Pseudomonas Lipopeptide-Mediated Biocontrol: Chemotaxonomy and Biological Activity
Abstract
:1. Introduction
2. Methodology
3. Network Analysis Showing the Distribution of Pseudomonas LP-Related Articles
4. Genome Comparison of Selected Lipopeptide-Producing Pseudomonas spp.
5. Chemical Diversity of Beneficial Pseudomonas LPs
6. Pseudomonas LPs: Broad Spectrum Arsenals for Biological Control of Plant Pathogens
6.1. The P. fluorescens SG: Houses the Viscosin Group, Certain Members of the Tolaasin Group and the Poaeamide Producer
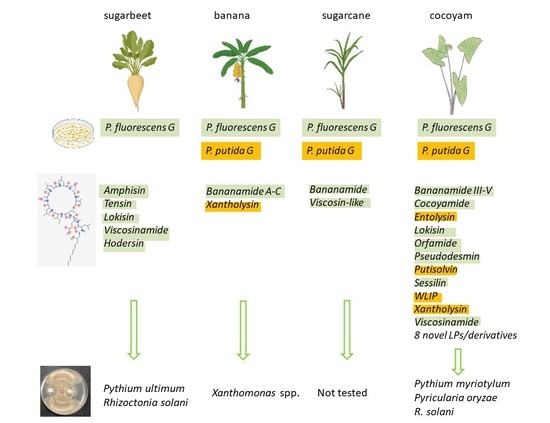
6.2. The P. koreensis SG: LP Cocktail Comprising Amphisin, Bananamide and Cocoyamide Groups
6.3. The P. protegens SG: Home to Multiple Orfamide Derivatives and Sessilins
6.4. P. chlororaphis SG: Pseudodesmin, WLIP and Uncharacterized Viscosin Group LPs
6.5. P. mandelii SG, P. asplenii SG and P. corrugata SG: Thin Borderline between Pathogenic and Beneficial LPs I
6.6. P. syringae G: Thin Borderline between Pathogenic and Beneficial LPs II
6.7. P. putida G: Beneficial LPs with Broad-Spectrum Targets
6.8. Mapping Strain Taxonomy to LP Chemistry and Antimicrobial Activity
7. Pseudomonas LPs: Emerging Broad-Spectrum Arsenals in Plant–Pathogen and Microbe–Microbe Interactions
7.1. LP-Mediated Induced Systemic Resistance (ISR)
7.2. Microbial Competition: Bacterial Mycophagy and White Line-in-Agar Interaction
8. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Strain | Lipopeptide | Accesssion Number | Reference |
|---|---|---|---|
| P. azadiae sp. nov. SWRI103 | Uncharacterized | NZ_JAHSTY010000001 | [92] |
| P. fluorescens SBW25 | Viscosin | NC_012660.1 | [35] |
| P. lactis SS101 | Massetolide | NZ_CM001513.1 | [135] |
| P. poae RE*1-1-14 | Poaeamide | NC_020209.1 | [41] |
| P. tolaasii strain 2192T | Tolaasiin | NZ_CP020369.1 | [136] |
| P. fluorescens Pf0-1 | Gacamide | NC_007492.2 | [53] |
| P. fluorescens MS82 | Putative Bananamide producer | NZ_CP028826.1 | [29,137] |
| P. kribbensis 46-2 T | Bananamide-like | NZ_CP029608.1 | [138] |
| P. granadensis LMG 27940 | MDN-066 (Bananamide-like) | NZ_LT629778.1 | [139,140] |
| P. chlororaphis PBSt-2 | WLIP | CP027716.1 | [69] |
| P. chlororaphis DSM 21509 | Viscosin group | NZ_LT629761.1 | [96] |
| P. chlororaphis Lzh-T5 | Viscosin group | NZ_CP025309.1 | [38] |
| P. protegens Cab57 | Orfamide | NZ_AP014522.1 | [94] |
| P. protegens Pf-5 | Orfamide | NC_004129.6 | [79] |
| P. protegens CHA0 | Orfamide | NC_021237.1 | [56] |
| P. sessilinigenes sp. nov. CMR12a | Orfamide and Sessilin | NZ_CP077074.1 | [59] |
| P. fuscovaginae LMG 2158T | Fuscopeptin, syringotoxin | NZ_LT629972.1 | [28] |
| P. asplenii ATCC 23835 | Fuscopeptin, syringostatin | NZ_LT629777.1 | [28] |
| P. mandelii LMG 21607 T = LMG 2210 | Uncharacterized | NZ_LT629796.1 | [26] |
| P. brassicacearum DF41 | Sclerosin, Thanamycin-var1 | NZ_CP007410.1 | [61,141] |
| P. mediterranea DSM 16733 T | Thanamycin, Peptin 22-var1 | NZ_LT629790.1 | [28] |
| P. viciae 11K1 | Brasmycin, Braspeptin | NZ_CP035088.1 | [62] |
