated azide-alkyne cycloaddition. J. Org. Chem. 2010, 75, 6540. [Google Scholar] [CrossRef] [Green Version]
- Ashok, U.P.; Kollur, S.P.; Anil, N.; Arun, B.P.; Jadhav, S.N.; Sarsamkar, S.; Helavi, V.B.; Srinivasan, A.; Kaulage, S.; Veerapur, R.; et al. Preparation, Spectroscopic Characterization, Theoretical Investigations, and In Vitro Anticancer Activity of Cd (II), Ni (II), Zn (II), and Cu (II) Complexes of 4(3H)-Quinazolinone-Derived Schiff Base. Molecules 2020, 25, 5973. [Google Scholar] [CrossRef]
- Wu, W.-N.; Jiang, Y.-M.; Fei, Q.; Du, H.-T. Synthesis and fungicidal activity of novel 1,2,4-triazole derivatives containing a pyrimidine moiety. Phosphorus Sulfur Silicon Relat. Elem. 2019, 194, 1171–1175. [Google Scholar] [CrossRef]
- Koparir, P. Synthesis, antioxidant and antitumor activities of some of new cyclobutane containing triazoles derivatives. Phosphorus Sulfur Silicon Relat. Elem. 2019, 194, 1028–1034. [Google Scholar] [CrossRef]
- Cui, X.-S.; Chen, J.; Chai, K.-Y.; Lee, J.S.; Quan, Z.-S. Synthesis and anticonvulsant evaluation of 3-substituted-4-(4-hexyloxyphenyl)-4H-1,2,4-triazoles. Med. Chem. Res. 2009, 18, 49–58. [Google Scholar] [CrossRef]
- Ahmed, Y.M.; Mahmoud, W.H.; Omar, M.M.; Mohamed, G.G. Synthesis, Characterization and Biological Activity of Transition Metals Schiff Base Complexes Derived from 4,6-Diacetylresorcinol and 1,8-Naphthalenediamine. J. Inorg. Organomet. Polym. Mater. 2021, 31, 2339–2359. [Google Scholar] [CrossRef]
- Kamble, U.V.; Patil, S.A.; Badami, P.S. DNA cleavage and antimicrobial studies of 17-membered Schiff base macrocyclictriazoles: Synthesis and spectroscopic approach. J. Inc. Phenom. Macrocycl. Chem. 2010, 68, 347. [Google Scholar] [CrossRef]
- Almasirad, A.; Shafiee, A.; Abdollahi, M.; Noeparast, A.; Shahrokhinejad, N.; Vousooghi, N.; Tabatabai, S.A.; Khorasani, R. Synthesis and analgesic activity of new 1,3,4-oxadiazoles and 1,2,4-triazoles. Med. Chem. Res. 2011, 20, 435. [Google Scholar] [CrossRef]
- Tyagi, P.; Chandra, S.; Saraswat, B.; Yadav, D. Design, spectral characterization, thermal, DFT studies and anticancer cell line activities of Co (II), Ni (II) and Cu (II) complexes of Schiff bases derived from 4-amino-5-(pyridin-4-yl)-4H-1,2,4-triazole-3-thiol. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 145, 155–164. [Google Scholar] [CrossRef]
- Abd-Rabou, A.A.; Abdel-Wahab, B.F.; Bekheit, M.S. Synthesis, molecular docking, and evaluation of novel bivalent pyrazolinyl-1,2,3-triazoles as potential VEGFR TK inhibitors and anti-cancer agents. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 72, 2225. [Google Scholar] [CrossRef]
- Kumar, P.; Narsimhan, B.; Sharma, D. Substituted benzoic acid benzylidene/furan-2-ylmethylenehydrazides: Synthesis, antimicrobial evaluation and QSAR analysis. Arkivoc 2008, 13, 159–178. [Google Scholar] [CrossRef] [Green Version]
- Kumar, D.; Judge, V.; Narang, R.; Sangwan, S.; De Clercq, E.; Balzarini, J.; Narasimhan, B. Benzylidene/2-chlorobenzylidene hydrazides: Synthesis, antimicrobial activity, QSAR studies and antiviral evaluation. Eur. J. Med. Chem. 2010, 45, 2806–2816. [Google Scholar] [CrossRef]
