Novel Unsymmetric 3,5-Bis(benzylidene)-4-piperidones That Display Tumor-Selective Toxicity
Abstract
:1. Introduction
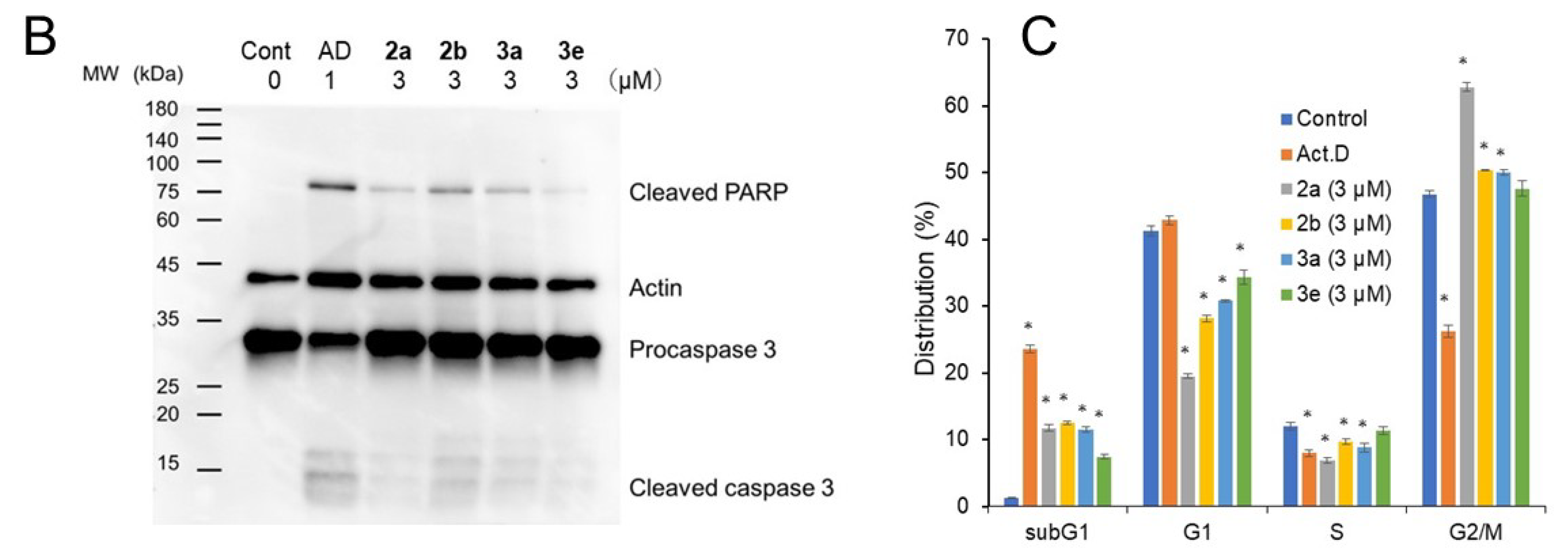
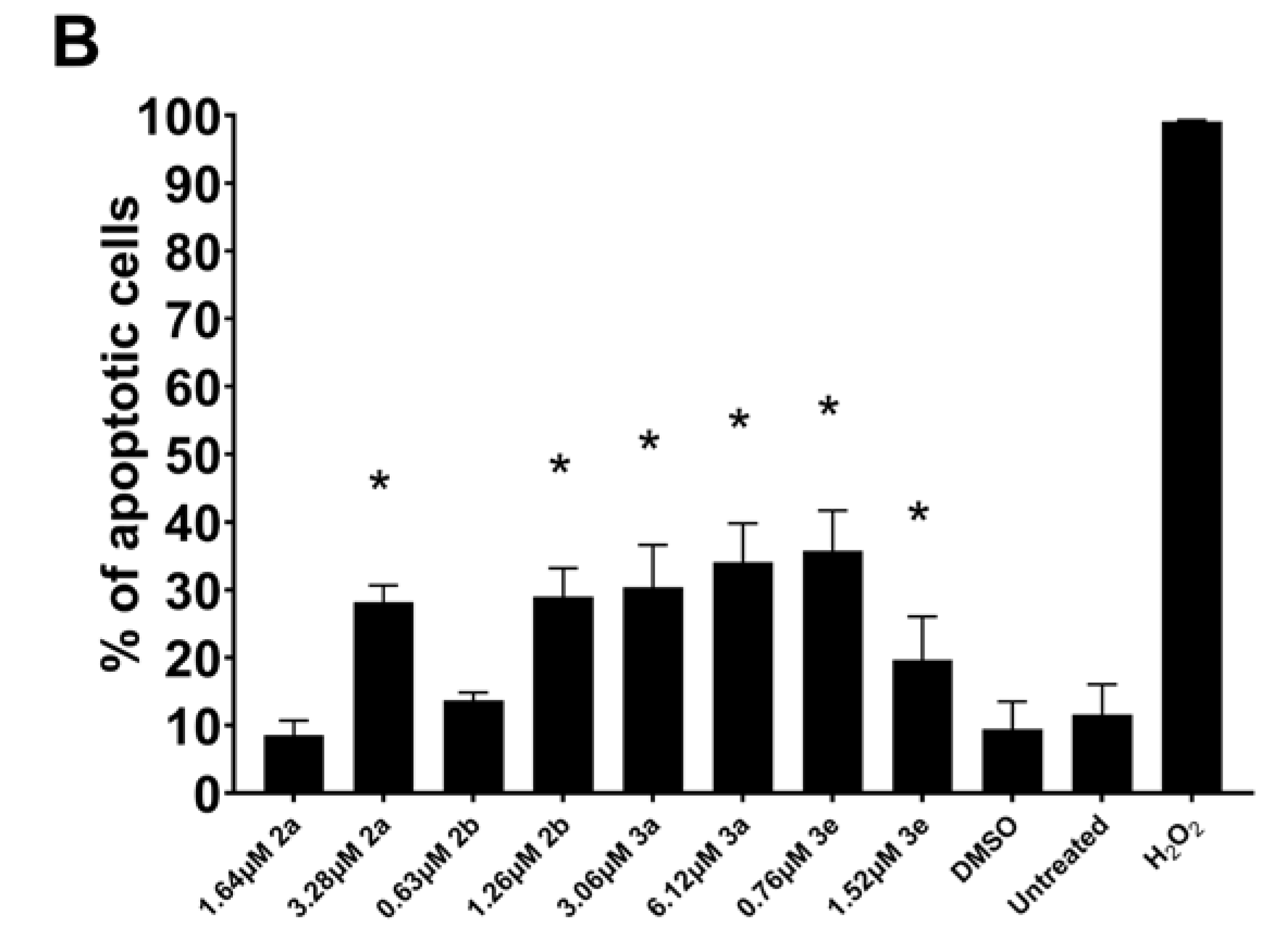
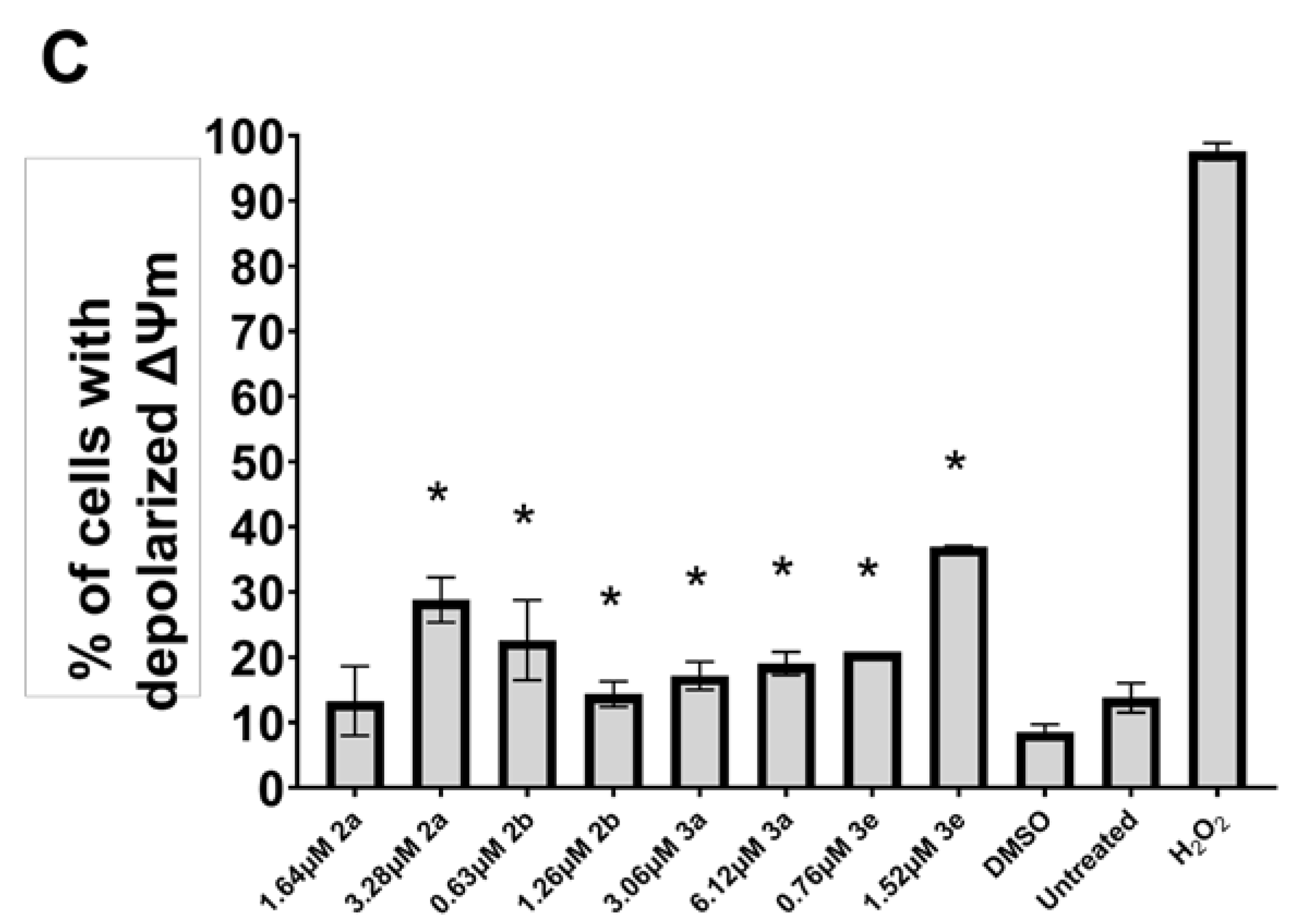
2. Results
3. Discussion
4. Materials and Methods
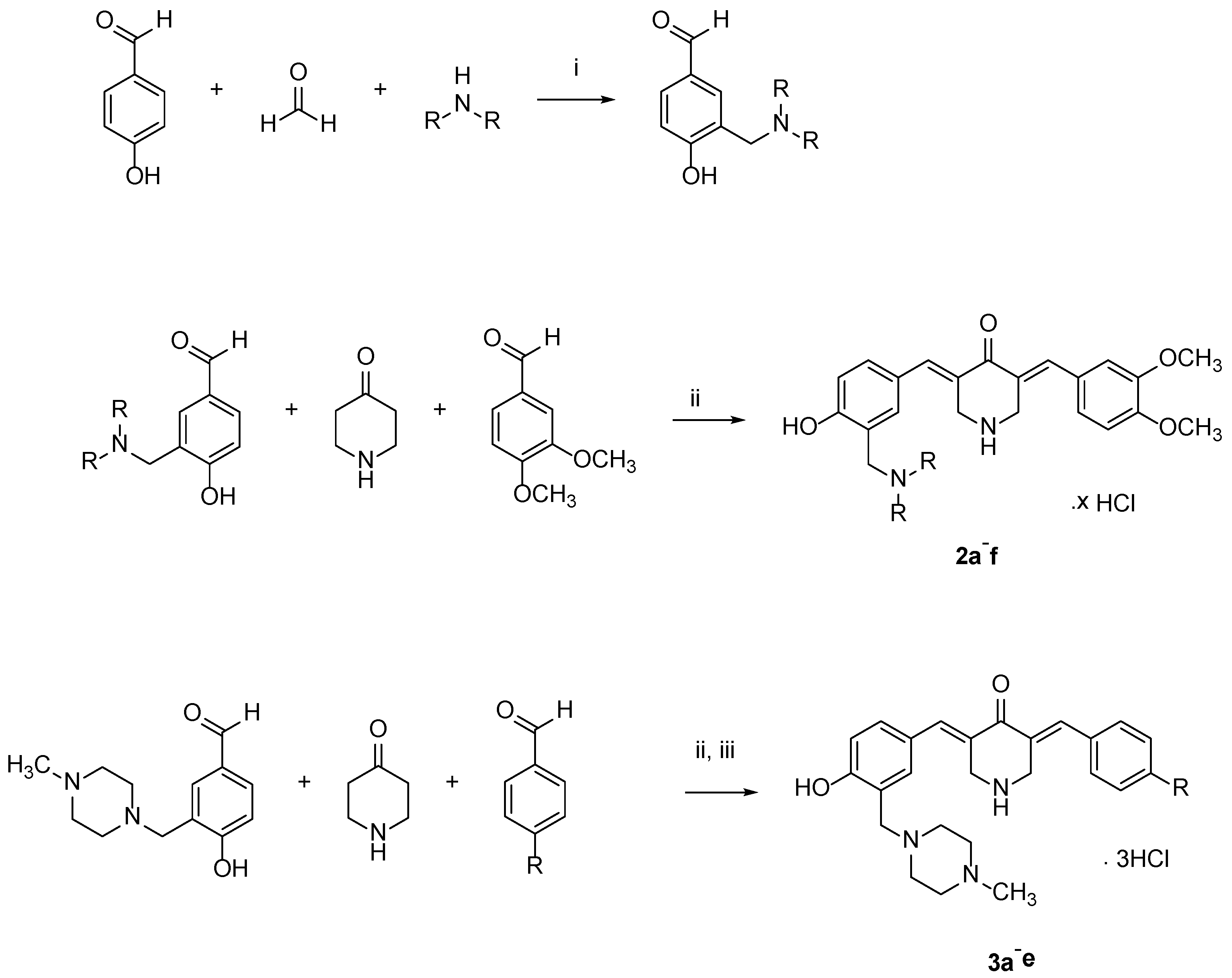
4.1. Syntheses of Compounds
4.2. Bioevaluations
4.2.1. Cytotoxicity Assays
4.2.2. Bioassays
4.3. ADME Determination
4.4. Statistical Treatment
5. Conclusions
- This study revealed that the compounds in series 2 and 3 are novel potent cytotoxins. In several instances, their potencies exceed that of established anticancer agents. A further positive feature is their greater toxicity to tumors than non-malignant cells in a number of bioassays.
- The modes of action whereby representative compounds exert their bioactivity was determined in some cases.
- The compounds are toxic to different clusters of cancers. In the future, this section of the project needs to be expanded in order to evaluate the efficacy of 2a–f and 3a–e on such important tumors as breast, lung, and prostate cancers.
- Some support was gathered for the hypothesis that SI values are influenced by differences in the atomic charges on the olefinic carbon atoms. In other words, sufficient encouragement has been found to continue evaluating this theory with other clusters of compounds and different biological targets.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Maydt, D.; De Spirt, S.; Muschelknautz, C.; Stahl, W.; Muller, T.J.J. Chemical reactivity and biological activity of chalcones and other α,β-unsaturated carbonyl compounds. Xenobiotica 2013, 43, 711–718. [Google Scholar] [CrossRef]
- Dimmock, J.R.; Shyam, K.; Hamon, N.W.; Logan, B.M.; Raghavan, S.K.; Smith, P.J. Evaluation of some Mannich bases derived from substituted acetophenones against P388 lymphocytic leukemia and on respiration in isolated rat liver mitochondria. J. Pharm. Sci. 1983, 72, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Mutus, B.; Wagner, J.D.; Talpas, C.J.; Dimmock, J.R.; Philips, O.A.; Reid, R.S. 1-p-Chlorophenyl-4,4-dimethyl-5-diethylamino-1-penten-3-one hydrobromide, a sulfhydryl-specific compound which reacts irreversibly with small molecular weight thiols. Anal. Biochem. 1989, 177, 237–243. [Google Scholar] [CrossRef]