| P. syringae B728a | Syringomycin, Syringopeptin SP22, Syringafactin | NC_007005.1 | [28,142] |
| P. syringae B301D | Syringomycin, Syringopeptin SP22, Syringafactin | NZ_CP005969.1 | [143,144] |
| P. syringae pv. syringae HS191 | Syringomycin, Syringopeptin SP25, Syringafactin | NZ_CP006256.1 | [143,144] |
| P. syringae pv. tomato DC3000 | Syringafactin | NC_004578.1 | [145] |
| P. cichorii JBC1 | Pseudomycin, Cichopeptin, cichofactin | NZ_CP007039.1 | [1,146] |
| P. entomophilia L48 T | Entolysin | NC_008027.1 | [114] |
| P. putida PC2 | Putative WLIP producer | NZ_CP011789.1 | [68] |
| P. soli SJ10 | Xantholysin-like | NZ_CP009365.1 | [147] |
| P. putida E41 | Putative Putisolvin producer | NZ_CP024085.1 | [11] |
| P. mosselii BS011 | Xantholysin | CP023299.1 | [64] |
References
- Götze, S.; Stallforth, P. Structure, properties, and biological functions of nonribosomal lipopeptides from pseudomonads. Nat. Prod. Rep. 2020, 37, 29–54. [Google Scholar] [CrossRef] [PubMed]
- Weller, D.M. Pseudomonas Biocontrol Agents of Soilborne Pathogens: Looking Back Over 30 Years. Phytopathology 2007, 97, 250–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loper, J.E.; Hassan, K.A.; Mavrodi, D.V.; Davis, E.W., II; Lim, C.K.; Shaffer, B.T.; Elbourne, L.D.H.; Stockwell, V.O.; Hartney, S.L.; Breakwell, K.; et al. Comparative Genomics of Plant-Associated Pseudomonas spp.: Insights into Diversity and Inheritance of Traits Involved in Multitrophic Interactions. PLoS Genet. 2012, 8, e1002784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lalucat, J.; Mulet, M.; Gomila, M.; García-Valdés, E. Genomics in Bacterial Taxonomy: Impact on the Genus Pseudomonas. Genes 2020, 11, 139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomila, M.; Peña, A.; Mulet, M.; Lalucat, J.; García-Valdés, E. Phylogenomics and systematics in Pseudomonas. Front. Microbiol. 2015, 6, 214. [Google Scholar] [CrossRef] [Green Version]
- Girard, L.; Lood, C.; Höfte, M.; Vandamme, P.; Rokni-Zadeh, H.; van Noort, V.; Lavigne, R.; De Mot, R. The Ever-Expanding Pseudomonas Genus: Description of 43 New Species and Partition of the Pseudomonas putida Group. Microorganisms 2021, 9, 1766. [Google Scholar] [CrossRef] [PubMed]
- Hesse, C.; Schulz, F.; Bull, C.T.; Shaffer, B.T.; Yan, Q.; Shapiro, N.; Hassan, K.A.; Varghese, N.; Elbourne, L.D.H.; Paulsen, I.T.; et al. Genome-based evolutionary history of Pseudomonas spp. Environ. Microbiol. 2018, 20, 2142–2159. [Google Scholar] [CrossRef]
- Behzadi, P.; Baráth, Z.; Gajdács, M. It’s Not Easy Being Green: A Narrative Review on the Microbiology, Virulence and Therapeutic Prospects of Multidrug-Resistant Pseudomonas aeruginosa. Antibiotics 2021, 10, 42. [Google Scholar] [CrossRef]
- Perneel, M.; Heyrman, J.; Adiobo, A.; De Maeyer, K.; Raaijmakers, J.M.; De Vos, P.; Höfte, M. Characterization of CMR5c and CMR12a, novel fluorescent Pseudomonas strains from the cocoyam rhizosphere with biocontrol activity. J. Appl. Microbiol. 2007, 103, 1007–1020. [Google Scholar] [CrossRef] [Green Version]