- Shaikh, A.B.; Barache, U.B.; Anuse, M.A.; Gaikwad, S.H. 4-(4′-Nitrobenzylideneimino)-3-methyl-5-mercapto-1,2,4-triazole, A new chromogenic reagent for extractive spectrophotometric determination of copper (II) in pharmaceutical and alloy samples. S. Afr. J. Chem. 2016, 69, 157. [Google Scholar] [CrossRef]
- Shaikh, A.B.; Barache, U.B.; Lokhande, T.N.; Kamble, G.S.; Anuse, M.A.; Gaikwad, S.H. Expeditious extraction and spectrophotometric determination of palladium (II) from catalysts and alloy samples using new chromogenic reagent. Rasayan J. Chem. 2017, 10, 967. [Google Scholar]
- Barache, U.B.; Shaikh, A.B.; Deodware, S.A.; Dhale, P.C.; Lokhande, T.N.; Gaikwad, S.H. A new experimental approach for liquid-liquid extractive spectrophotometric determination of chromium (VI) in tannery wastewater and alloy samples. Int. J. Environ. Anal. Chem. 2019, 99, 621–640. [Google Scholar] [CrossRef]
- Barache, U.B.; Shaikh, A.B.; Lokhande, T.N.; Kamble, G.S.; Anuse, M.A.; Gaikwad, S.H. An efficient, cost effective, sensing behaviour liquid-liquid extraction and spectrophotometric determination of copper (II) incorporated with 4-(4′-chlorobenzylideneimino)-3-methyl-5-mercapto-1,2,4-triazole: Analysis of food samples, leafy vegetables, fertilizers and environmental samples. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 189, 443–453. [Google Scholar]
- Barache, U.B.; Shaikh, A.B.; Lokhande, T.N.; Anuse, M.A.; Kamble, G.S.; Gurame, V.M.; Gaikwad, S.H. Acid switched efficient, cost effective, selective separation and determination of selenium (IV). J. Environ. Chem. Eng. 2017, 5, 4828. [Google Scholar] [CrossRef]
- Barache, U.B.; Khogare, B.T.; Shaikh, A.B.; Deodware, S.A.; Kokare, B.N.; Rodriguez, A.G.P.; Lokhande, T.N.; Gaikwad, S.H. Selective and sensitive liquid-liquid extraction and spectrophotometric determination of tellurium (IV) using sulfur containing reagent. Chem. Data Collect. 2019, 19, 100173. [Google Scholar] [CrossRef]
- Barache, U.B.; Shaikh, A.B.; Deodware, S.A.; Dhale, P.C.; Kamble, G.S.; Lokhande, T.N.; Gaikwad, S.H. Sensitive and selective liquid-liquidextractive spectrophotometric determination of Bismuth (III) from water, pharmaceuticals and synthetic mixtures. Groundw. Sust. Dev. 2019, 9, 100221. [Google Scholar] [CrossRef]
- Zagotto, G.; Palumbo, M. Development of DNA Topoisomerase-Related Therapeutics: A Short Perspective of New Challenges. Curr. Med. Chem. Agents 2004, 4, 335–345. [Google Scholar] [CrossRef]
- Bryant, S.G.; Ereshefsky, L. Antidepressant properties of trazodone. Clin. Pharm. 1982, 1, 406–417. [Google Scholar]
- Stahl, S.M. Selective Histamine H1 Antagonism: Novel Hypnotic and Pharmacologic Actions Challenge Classical Notions of Antihistamines. CNS Spectr. 2008, 13, 1027–1038. [Google Scholar] [CrossRef] [Green Version]
- Deodware, S.A.; Sathe, D.J.; Choudhari, P.B.; Lokhande, T.N.; Gaikwad, S.H. Development and molecular modelling of Co (II), Ni (II) and Cu (II) complexes as high acting antibreast cancer agents. Arabian J. Chem. 2017, 10, 262. [Google Scholar] [CrossRef] [Green Version]