- Karki, S.S.; Das, U.; Umemura, N.; Sakagami, H.; Iwamoto, S.; Kawase, M.; Balzarini, J.; De Clercq, E.; Dimmock, S.G.; Dimmock, J.R. 3,5-Bis(3-alkylaminomethyl-4-hydroxybenzylidene)-4-piperidones: A novel class of potent tumor-selective cytotoxins. J. Med. Chem. 2016, 59, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Addala, E.; Raffiei, H.; Das, S.; Bandy, B.; Das, U.; Karki, S.S.; Dimmock, J.R. 3,5-Bis(dimethyl aminomethyl-4-hydroxybenzylidene)-4-piperidone and related compounds induce glutathione oxidation and mitochondria-mediated cell death in HCT-116 colon cancer cells. Bioorg. Med. Chem. Lett. 2017, 27, 3669–3673. [Google Scholar] [CrossRef]
- Lu, Y.; Li, C.-M.; Wang, Z.; Ross, C.R., II; Chen, J.; Dalton, J.T.; Li, W.; Miller, D.D. Discovery of 4-substituted methoxybenzoyl-aryl-thiazole as novel anticancer agents: Synthesis, biological evaluation and structure-activity relationships. J. Med. Chem. 2009, 52, 1701–1711. [Google Scholar] [CrossRef] [Green Version]
- Roayapalley, P.K.; Sakagami, H.; Satoh, K.; Amano, S.; Bandow, K.; Aguilera, R.J.; Hernandaz, K.G.C.; Bustamante, A.Y.S.; Dimmock, S.G.; Sharma, R.K.; et al. Cytotoxic tumor-selective 1,5-diaryl-3-oxo-1,4-pentadienes mounted on a piperidine ring. Medicines 2021, 8, 78. [Google Scholar] [CrossRef]
- Das, S.; Das, U.; Michel, D.; Gorecki, D.K.J.; Dimmock, J.R. Novel 3,5-bis(arylidene)-4-piperidone dimers: Potent cytotoxins against colon cancer cells. Eur. J. Med. Chem. 2013, 64, 321–328. [Google Scholar] [CrossRef]
- Das, U.; Doroudi, A.; Das, S.; Bandy, B.; Balzarini, J.; De Clercq, E.; Dimmock, J.R. E, E-2- Benzylidene-6-(nitrobenzylidene)cyclohexanones: Syntheses, cytotoxicity and an examination of some of their electronic, steric and hydrophobic properties. Bioorg. Med. Chem. 2008, 16, 6261–6268. [Google Scholar] [CrossRef] [Green Version]
- Das, U.; Gul, H.I.; Alcorn, J.; Shrivastav, A.; George, T.; Sharma, R.K.; Nienaber, K.H.; De Clercq, E.; Balzarini, J.; Kawase, M.; et al. Cytotoxic 5-aryl-1-(nitrophenyl)-3-oxo-1,4-pentadienes mounted on alicyclic scaffolds. Eur. J. Med. Chem. 2006, 41, 577–585. [Google Scholar] [CrossRef]