- Oni, F.E.; Geudens, N.; Omoboye, O.O.; Bertier, L.; Hua, H.G.K.; Adiobo, A.; Sinnaeve, D.; Martins, J.C.; Höfte, M. Fluorescent Pseudomonas and cyclic lipopeptide diversity in the rhizosphere of cocoyam (Xanthosoma sagittifolium). Environ. Microbiol. 2019, 21, 1019–1034. [Google Scholar] [CrossRef] [Green Version]
- Oni, F.E.; Geudens, N.; Onyeka, J.T.; Olorunleke, O.F.; Salami, A.E.; Omoboye, O.O.; Arias, A.A.; Adiobo, A.; De Neve, S.; Ongena, M.; et al. Cyclic lipopeptide-producing Pseudomonas koreensis group strains dominate the cocoyam rhizosphere of a Pythium root rot suppressive soil contrasting with P. putida prominence in conducive soils. Environ. Microbiol. 2020, 22, 5137–5155. [Google Scholar] [CrossRef]
- Vlassak, K.; Van Holm, L.; Duchateau, L.; Vanderleyden, J.; De Mot, R. Isolation and characterization of fluorescent Pseudomonas associated with the roots of rice and banana grown in Sri Lanka. Plant Soil 1992, 145, 51–63. [Google Scholar] [CrossRef]
- Tran, H.; Kruijt, M.; Raaijmakers, J.M. Diversity and activity of biosurfactant-producing Pseudomonas in the rhizosphere of black pepper in Vietnam. J. Appl. Microbiol. 2008, 104, 839–851. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, T.H.; Sørensen, D.; Tobiasen, C.; Andersen, J.B.; Christophersen, C.; Givskov, M.; Sørensen, J. Antibiotic and Biosurfactant Properties of Cyclic Lipopeptides Produced by Fluorescent Pseudomonas spp. from the Sugar Beet Rhizosphere. Appl. Environ. Microbiol. 2002, 68, 3416–3423. [Google Scholar] [CrossRef] [Green Version]
- Lopes, L.D.; Davis, E.W.; Pereira e Silva, M.d.C.; Weisberg, A.J.; Bresciani, L.; Chang, J.H.; Loper, J.E.; Andreote, F.D. Tropical soils are a reservoir for fluorescent Pseudomonas spp. biodiversity. Environ. Microbiol. 2018, 20, 62–74. [Google Scholar] [CrossRef]
- Berry, C.; Fernando, W.G.D.; Loewen, P.C.; de Kievit, T.R. Lipopeptides are essential for Pseudomonas sp. DF41 biocontrol of Sclerotinia sclerotiorum. Biol. Control. 2010, 55, 211–218. [Google Scholar] [CrossRef]
- Müller, T.; Behrendt, U.; Ruppel, S.; Von Der Waydbrink, G.; Müller, M.E.H. Fluorescent Pseudomonads in the Phyllosphere of Wheat: Potential Antagonists Against Fungal Phytopathogens. Curr. Microbiol. 2015, 72, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Michelsen, C.F.; Watrous, J.; Glaring, M.A.; Kersten, R.; Koyama, N.; Dorrestein, P.C.; Stougaard, P. Nonribosomal Peptides, Key Biocontrol Components for Pseudomonas fluorescens In5, Isolated from a Greenlandic Suppressive Soil. mBio 2015, 6, e00079. [Google Scholar] [CrossRef] [Green Version]
- Höfte, M. The Use of Pseudomonas spp. as Bacterial Biocontrol Agents to Control Plant Disease. In Microbial Bioprotectants for Plant Disease Management; Burleigh Dodds Science Publishing: Cambridge, UK, 2021; p. 75. [Google Scholar]
- Olorunleke, F.E.; Kieu, N.P.; Höfte, M. Recent Advances in Pseudomonas Biocontrol. In Bacterial-Plant Interactions: Advance Research and Future Trends; Caister Academic Press: Cambridge, UK, 2015; Volume 1, pp. 167–198. [Google Scholar]