- Bala, S.; Gupta, R.P.; Sachdeva, M.L.; Singh, A.; Pujari, H.K. Heterocyclic system containing bridge head nitrogen atom: Part XXXIII-sysnthesis of s-Triazolo[3,4-b] [1,3,4] thiadiazino [6,7-b] quinoxaline and as-triazino-[3,4-b][1,3,4] thiadiazino[6,7-b] quinoxaline and astriazino-[3,4-b][1,3,4] thiadiazines. Indian J. Chem. 1978, 16, 481. [Google Scholar]
- Durairaja, S.; Srinivasan, S.; Perumalsamy, P.L. In vitro antibacterial activity and stability of garlic extract at different pH and temperature. Electron. J. Biol. 2009, 5, 5. [Google Scholar]
- Bowers, E.F.; Jeffries, L.R. Optochin in the Identification of Str. pneumoniae. J. Clin. Pathol. 1955, 8, 58. [Google Scholar] [CrossRef] [Green Version]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular Docking: A Powerful Approach for Structure-Based Drug Discovery. Curr. Comput. Aided Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef]
- Lopez-Vallejo, F.; Caulfield, T.; Martinez-Mayorga, K.; Giulianotti, M.A.; Nefzi, A.; Houghten, R.A.; Medina-Franco, J.L. Integrating Virtual Screening and Combinatorial Chemistry for Accelerated Drug Discovery. Comb. Chem. High Throughput Screen. 2011, 14, 475–487. [Google Scholar] [CrossRef]
- Huang, S.-Y.; Zou, X. Advances and Challenges in Protein-Ligand Docking. Int. J. Mol. Sci. 2010, 11, 3016–3034. [Google Scholar] [CrossRef]
- Holla, B.S.; Rao, B.S.; Sarojini, B.K. Synthesis, characterization andanticancer activity studies on some Mannich bases derived from1,2,4-triazoles. Eur. J. Med. Chem. 2006, 41, 657. [Google Scholar]
- Chermette, H. Chemical Reactivity Indexes in Density Functional Theory. J. Comput. Chem. 1999, 20, 129–154. [Google Scholar] [CrossRef]
- Geerlings, P.; de Proft, F.; Langenaeker, W. Conceptual Density Functional Theory. Chem. Rev. 2003, 103, 1793–1874. [Google Scholar] [CrossRef]
- Geerlings, P.; Chamorro, E.; Chattaraj, P.K.; de Proft, F.; Gázquez, J.L.; Liu, S.; Morell, C.; Toro-Labbé, A.; Vela, A.; Ayers, P. Conceptual Density Functional Theory: Status, Prospects, Issues. Theor. Chem. Acc. 2020, 139, 36. [Google Scholar] [CrossRef]
- Toro-Labbé, A. (Ed.) Theoretical Aspects of Chemical Reactivity; Elsevier Science: Amsterdam, The Netherlands, 2007. [Google Scholar]
- Chattaraj, P.K. (Ed.) Chemical Reactivity Theory—A Density Functional View; CRC Press: Boca Raton, FL, USA; Taylor & Francis Group: Abingdon, UK, 2009. [Google Scholar]
- Chakraborty, D.; Chattaraj, P.K. Conceptual Density Functional Theory Based Electronic Structure Principles. Chem. Sci. 2021, 12, 6264–6279. [Google Scholar] [CrossRef]
- Halgren, T.A. Merck Molecular Force Field. I. Basis, Form, Scope, Parameterization, and Performance of MMFF94. J. Comput. Chem. 1996, 17, 490–519. [Google Scholar] [CrossRef]
- Halgren, T.A. Merck Molecular Force Field. II. MMFF94 van der Waals and Electrostatic Parameters for Intermolecular Interactions. J. Comput. Chem. 1996, 17, 520–552. [Google Scholar] [CrossRef]
- Halgren, T.A. MMFF VI. MMFF94s Option for Energy Minimization Studies. J. Comput. Chem. 1999, 20, 720–729. [Google Scholar] [CrossRef]
- Halgren, T.A.; Nachbar, R.B. Merck Molecular Force Field. IV. Conformational Energies and Geometries for MMFF94. J. Comput. Chem. 1996, 17, 587–615. [Google Scholar] [CrossRef]