- Dimmock, J.R.; Padmanilayam, M.P.; Zello, G.A.; Quail, J.W.; Oloo, E.O.; Prisciak, J.S.; Kraatz, H.-B.; Cherkasov, A.; Lee, J.S.; Allen, T.M.; et al. Cytotoxic 1,3-diarylidene-2-tetralones and related compounds. Eur. J. Med. Chem. 2002, 37, 813–824. [Google Scholar] [CrossRef]
- Dimmock, J.R.; Kumar, P.; Nazarali, A.J.; Motaganahalli, N.L.; Kowalchuk, T.P.; Beazely, M.A.; Quail, J.W.; Oloo, E.O.; Allen, T.M.; Szydlowski, J.; et al. Cytotoxic 2,6-bis(arylidene)cyclohexanones and related compounds. Eur. J. Med. Chem. 2000, 35, 967–977. [Google Scholar] [CrossRef]
- Yao, B.-R.; Sun, Y.; Chen, S.-L.; Suo, H.-D.; Zhang, Y.-L.; Wei, H.; Wang, C.H.; Zhou, F.; Cong, W.; Xin, W.Y.; et al. Disymmetric pyridyl-substituted 3,5-bis(arylidene)-4-piperidones as anti-hepatoma agents by inhibiting NF-kB pathway activation. Eur. J. Med. Chem. 2019, 167, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xin, W.-Y.; Yao, B.-R.; Cong, W.; Wang, C.-H.; Hou, G.G. N-Phenylsulfonyl-3,5-bis(arylidene)-4-piperidone derivatives as activation NF-kB inhibitors in hepatic carcinoma cell lines. Eur. J. Med. Chem. 2018, 155, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xin, W.-Y.; Yao, B.-R.; Wang, C.-H.; Cong, W.; Zhao, F.; Li, H.-J.; Hou, Y.; Meng, Q.-G.; Hou, G.G. Novel dissymmetric 3,5-bis(arylidene)-4-piperidones as potential antitumor agents with biological evaluation in vitro and in vivo. Eur. J. Med. Chem. 2018, 147, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.; Hall, S.C.; Wiggington, P.J.; Roth, S.M.; Das, S.; Das, U.; Roayapalley, P.K.; Dimmock, J.R. Cytotoxic benzylidene hydrazides of terephthalic acid and related compounds. Pharmazie 2022, 77, 90–94. [Google Scholar]
- Azwar, S.; Seow, H.F.; Abdullah, M.; Jabar, M.F.; Mohtaprubin, N. Recent updates in mechanisms of resistance to 5-fluorouracil and reversal strategies in colon cancer treatment. Biology 2021, 10, 854. [Google Scholar] [CrossRef]
- Albert, A.; Serjeant, E.P. The Determination of Ionization Constants: A Laboratory Manual, 3rd ed.; Chapman and Hall: London, UK, 1984; pp. 151–152. [Google Scholar]
- Bayr, H. Reactive oxygen species. Crit. Care Med. 2005, 33, S498–S501. [Google Scholar] [CrossRef]