- Geudens, N.; Martins, J.C. Cyclic Lipodepsipeptides From Pseudomonas spp.—Biological Swiss-Army Knives. Front. Microbiol. 2018, 9, 1–18. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; De Bruijn, I.; Nybroe, O.; Ongena, M. Natural functions of lipopeptides from Bacillus and Pseudomonas: More than surfactants and antibiotics. FEMS Microbiol. Rev. 2010, 34, 1037–1062. [Google Scholar] [CrossRef] [Green Version]
- Calderón, C.E.; Pérez-García, A.; de Vicente, A.; Cazorla, F.M. The dar Genes of Pseudomonas chlororaphis PCL1606 Are Crucial for Biocontrol Activity via Production of the Antifungal Compound 2-Hexyl, 5-Propyl Resorcinol. Mol. Plant Microbe Interact. 2013, 26, 554–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flury, P.; Aellen, N.; Ruffner, B.; Péchy-Tarr, M.; Fataar, S.; Metla, Z.; Ferreras, A.D.; Bloemberg, G.; Frey, J.; Goesmann, A.; et al. Insect pathogenicity in plant-beneficial pseudomonads: Phylogenetic distribution and comparative genomics. ISME J. 2016, 10, 2527–2542. [Google Scholar] [CrossRef]
- Flury, P.; Vesga, P.; Péchy-Tarr, M.; Aellen, N.; Dennert, F.; Hofer, N.; Kupferschmied, K.P.; Kupferschmied, P.; Metla, Z.; Ma, Z.; et al. Antimicrobial and Insecticidal: Cyclic Lipopeptides and Hydrogen Cyanide Produced by Plant-Beneficial Pseudomonas Strains CHA0, CMR12a, and PCL1391 Contribute to Insect Killing. Front. Microbiol. 2017, 8, 100. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Sanz, D.; Meier-Kolthoff, J.P.; Göker, M.; Martín, M.; Rivilla, R.; Redondo-Nieto, M. Genomic and Genetic Diversity within the Pseudomonas fluorescens Complex. PLoS ONE 2016, 11, e0150183. [Google Scholar] [CrossRef] [Green Version]
- Garrido-Sanz, D.; Arrebola, E.; Martínez-Granero, F.; García-Méndez, S.; Muriel, C.; Blanco-Romero, E.; Martín, M.; Rivilla, R.; Redondo-Nieto, M. Classification of Isolates from the Pseudomonas fluorescens Complex into Phylogenomic Groups Based in Group-Specific Markers. Front. Microbiol. 2017, 8, 413. [Google Scholar] [CrossRef] [Green Version]
- Girard, L.; Höfte, M.; De Mot, R. Lipopeptide families at the interface between pathogenic and beneficial Pseudomonas-plant interactions. Crit. Rev. Microbiol. 2020, 46, 397–419. [Google Scholar] [CrossRef]
- Omoboye, O.O.; Geudens, N.; Duban, M.; Chevalier, M.; Flahaut, C.; Martins, J.C.; Leclère, V.; Oni, F.E.; Höfte, M. Pseudomonas sp. COW3 Produces New Bananamide-Type Cyclic Lipopeptides with Antimicrobial Activity against Pythium myriotylum and Pyricularia oryzae. Molecules 2019, 24, 4170. [Google Scholar] [CrossRef] [Green Version]
- Crouzet, J.; Arguelles-Arias, A.; Dhondt-Cordelier, S.; Cordelier, S.; Pršić, J.; Hoff, G.; Mazeyrat-Gourbeyre, F.; Baillieul, F.; Clément, C.; Ongena, M.; et al. Biosurfactants in Plant Protection Against Diseases: Rhamnolipids and Lipopeptides Case Study. Front. Bioeng. Biotechnol. 2020, 8, 1014. [Google Scholar] [CrossRef]
- Pršić, J.; Ongena, M. Elicitors of Plant Immunity Triggered by Beneficial Bacteria. Front. Plant Sci. 2020, 11, 594530. [Google Scholar] [CrossRef]