- Halgren, T.A. Merck Molecular Force Field. V. Extension of MMFF94 Using Experimental Data, Additional Computational Data, and Empirical Rules. J. Comput. Chem. 1996, 17, 616–641. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16 Revision C.01; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Peverati, R.; Truhlar, D.G. Screened-Exchange Density Functionals with Broad Accuracy for Chemistry and Solid-State Physics. Phys. Chem. Chem. Phys. 2012, 14, 16187. [Google Scholar] [CrossRef]
- Weigend, F.; Ahlrichs, R. Balanced Basis Sets of Split Valence, Triple Zeta Valence and Quadruple Zeta Valence Quality for H to Rn: Design and Assessment of Accuracy. Phys. Chem. Chem. Phys. 2005, 7, 3297. [Google Scholar] [CrossRef] [PubMed]
- Weigend, F. Accurate Coulomb-fitting basis sets for H to Rn. Phys. Chem. Chem. Phys. 2006, 8, 1057–1065. [Google Scholar] [CrossRef] [PubMed]
- Lewars, E. Computational Chemistry—Introduction to the Theory and Applications of Molecular and Quantum Mechanics; Kluwer Academic: Dordrecht, The Netherlands, 2003. [Google Scholar]
- Young, D.C. Computational Chemistry—A Practical Guide for Applying Techniques to Real-World Problems; John Wiley & Sons: New York, NY, USA, 2001. [Google Scholar]
- Jensen, F. Introduction to Computational Chemistry, 2nd ed.; John Wiley & Sons: Chichester, UK, 2007. [Google Scholar]
- Cramer, C.J. Essentials of Computational Chemistry—Theories and Models, 2nd ed.; John Wiley & Sons: Chichester, UK, 2004. [Google Scholar]
- Marenich, A.V.; Cramer, C.J.; Truhlar, D.G. Universal Solvation Model Based on Solute Electron Density and on a Continuum Model of the Solvent Defined by the Bulk Dielectric Constant and Atomic Surface Tensions. J. Phys. Chem. B 2009, 113, 6378–6396. [Google Scholar] [CrossRef] [PubMed]
- Flores-Holguín, N.; Frau, J.; Glossman-Mitnik, D. A Fast and Simple Evaluation of the Chemical Reactivity Properties of the Pristinamycin Family of Antimicrobial Peptides. Chem. Phys. Lett. 2020, 739, 137021. [Google Scholar] [CrossRef]
- Flores-Holguín, N.; Frau, J.; Glossman-Mitnik, D. Conceptual DFT-Based Computational Peptidology of Marine Natural Compounds: Discodermins A–H. Molecules 2020, 25, 4158. [Google Scholar] [CrossRef]
- Flores-Holguín, N.; Frau, J.; Glossman-Mitnik, D. Virtual Screening of Marine Natural Compounds by Means of Chemoinformatics and CDFT-Based Computational Peptidology. Mar. Drugs 2020, 18, 478. [Google Scholar] [CrossRef]
- Flores-Holguín, N.; Frau, J.; Glossman-Mitnik, D. Conceptual DFT as a Helpful Chemoinformatics Tool for the Study of the Clavanin Family of Antimicrobial Marine Peptides. In Density Functional Theory; de Lazaro, S.R., Lacerda, L.H.D., Ribeiro, R.A.P., Eds.; IntechOpen: London, UK, 2021; Chapter 3; pp. 57–67. [Google Scholar]
- Gudasi, K.B.; Patil, S.A.; Vadavi, R.S.; Shenoy, R.V.; Patil, M.S. Synthesis and Spectral Characterization of Some Transition Metal Complexes Containing Pentadentate SNNNS Donor Heterocyclic Schiff Base Ligands. Transit. Met. Chem. 2005, 30, 1014–1019. [Google Scholar] [CrossRef]
- Chandra, S.; Gupta, K. Chromium (III), Manganese (II), Iron (III), Cobalt (II), Nickel (II) and Copper (II) Complexes with a Pentadentate, 15-Membered New Macrocyclic Ligand. Transit. Met. Chem. 2002, 27, 196–199. [Google Scholar] [CrossRef]