- Spartan ’18; Wavefunction, Inc.: Irvine, CA, USA, 2019; Available online: https://downloads.wavefun.com/FAQ/Spartan18Manual.pdf (accessed on 3 May 2022).
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Sugita, Y.; Takao, K.; Uesawa, Y.; Nagai, J.; Iijima, Y.; Sano, M.; Sakagami, H. Development of newly synthesized chromone derivatives with high tumor specificity against human oral squamous cell carcinoma. Medicines 2020, 7, 50. [Google Scholar] [CrossRef]
- Takao, K.; Hoshi, K.; Sakagami, H.; Shi, H.; Bandow, K.; Nagai, J.; Uesawa, Y.; Tomomura, A.; Tomomura, M.; Sugita, Y. Further quantitative structure-cytotoxicity relationship analysis of 3-styrylchromones. Anticancer Res. 2020, 40, 87–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vichai, V.; Kirtikara, K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protocols 2006, 1, 1112–1116. [Google Scholar] [CrossRef] [PubMed]
- Lema, C.; Varela-Ramirez, A.; Aguilera, R.J. Differential nuclear staining assay for high-throughput screening to identify cytotoxic compounds. Curr. Cell Biochem. 2022, 1, 1–14. [Google Scholar]
- Santiago-Vazquez, Y.; Das, U.; Varela-Ramirez, A.; Baca, S.T.; Ayala-Marin, Y.; Lema, C.; Das, U.; Baryyan, A.; Dimmock, J.R.; Aguilera, R.J. Tumor-selective cytotoxicity of a novel pentadiene analogue on human leukemia/lymphoma cells. Clin. Cancer Drugs 2016, 3, 138–146. [Google Scholar] [CrossRef] [Green Version]
- Varela-Ramirez, A.; Costanzo, M.; Carrasco, Y.P.; Pannel, K.H.; Aguilera, R.J. Cytotoxic effects of two organotin compounds and their mode of inflicting cell death on four mammalian cancer cells. Cell Biol. Toxicol. 2011, 27, 159–168. [Google Scholar] [CrossRef] [Green Version]
- Contreras, L.; Medina, S.; Bustamante, A.Y.S.; Borrego, E.A.; Valenzuela, C.A.; Das, U.; Karki, S.S.; Dimmock, J.R.; Aguilera, R.J. Three novel piperidones exhibit tumor-selective cytotoxicity on leukemia cells via protein degradation and stress-mediated mechanisms. Pharmacol. Rep. 2022, 74, 159–174. [Google Scholar] [CrossRef]
- SwissADME Software Program, Swiss Institute of Bioinformatics, Lausanne, Switzerland. Available online: http://www-swissadme.ch/ (accessed on 3 May 2022).