- Behzadi, P.; Gajdács, M. Writing a strong scientific paper in medicine and the biomedical sciences: A checklist and recommendations for early career researchers. Biol. Futur. 2021, 72, 395–407. [Google Scholar] [CrossRef]
- Petkau, A.; Stuart-Edwards, M.; Stothard, P.; Van Domselaar, G. Interactive microbial genome visualization with GView. Bioinformatics 2010, 26, 3125–3126. [Google Scholar] [CrossRef] [PubMed]
- de Souza, J.T.; de Boer, M.; de Waard, P.; van Beek, T.A.; Raaijmakers, J.M. Biochemical, Genetic, and Zoosporicidal Properties of Cyclic Lipopeptide Surfactants Produced by Pseudomonas fluorescens. Appl. Environ. Microbiol. 2003, 69, 7161–7172. [Google Scholar] [CrossRef] [Green Version]
- de Bruijn, I.; de Kock, M.J.D.; Yang, M.; de Waard, P.; van Beek, T.A.; Raaijmakers, J.M. Genome-based discovery, structure prediction and functional analysis of cyclic lipopeptide antibiotics in Pseudomonas species. Mol. Microbiol. 2007, 63, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Thrane, C.; Olsson, S.; Nielsen, T.H.; Sørensen, J. Vital fluorescent stains for detection of stress in Pythium ultimum and Rhizoctonia solani challenged with viscosinamide from Pseudomonas fluorescens DR54. FEMS Microbiol. Ecol. 1999, 30, 11–23. [Google Scholar] [CrossRef]
- Nielsen, M.N.; Sørensen, J.; Fels, J.; Pedersen, H.C. Secondary Metabolite- and Endochitinase-Dependent Antagonism toward Plant-Pathogenic Microfungi of Pseudomonas fluorescens Isolates from Sugar Beet Rhizosphere. Appl. Environ. Microbiol. 1998, 64, 3563–3569. [Google Scholar] [CrossRef] [Green Version]
- Oni, F.E.; Geudens, N.; Adiobo, A.; Omoboye, O.O.; Enow, E.A.; Onyeka, J.T.; Salami, A.E.; De De Mot, R.; Martins, J.C.; Höfte, M. Biosynthesis and Antimicrobial Activity of Pseudodesmin and Viscosinamide Cyclic Lipopeptides Produced by Pseudomonads Associated with the Cocoyam Rhizosphere. Microorganisms 2020, 8, 1079. [Google Scholar] [CrossRef]
- Pedras, M.S.C.; Ismail, N.; Quail, J.W.; Boyetchko, S.M. Structure, chemistry, and biological activity of pseudophomins A and B, new cyclic lipodepsipeptides isolated from the biocontrol bacterium Pseudomonas fluorescens. Phytochemistry 2003, 62, 1105–1114. [Google Scholar] [CrossRef]
- Zachow, C.; Tilcher, R.; Berg, G. Sugar Beet-Associated Bacterial and Fungal Communities Show a High Indigenous Antagonistic Potential Against Plant Pathogens. Microb. Ecol. 2007, 55, 119–129. [Google Scholar] [CrossRef]
- Zachow, C.; Jahanshah, G.; de Bruijn, I.; Song, C.; Ianni, F.; Pataj, Z.; Gerhardt, H.; Pianet, I.; Lämmerhofer, M.; Berg, G.; et al. The Novel Lipopeptide Poaeamide of the Endophyte Pseudomonas poae RE*1-1-14 Is Involved in Pathogen Suppression and Root Colonization. Mol. Plant Microbe Interact. 2015, 28, 800–810. [Google Scholar] [CrossRef] [Green Version]