- Singh, K. Antibacterial Co (II), Ni (II), Cu (II) and Zn (II) Complexes of Schiff bases Derived from Fluorobenzaldehyde and Triazoles. J. Enzym. Inhib. Med. Chem. 2006, 21, 557–562. [Google Scholar] [CrossRef]
- Nakamoto, K. Infrared and Raman Spectra of Inorganic and Coordination Compounds; Wiley: New York, NY, USA, 1997. [Google Scholar]
- Ashok, U.P.; Kollur, S.P.; Arun, B.P.; Sanjay, C.; Suresh, K.S.; Anil, N.; Baburao, H.V.; Markad, D.; Castro, J.O.; Frau, J.; et al. In vitro Anticancer activity of 4(3H)-Quinazolinone Derived Schiff Base and its Cu (II), Zn (II) and Cd (II) Complexes: Preparation, X-ray Structural, Spectral Characterization and Theoretical Investigations. Inorg. Chim. Acta 2020, 511, 119846. [Google Scholar] [CrossRef]
- Abou-Melha, K.S. Octahedral Co (II) and Ni (II) Complexes of Schiff Bases, Semicarbazone and Thiosemicarbazone, Synthesis, Biological, Spectral, and Thermal Studies. J. Coord. Chem. 2008, 61, 2053–2067. [Google Scholar] [CrossRef]
- Vinusha, H.M.; Kollur, S.P.; Revanasiddappa, H.D.; Ramu, R.; Shirahatti, P.S.; Prasad, M.N.; Chandrashekar, S.; Begum, M. Preparation, Spectral Characterization and Biological Applications of Schiff Base Ligand and its Transition Metal Complexes. Results Chem. 2019, 1, 100012. [Google Scholar] [CrossRef]
- Cotton, F.A.; Wilkinson, G.; Murillo, C.A.; Bochmann, M. Advanced Inorganic Chemistry; Wiley: New York, NY, USA, 1999. [Google Scholar]
- Bagihalli, G.B.; Patil, S.A. Synthesis, Physico-Chemical investigations of Co (II), Ni (II) and Cu (II) complexes and their in vitro microbial, cytotoxic, DNA cleavage studies. J. Enzym. Inhib. Med. Chem. 2010, 25, 430–439. [Google Scholar] [CrossRef]
- Singh, K.; Barwa, M.; Tyagi, P. Synthesis, Characterization and Biological Studies of Co (II), Ni (II), Cu (II) and Zn (II) Complexes with Bidentate Schiff Bases Derived by Heterocyclic Ketone. Eur. J. Med. Chem. 2006, 41, 147–153. [Google Scholar] [CrossRef]
- Shelke, V.A.; Jadhav, S.M.; Patharkar, V.R.; Shankarwar, S.G.; Munde, A.S.; Chondhekar, T.K. Synthesis, Spectroscopic Characterization and Thermal Studies of Some Rare Earth Metal Complexes of Unsymmetrical Tetradentate Schiff Base Ligand. Arab. J. Chem. 2012, 5, 501–507. [Google Scholar] [CrossRef] [Green Version]
- Shoemaker, D.P.; Garland, C.W. Experiments in Physical Chemistry; McGraw Hill International Edition: New York, NY, USA, 1989. [Google Scholar]
- Deshmukh, M.B.; Desai, S.D.; Chavan, S.S. Synthesis, X-ray Diffraction Study and Biological Activity of 7-hydroxy-4-methylquinolin-2(1H)-one. Indian J. Chem. B 2005, 44, 1659. [Google Scholar]
- Cullity, B.D. Elements of X-ray Diffraction; Addison-Wesley Pub. Co.: Reading, MA, USA, 1956. [Google Scholar]
- Janak, J.F. Proof that ∂E/∂ni = E in Density Functional Theory. Phys. Rev. B 1978, 18, 7165–7168. [Google Scholar] [CrossRef]
- Kar, R.; Song, J.; Hirao, K. Long-Range Corrected Functionals Satisfy Koopmans’ Theorem: Calculation of Correlation and Relaxation Energies. J. Comput. Chem. 2013, 34, 958–964. [Google Scholar] [CrossRef]
- Tsuneda, T.; Song, J.; Suzuki, S.; Hirao, K. On Koopmans’ Theorem in Density Functional Theory. J. Chem. Phys. 2010, 133, 174101. [Google Scholar] [CrossRef]