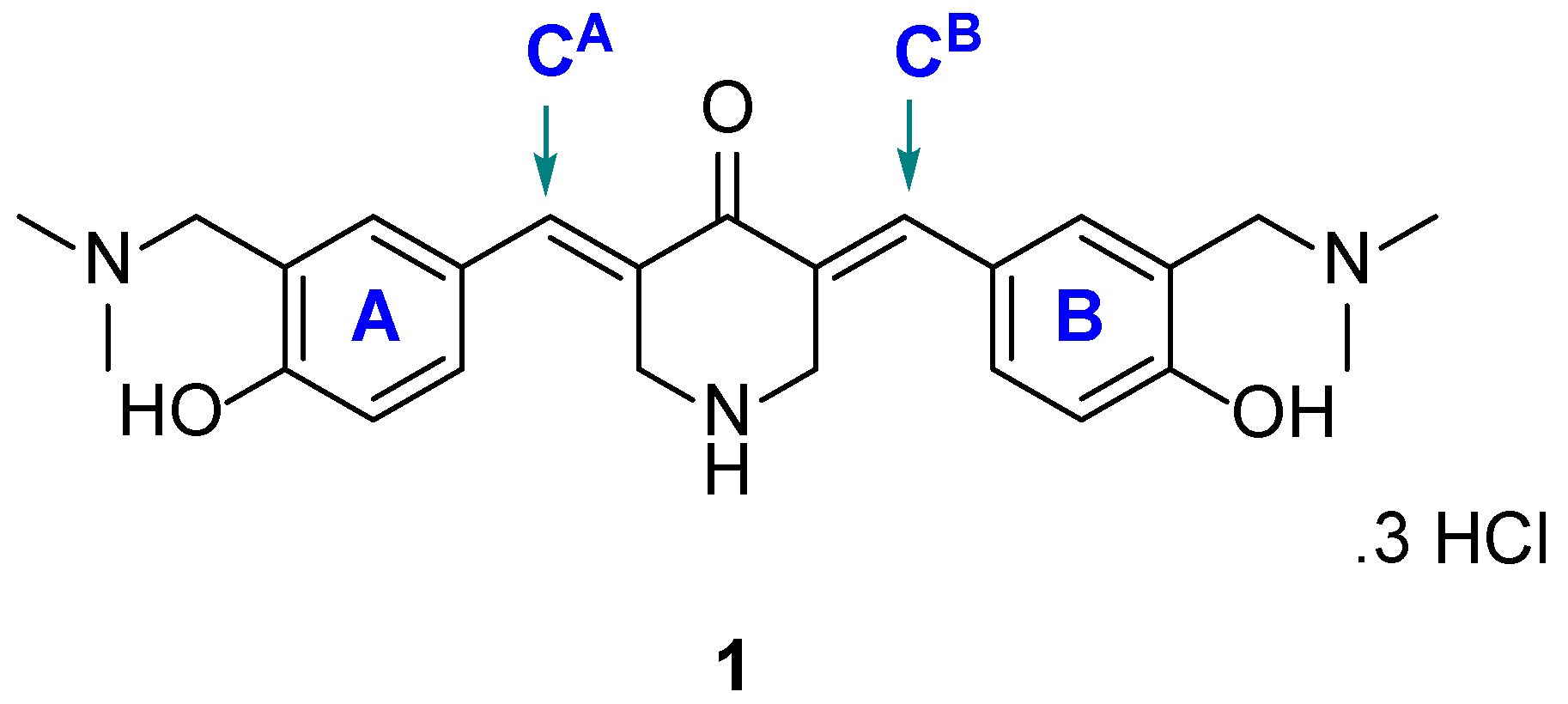
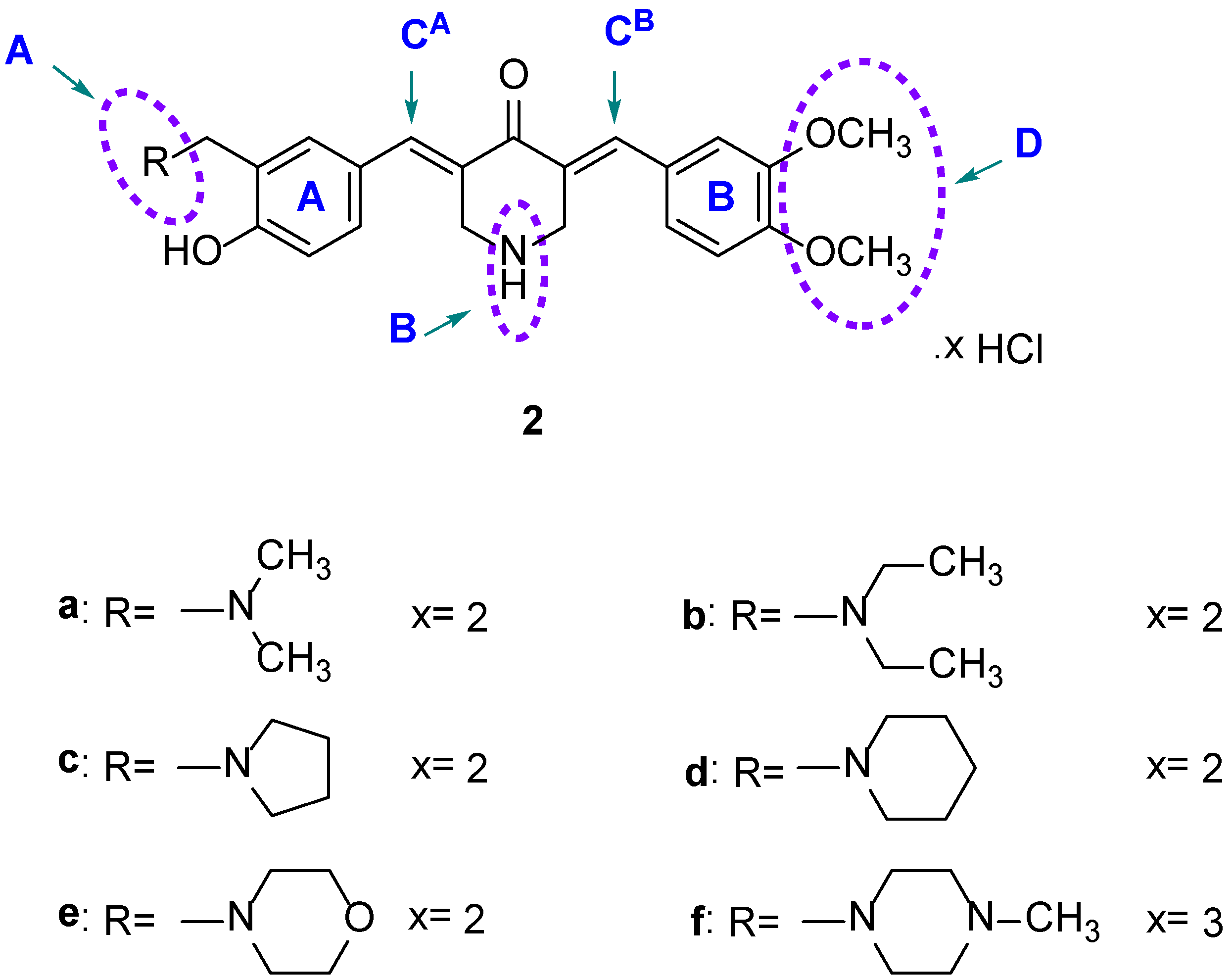
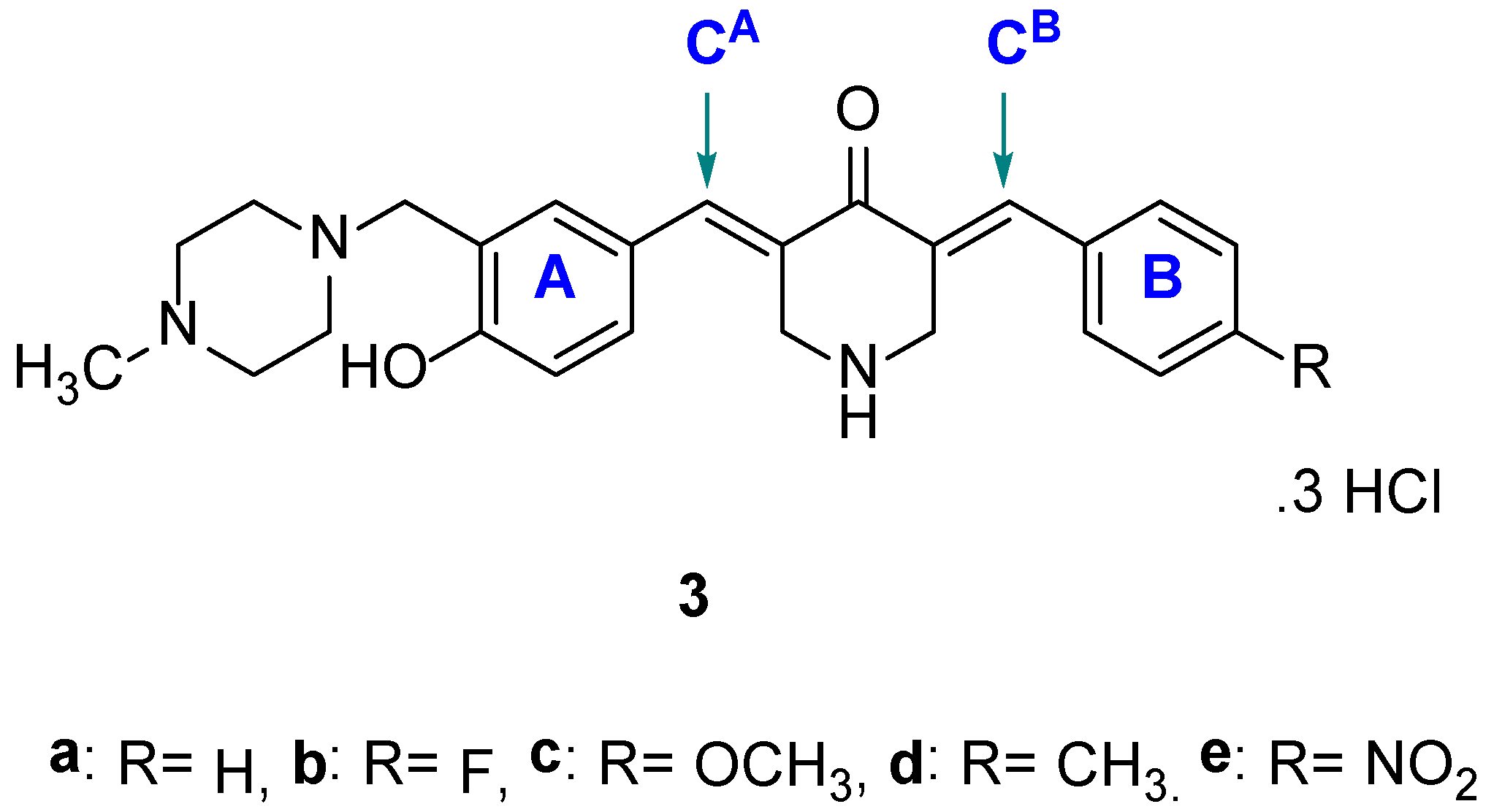

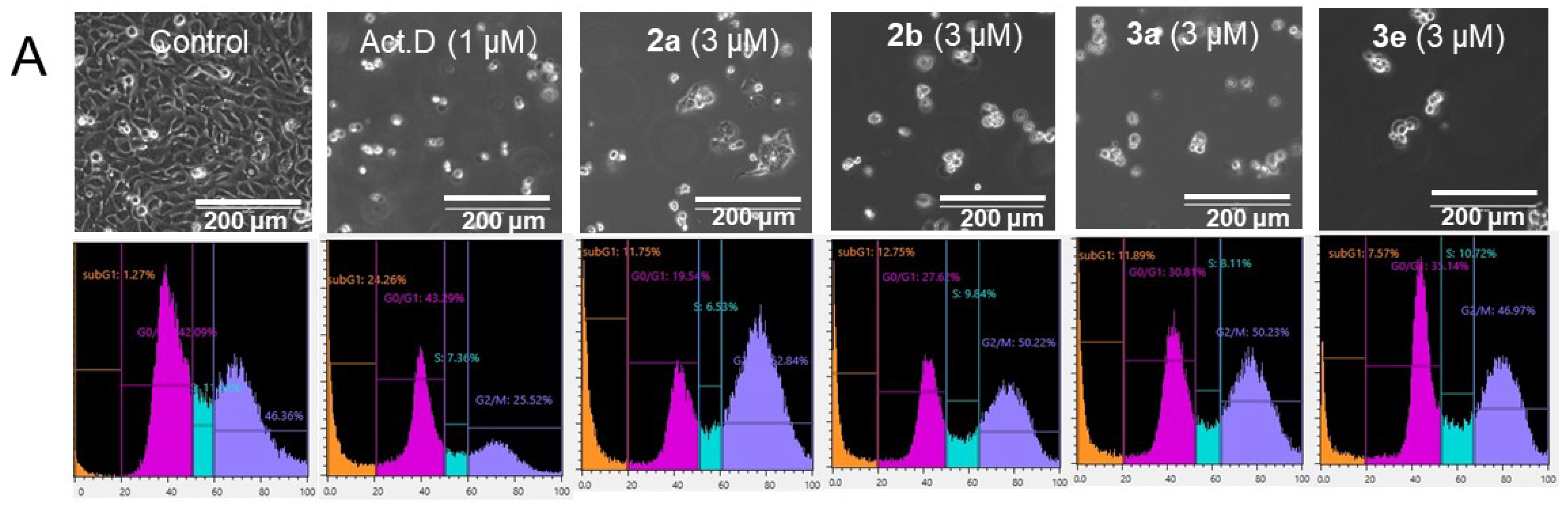

| Compound | HCT116 | HT29 | Average | CRL1790 | PSE b | |||
|---|---|---|---|---|---|---|---|---|
| IC50 (µM) | SI a | IC50 (µM) | SI a | IC50 | SI | IC50 (µM) | ||
| 1 | 3.27 ± 0.10 | 1.07 | 0.61 ± 0.06 | 5.74 | 1.94 | 3.41 | 3.50 ± 0.28 | 176 |
| 2a | 1.73 ± 0.48 | 10.1 | 1.80 ± 1.24 | 9.72 | 1.77 | 9.91 | 17.5 ± 0.58 | 560 |
| 2b | 0.20 ± 0.03 | 76.0 | 0.29 ± 0.08 | 52.4 | 0.25 | 64.2 | 15.2 ± 0.15 | 25,680 |
| 2c | 3.44 ± 0.42 | 7.67 | 1.57 ± 0.54 | 16.8 | 2.51 | 12.2 | 26.4 ± 0.28 | 486 |
| 2d | 0.45 ± 0.15 | 52.2 | 0.54 ± 0.17 | 43.5 | 0.50 | 47.9 | 23.5 ± 0.26 | 9580 |
| 2e | 3.37 ± 0.68 | 8.13 | 3.75 ± 0.78 | 7.31 | 3.56 | 7.72 | 27.4 ± 0.42 | 217 |
| 2f | 0.68 ± 0.31 | 18.7 | 0.76 ± 0.30 | 16.7 | 0.72 | 17.7 | 12.7 ± 0.31 | 2458 |
| 3a | 0.33 ± 0.11 | 65.8 | 0.40 ± 0.03 | 54.3 | 0.37 | 60.1 | 21.7 ± 0.25 | 16,243 |
| 3b | 0.28 ± 0.16 | 45.0 | 0.26 ± 0.08 | 48.5 | 0.27 | 46.8 | 12.6 ± 0.24 | 17,333 |
| 3c | 1.04 ± 0.48 | 12.7 | 0.76 ± 0.26 | 17.4 | 0.90 | 15.1 | 13.2 ± 0.36 | 1678 |
| 3d | 3.91 ± 1.69 | 5.78 | 0.71 ± 0.29 | 31.8 | 2.31 | 18.8 | 22.6 ± 0.59 | 814 |
| 3e | 0.34 ± 0.09 | 46.5 | 0.32 ± 0.07 | 49.4 | 0.33 | 48.0 | 15.8 ± 0.27 | 14,546 |
| 5-FU | 3.54 ± 0.72 | 4.97 | 14.4 ± 0.65 | 1.22 | 8.97 | 3.10 | 17.6 ± 0.82 | 34.6 |
| Compound | Ca9-22 | HSC-2 | HSC-3 | HSC-4 | Average | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| CC50 a | SI b | CC50 a | SI b | CC50 a | SI b | CC50 a | SI b | CC50 a | SI b | |