- Tsuneda, T.; Hirao, K. Long-Range Correction for Density Functional Theory. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2014, 4, 375–390. [Google Scholar] [CrossRef]
- Flores-Holguín, N.; Ortega-Castro, J.; Frau, J.; Glossman-Mitnik, D. Conceptual DFT-Based Computational Peptidology, Pharmacokinetics Study and ADMET Report of the Veraguamides A-G Family of Marine Natural Drugs. Mar. Drugs 2022, 20, 97. [Google Scholar] [CrossRef]
- Gázquez, J.L.; Cedillo, A.; Vela, A. Electrodonating and Electroaccepting Powers. J. Phys. Chem. A 2007, 111, 1966–1970. [Google Scholar] [CrossRef]
- Chattaraj, P.K.; Chakraborty, A.; Giri, S. Net Electrophilicity. J. Phys. Chem. A 2009, 113, 10068–10074. [Google Scholar] [CrossRef]
- Domingo, L.R.; Chamorro, E.; Perez, P. Understanding the Reactivity of Captodative Ethylenes in Polar Cycloaddition Reactions. A Theoretical Study. J. Org. Chem. 2008, 73, 4615–4624. [Google Scholar] [CrossRef]
- Jaramillo, P.; Domingo, L.R.; Chamorro, E.; Pérez, P. A Further Exploration of a Nucleophilicity Index Based on the Gas-Phase Ionization Potentials. J. Mol. Struct. THEOCHEM 2008, 865, 68–72. [Google Scholar] [CrossRef]
- Domingo, L.R.; Sáez, J.A. Understanding the Mechanism of Polar Diels-Alder Reactions. Org. Biomol. Chem. 2009, 7, 3576–3583. [Google Scholar] [CrossRef] [PubMed]
- Domingo, L.R.; Perez, P. The Nucleophilicity N Index in Organic Chemistry. Org. Biomol. Chem. 2011, 9, 7168–7175. [Google Scholar] [CrossRef] [PubMed]
- Domingo, L.R.; Ríos-Gutiérrez, M.; Pérez, P. Applications of the Conceptual Density Functional Theory Indices to Organic Chemistry Reactivity. Molecules 2016, 21, 748. [Google Scholar] [CrossRef] [Green Version]
- Schepartz, S.A.; Graver, M.R.; Chabner, B.A. The National Cancer Institute: Cancer Drug Discovery and Development Program. Semin. Oncol. 1992, 19, 622–638. [Google Scholar]
- Boyd, M.R. Anticancer Drug Development Guide: Preclinical Screening, Clinical Trials and Approval; Human Press Inc.: NY, USA, 2004. [Google Scholar]
- Vichai, V.; Kirtikara, K. Sulforhodamine B Colorimetric Assay for Cytotoxicity Screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef]

| Sl. No. | Name of Comp. | Mol. Formula | Mol. Wt | Color | Elemental Analysis | |||
|---|---|---|---|---|---|---|---|---|
| C% Found (Calc.) | H% Found (Calc.) | N% Found (Calc.) | M% Found (Calc.) | |||||
| 1 | HL1 | C10H9N5SO2 | 263 | Pale yellow | 45.20 (45.63) | 3.52 (3.42) | 26.48 (26.61) | - |
| 2 | C1 | C20H20N10S2O6Ni [Ni(L1)22H2O] | 618.69 | green | 38.71 (38.78) | 3.30 (3.23) | 22.58 (22.62) | 9.59 (9.52) |
| 3 | HL2 | C10H9N5SO2 | 263 | Yellow | 45.78 (45.63) | 3.45 (3.42) | 26.70 (26.61) | - |
| 4 | C2 | C20H20N10S2O6Ni [Ni(L2)22H2O] | 618.69 | green | 38.66 (38.78) | 3.31 (3.23) | 22.71 (22.62) | 9.61 (9.52) |
| 5 | HL3 | C10H9N5SO2 | 263 | Dark yellow | 45.52 (45.63) | 3.55 (3.42) | 26.65 (26.61) | - |
| 6 | C3 | C20H20N10S2O6Ni [Ni(L3)22H2O] | 618.69 | Yellow green | 38.70 (38.78) | 3.28 (3.23) | 22.49 (22.62) | 9.40 (9.52) |
| Sl. No. | Compd. | νC = N | νSH | νC = S | νN-H | νH2O | νM-S | νM-N |
|---|---|---|---|---|---|---|---|---|
| 1 | HL1 | 1590 | 2760 | 1114 | 3067 | - | - | - |