| 2a | 0.68 | 11.4 | 1.20 | 6.44 | 1.06 | 7.29 | 1.39 | 5.56 | 1.08 | 7.67 |
| 2b | 0.65 | 10.8 | 1.01 | 6.96 | 0.76 | 9.25 | 1.16 | 6.06 | 0.90 | 8.27 |
| 2c | 8.20 | 6.54 | 14.0 | 3.83 | 11.7 | 4.58 | 14.1 | 3.80 | 12.0 | 4.69 |
| 2d | 0.42 | <8.98 | 0.61 | <6.18 | 0.39 | <9.67 | 0.75 | 5.03 | 0.54 | 7.47 |
| 2e | 41.8 | >4.79 | 88.5 | >2.26 | 73.2 | >2.73 | >3.7 | 2.71 | 69.3 | 3.12 |
| 2f | 1.94 | 8.81 | 2.80 | 6.11 | 2.47 | 6.92 | 2.73 | 6.26 | 2.49 | 7.03 |
| 3a | 0.43 | 13.6 | 0.67 | 8.70 | 0.49 | 11.9 | 0.71 | 8.21 | 0.58 | 10.6 |
| 3b | 0.55 | 18.4 | 0.91 | 11.1 | 0.95 | 10.6 | 1.14 | 8.86 | 0.89 | 12.2 |
| 3c | 0.66 | 8.33 | 1.10 | 5.00 | 1.05 | 5.24 | 1.18 | 4.66 | 1.00 | 5.81 |
| 3d | 1.30 | 7.85 | 2.20 | 4.64 | 1.91 | 5.34 | 2.29 | 4.45 | 1.93 | 5.57 |
| 3e | 0.33 | 17.3 | 0.58 | 9.83 | 0.47 | 12.1 | 0.60 | 9.50 | 0.50 | 12.2 |
| 5-FU c | 141 | >7.09 | 359 | >2.79 | 450 | >2.22 | 7.8 | >128 | >240 | >35.0 |
| MPN c | 42.7 | >4.68 | 17.0 | >11.8 | 20.8 | >9.62 | 10.4 | >19.2 | 22.7 | >11.3 |
| CDDP c | 212 | >4.72 | 204 | >4.90 | 46.9 | >21.3 | 60.4 | >16.6 | 131 | >11.1 |
| DXR c | 0.33 | >30.3 | 0.20 | >50.0 | 0.24 | >41.7 | 0.14 | >71.4 | 0.23 | >48.4 |
| Compound | CC50 (µM) a | PSE b | |||
|---|---|---|---|---|---|
| HGF | HPLF | HPC | Average | ||
| 2a | 15.9 | 5.0 | 2.3 | 7.73 | 710 |
| 2b | 13.3 | 5.5 | 2.3 | 7.03 | 951 |
| 2c | 68.6 | 48.9 | 43.3 | 53.6 | 39.1 |
| 2d | 7.0 | 2.7 | <1.6 | <3.77 | 1383 |
| 2e | >200 | >200 | >200 | >200 | >4.50 |