| 2 | C1 | 1525 | - | - | - | 3206 | 379 | 483 |
| 3 | HL2 | 1589 | 2753 | 1176 | 3072 | - | - | - |
| 4 | C2 | 1568 | - | - | - | 3311 | 332 | 491 |
| 5 | HL3 | 1579 | 2770 | 1170 | 3096 | - | - | - |
| 6 | C3 | 1552 | - | - | - | 3200 | 379 | 483 |
| Complex | γ1 (nm) | γ2 (nm) | γ3 (nm) | µ Eff. (BM) |
|---|---|---|---|---|
| C1 | 1011 | 620 | 400 | 3.45 |
| C2 | 990 | 580 | 400 | 3.23 |
| C3 | 1012 | 616 | 401 | 3.34 |
| Compound | Stages | Temp. (°C) | TG Mass % | Assignment | |
|---|---|---|---|---|---|
| Calc. | Found | ||||
| C1 | 1 2 3 | 50–180 180–400 400–550 >550 | 5.81 48.14 36.51 12.10 | 6.92 46.11 34.09 12.94 | -2.H2O (water molecules) -C14H10N4O4 (organic moiety) -C6H6N6S2 (triazole ring) -NiO (residue) |
| C2 | 1 2 3 | 50–190 190–405 405–555 >555 | 5.81 48.14 36.51 12.10 | 6.90 46.16 34.11 12.90 | -2.H2O (water molecules) -C14H10N4O4 (organic moiety) -C6H6N6S2 (triazole ring) -NiO (residue) |
| C3 | 1 2 3 | 50–165 165–435 435–560 >560 | 5.81 48.14 36.51 12.10 | 6.71 47.07 34.70 12.90 | -2.H2O (water molecules) -C14H10N4O4 (organic moiety) -C6H6N6S2 (triazole ring) -NiO (residue) |
| Sl. No. | Parameter | HL1 | HL2 | HL3 | C3 |
|---|---|---|---|---|---|
| 1 | Structure | Orthorhombic | Orthorhombic | Orthorhombic | Orthorhombic |
| 2 | Space group | I mam | I mam | I mam | I mam |
| 3 | Symmetry of lattice | Primitive | Primitive | Body centered | Primitive |
| 4 | Bond angle | α = β = γ = 90° | α = β = γ = 90° | α = β = γ = 90° | α = β = γ = 90° |
| 5 | Lattice parameters (Å) | a = 16.61 b = 08.16 c = 03.91 | a = 14.40 b = 18.52 c = 07.22 | a = 10.83 b = 16.08 c = 07.93 | a = 11.89 b = 13.24 c = 12.63 |
| 6 | Volume of the cell (V) (Å3) | 529.96 | 1927.23 | 1343.12 | 1987.79 |
| 7 | Density (g/cc) Theoretical Experimental | 0.8239 0.7512 | 0.2265 0.2016 | 0.65 0.61 | 0.52 0.47 |
| 8 | No. of molecules/unit cell (η) | 1 | 1 | 2 | 1 |
| 9 | Pore fraction | 0.0853 | 0.1099 | 0.0599 | 0.0961 |
| 10 | Crystallite size (Å) | 337 | 169 | 337 | 235 |
| 11 | Micro strain (×10−3) | 1.2649 | 219 | 1.024 | 1.376 |
| Compound | HOMO | LUMO | SOMO | H-L Gap | JI | JA | JHL | ∆SL |
|---|---|---|---|---|---|---|---|---|
| HL1 | −6.31 | −3.12 | −2.69 | 3.19 | 0.25 | 0.23 | 0.34 | 0.43 |
| HL2 | −6.28 | −2.89 | −2.56 | 3.39 | 0.24 | 0.19 | 0.31 | 0.33 |
| HL3 | −6.30 | −3.05 | −2.73 | 3.25 | 0.24 | 0.17 | 0.21 | 0.31 |
| C1 | −5.17 | −3.19 | −2.87 | 1.98 | 0.45 | 0.18 | 0.48 | 0.32 |
| C2 | −5.66 | −3.03 | −2.83 | 2.63 | 0.13 | 0.11 | 0.17 | 0.21 |
| C3 | −5.77 | −3.04 | −2.85 | 2.73 | 0.14 | 0.10 | 0.17 | 0.19 |
| Compound | X | η | ω | S | N | ω− | ω+ | Δ ω± |
|---|---|---|---|---|---|---|---|---|
| HL1 | 4.72 | 3.19 | 3.47 | 0.31 | 2.48 | 9.53 | 4.81 | 14.35 |
| HL2 | 4.59 | 3.39 | 3.10 | 0.29 | 2.51 | 8.70 | 4.12 | 12.82 |
| HL3 | 4.67 | 3.25 | 3.36 | 0.31 | 2.50 | 9.25 | 4.58 | 13.84 |
| C1 | 4.18 | 1.98 | 4.41 | 0.51 | 3.63 | 11.03 | 6.85 | 17.88 |
| C2 | 4.35 | 2.63 | 3.60 | 0.38 | 3.13 | 9.53 | 5.18 | 14.71 |
| C3 | 4.40 | 2.73 | 3.55 | 0.37 | 3.02 | 9.47 | 5.06 | 14.53 |
| Sr. No. | Compd. | Conc.mg mL−1 | Pseudomonas Aeruginosa | Staphylococcus aureus | Aspergillus niger | Candida albicans |
|---|---|---|---|---|---|---|