| 2f | 33.1 | 12.0 | 6.1 | 17.1 | 282 |
| 3a | 10.2 | 4.7 | 2.6 | 5.83 | 1828 |
| 3b | 18.5 | 7.4 | 4.3 | 10.1 | 1371 |
| 3c | 8.8 | 4.9 | 2.8 | 5.50 | 593 |
| 3d | 14.6 | 10.1 | 6.0 | 10.2 | 289 |
| 3e | 12.5 | 2.7 | 1.9 | 5.70 | 2440 |
| 5-FU c | >1000 | >1000 | >1000 | >1000 | >14.6 |
| MPN c | >200 | >200 | >200 | >200 | >49.8 |
| CDDP c | >1000 | >1000 | >1000 | >1000 | >8.47 |
| DXR c | >10 | >10 | >10 | >10 | >21,044 |
| Compound | CC50 (µM) CEM Cells | CC50 (µM) HL-60 Cells | ||
|---|---|---|---|---|
| 24 h | 48 h | 24 h | 48 h | |
| 2a | 1.64 ± 0.68 | 0.82 ± 0.02 | 1.99 ± 0.92 | 2.21 ± 0.37 |
| 2b | 0.63 ± 0.08 | 0.82 ± 0.03 | 7.75 ± 0.26 | 4.82 ± 0.91 |
| 2c | >10 | 7.92 ± 0.98 | >10 | >10 |
| 2f | 8.91 ± 1.32 | 2.89 ± 0.20 | 3.75 ± 1.07 | 2.73 ± 0.57 |
| 3a | 3.06 ± 0.08 | 3.32 ± 0.30 | 8.27 ± 0.40 | 1.67 ± 0.54 |
| 3b | 3.15 ± 1.29 | 0.40 ± 0.03 | 5.90 ± 0.76 | 2.65 ± 0.43 |
| 3e | 0.76 ± 0.15 | 0.83 ± 0.03 | 4.11 ± 24.5 | 6.63 ± 0.68 |
| Average SI Data | Charges | Plots a | Correlations (p < 0.05) and Trends to a Correlation (p < 0.1) |
|---|---|---|---|
| 1, 2a–f, 3a–e | Mulliken | l | p < 0.05 |
| Mulliken | sl | p < 0.10 | |
| Natural | log | p < 0.10 | |
| 2a–f, 3a–e | Electrostatic | l | p < 0.05 |
| Natural | l | p < 0.05 | |
| Electrostatic | sl | p < 0.05 | |
| Natural | sl | p < 0.10 |
| Compound | MW (g/mol) a | log P a | HBA a | HBD a | RB a | TPSA (Å2) a |
|---|---|---|---|---|---|---|
| 2b | 436.54 | 3.14 | 6 | 2 | 8 | 71.03 |
| 2d | 448.55 | 2.90 | 6 | 2 | 6 | 71.03 |
| 3a | 403.52 | 1.27 | 5 | 2 | 4 | 55.81 |
| 3b | 421.51 | 1.83 | 6 | 2 | 4 | 55.81 |
| 3c | 433.54 | 1.28 | 6 | 2 | 5 | 65.04 |
| Ideal Compound | ≯500 | ≯5 | ≯10 | ≯5 | <10 | <140 Å2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chhikara, A.; Roayapalley, P.K.; Sakagami, H.; Amano, S.; Satoh, K.; Uesawa, Y.; Das, U.; Das, S.; Borrego, E.A.; Guerena, C.D.; et al. Novel Unsymmetric 3,5-Bis(benzylidene)-4-piperidones That Display Tumor-Selective Toxicity. Molecules 2022, 27, 6718. https://doi.org/10.3390/molecules27196718
Chhikara A, Roayapalley PK, Sakagami H, Amano S, Satoh K, Uesawa Y, Das U, Das S, Borrego EA, Guerena CD, et al. Novel Unsymmetric 3,5-Bis(benzylidene)-4-piperidones That Display Tumor-Selective Toxicity. Molecules. 2022; 27(19):6718. https://doi.org/10.3390/molecules27196718
Chicago/Turabian StyleChhikara, Aruna, Praveen K. Roayapalley, Hiroshi Sakagami, Shigeru Amano, Keitaro Satoh, Yoshihiro Uesawa, Umashankar Das, Swagatika Das, Edgar A. Borrego, Cristina D. Guerena, and et al. 2022. "Novel Unsymmetric 3,5-Bis(benzylidene)-4-piperidones That Display Tumor-Selective Toxicity" Molecules 27, no. 19: 6718. https://doi.org/10.3390/molecules27196718
APA StyleChhikara, A., Roayapalley, P. K., Sakagami, H., Amano, S., Satoh, K., Uesawa, Y., Das, U., Das, S., Borrego, E. A., Guerena, C. D., Hernandez, C. R., Aguilera, R. J., & Dimmock, J. R. (2022). Novel Unsymmetric 3,5-Bis(benzylidene)-4-piperidones That Display Tumor-Selective Toxicity. Molecules, 27(19), 6718. https://doi.org/10.3390/molecules27196718