| 1 | HL1 | 0.2 0.4 0.6 0.8 1.0 | +++ +++ +++ +++ +++ | +++ +++ +++ +++ +++ | - - - - - | - - - - - |
| 2 | HL1-Ni | 0.2 0.4 0.6 0.8 1.0 | +++ +++ +++ +++ +++ | +++ +++ +++ +++ +++ | - - - - - | - + ++ +++ +++ |
| 3 | HL2 | 0.2 0.4 0.6 0.8 1.0 | - ++ ++ - ++ | ++ +++ +++ +++ ++ | - - - - - | ++ ++ ++ ++ +++ |
| 4 | HL2-Ni | 0.2 0.4 0.6 0.8 1.0 | - - ++ - +++ | ++ ++ +++ +++ +++ | - - - - - | - - - ++ - |
| 5 | HL3 | 0.2 0.4 0.6 0.8 1.0 | + ++ ++ +++ +++ | ++ ++ +++ +++ +++ | - - - - - | - - - +++ +++ |
| 6 | HL3-Ni | 0.2 0.4 0.6 0.8 1.0 | - - - ++ ++ | ++ +++ +++ +++ +++ | - - - - - | - - - + + |
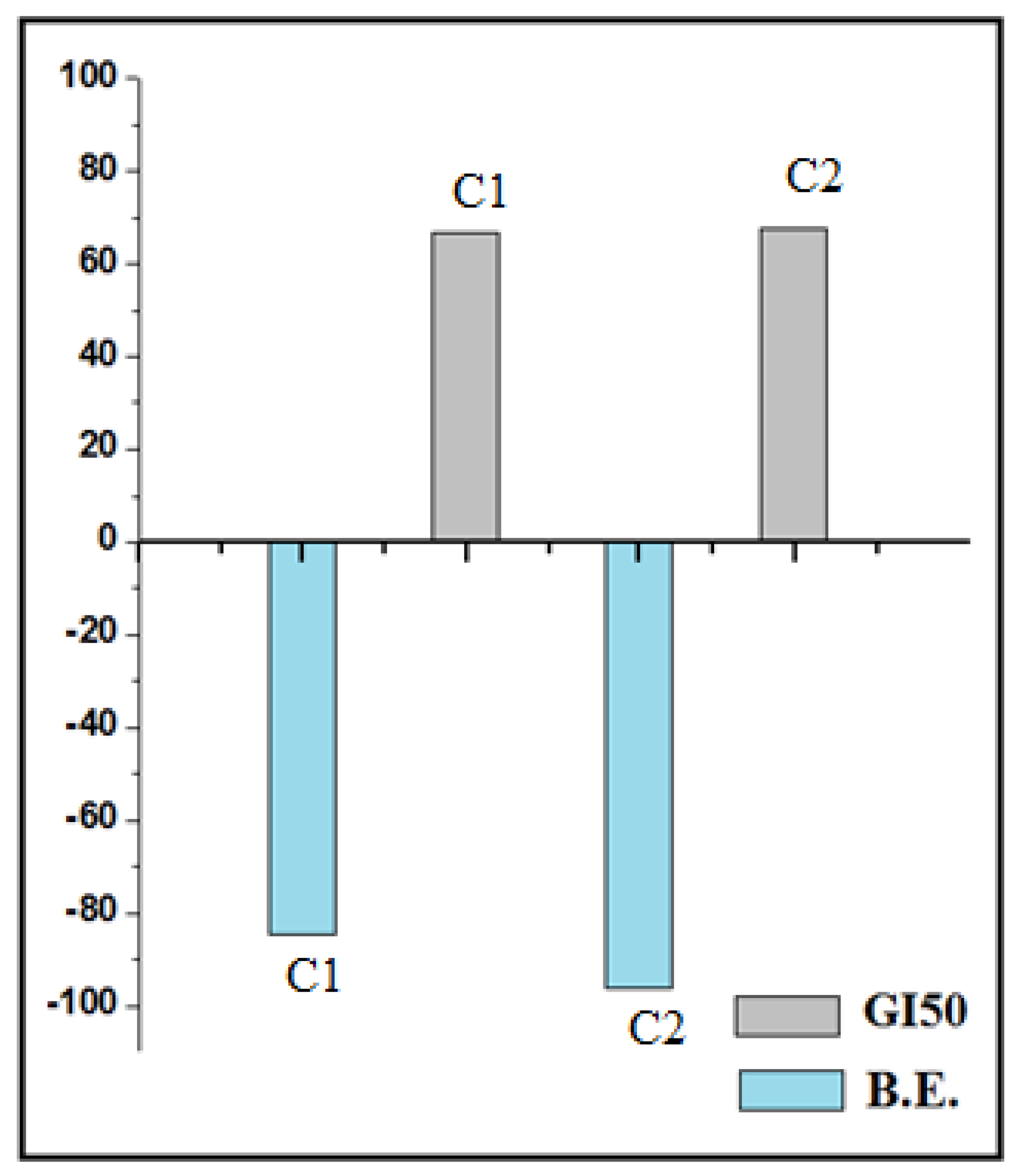
| Sr. No. | Molecule Code | B. E. (k Cal mol−1) | Interaction with Amino Acid | ||
|---|---|---|---|---|---|
| H Bond | Charge | Hydrophobic | |||
| 1 | HL1 | −40.52 | GLN22 | --- | LYS41, VAL36 |
| 2 | C1 | −84.53 | ASN142 | GLU99, ASP97 | GLY21, GLU99 |
| 3 | HL2 | −47.89 | PHE23, SER97 | π stackang-TRP43, PHE51, PHE122 VDW*- TYR18, PHE40, TRP43, SER44 etc. | LEU144, GLU121, PHE122, ALA125 |
| 4 | C2 | −96.35 | ASN142 | GLU99, | LYS103, ASN144 |
| 5 | HL3 | −47.89 | PHE23 | --- | LEU144, LYS41 |
| 6 | C3 | −88.76 | LYS139 | ASP97 | LYS139, VAL100 |
| Conc. (Molar) | Average Values for % Control Growth | ||||||
|---|---|---|---|---|---|---|---|
| HL1 | C1 | HL2 | C2 | HL3 | C3 | Adriamycin | |
| 10−7 | 124.1 | 122.4 | 157.9 | 139.2 | 51.1 | 109.7 | 43.2 |
| 10−6 | 144 | 125.3 | 173.4 | 127.6 | 129.1 | 101.8 | 3.6 |
| 10−5 | 130.6 | 113.9 | 158.3 | 144.4 | 119.2 | 93.5 | −53.9 |
| 10−4 | 73.4 | 12.7 | 90.9 | 4.8 | 85.4 | 55.1 | −55.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deodware, S.A.; Barache, U.B.; Dhale, P.C.; Gaikwad, K.D.; Shivamallu, C.; Ubale, P.A.; Shati, A.A.; Alfaifi, M.Y.; Elbehairi, S.E.I.; Achar, R.R.; et al. In Vitro Anticancer Screening, Molecular Docking and Antimicrobial Studies of Triazole-Based Nickel(II) Metal Complexes. Molecules 2022, 27, 6548. https://doi.org/10.3390/molecules27196548
Deodware SA, Barache UB, Dhale PC, Gaikwad KD, Shivamallu C, Ubale PA, Shati AA, Alfaifi MY, Elbehairi SEI, Achar RR, et al. In Vitro Anticancer Screening, Molecular Docking and Antimicrobial Studies of Triazole-Based Nickel(II) Metal Complexes. Molecules. 2022; 27(19):6548. https://doi.org/10.3390/molecules27196548
Chicago/Turabian StyleDeodware, Sachin A., Umesh B. Barache, Pratibha C. Dhale, Kundalkesha D. Gaikwad, Chandan Shivamallu, Panchsheela A. Ubale, Ali A. Shati, Mohammad Y. Alfaifi, Serag Eldin I. Elbehairi, Raghu Ram Achar, and et al. 2022. "In Vitro Anticancer Screening, Molecular Docking and Antimicrobial Studies of Triazole-Based Nickel(II) Metal Complexes" Molecules 27, no. 19: 6548. https://doi.org/10.3390/molecules27196548
APA StyleDeodware, S. A., Barache, U. B., Dhale, P. C., Gaikwad, K. D., Shivamallu, C., Ubale, P. A., Shati, A. A., Alfaifi, M. Y., Elbehairi, S. E. I., Achar, R. R., Silina, E., Stupin, V., Frau, J., Flores-Holguín, N., Gaikwad, S. H., Kollur, S. P., & Glossman-Mitnik, D. (2022). In Vitro Anticancer Screening, Molecular Docking and Antimicrobial Studies of Triazole-Based Nickel(II) Metal Complexes. Molecules, 27(19), 6548. https://doi.org/10.3390/molecules27196548