- Cantore, P.L.; Lazzaroni, S.; Coraiola, M.; Serra, M.D.; Cafarchia, C.; Evidente, A.; Iacobellis, N.S. Biological Characterization of White Line-Inducing Principle (WLIP) Produced by Pseudomonas reactans NCPPB1311. Mol. Plant Microbe Interact. 2006, 19, 1113–1120. [Google Scholar] [CrossRef] [Green Version]
- Andersen, J.B.; Koch, B.; Nielsen, T.H.; Sørensen, D.; Hansen, M.; Nybroe, O.; Christophersen, C.; Sørensen, J.; Molin, S.; Givskov, M. Surface motility in Pseudomonas sp. DSS73 is required for efficient biological containment of the root-pathogenic microfungi Rhizoctonia solani and Pythium ultimum. Microbiology 2003, 149, 37–46. [Google Scholar] [CrossRef] [Green Version]
- Götze, S.; Herbst-Irmer, R.; Klapper, M.; Görls, H.; Schneider, K.R.A.; Barnett, R.; Burks, T.; Neu, U.; Stallforth, P. Structure, Biosynthesis, and Biological Activity of the Cyclic Lipopeptide Anikasin. ACS Chem. Biol. 2017, 12, 2498–2502. [Google Scholar] [CrossRef] [PubMed]
- Klapper, M.; Götze, S.; Barnett, R.; Willing, K.; Stallforth, P. Bacterial Alkaloids Prevent Amoebal Predation. Angew. Chem.—Int. Ed. 2016, 55, 8944–8947. [Google Scholar] [CrossRef]
- Nielsen, T.H.; Sørensen, J. Production of Cyclic Lipopeptides by Pseudomonas fluorescens Strains in Bulk Soil and in the Sugar Beet Rhizosphere. Appl. Environ. Microbiol. 2003, 69, 861–868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sørensen, D.; Nielsen, T.H.; Sørensen, J.; Christophersen, C. Cyclic lipoundecapeptide lokisin from Pseudomonas sp. strain DSS41. Tetrahedron Lett. 2002, 43, 4421–4423. [Google Scholar] [CrossRef]
- Hultberg, M.; Alsberg, T.; Khalil, S.; Alsanius, B. Suppression of disease in tomato infected by Pythium ultimum with a biosurfactant produced by Pseudomonas koreensis. Entomophaga 2009, 55, 435–444. [Google Scholar] [CrossRef]
- Gu, Y.; Ma, Y.; Wang, J.; Xia, Z.; Wei, H. Genomic insights into a plant growth-promoting Pseudomonas koreensis strain with cyclic lipopeptide-mediated antifungal activity. MicrobiologyOpen 2020, 9, e1092. [Google Scholar] [CrossRef]
- Schlusselhuber, M.; Godard, J.; Sebban, M.; Bernay, B.; Garon, D.; Seguin, V.; Oulyadi, H.; Desmasures, N. Characterization of Milkisin, a Novel Lipopeptide With Antimicrobial Properties Produced By Pseudomonas sp. UCMA 17988 Isolated From Bovine Raw Milk. Front. Microbiol. 2018, 9, 1030. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, T.; Thrane, C.; Christophersen, C.; Anthoni, U.; Sorensen, J. Structure, production characteristics and fungal antagonism of tensin—a new antifungal cyclic lipopeptide from Pseudomonas fluorescens strain 96.578. J. Appl. Microbiol. 2000, 89, 992–1001. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.D.; Melnik, A.V.; Koyama, N.; Lu, X.; Schorn, M.; Fang, J.; Aguinaldo, K.; Lincecum, T.L., Jr.; Ghequire, M.G.; Carrion, V.J.; et al. Indexing the Pseudomonas specialized metabolome enabled the discovery of poaeamide B and the bananamides. Nat. Microbiol. 2016, 2, 16197. [Google Scholar] [CrossRef] [Green Version]
- Jahanshah, G.; Yan, Q.; Gerhardt, H.; Pataj, Z.; Lämmerhofer, M.; Pianet, I.; Josten, M.; Sahl, H.-G.; Silby, M.W.; Loper, J.E.; et al. Discovery of the Cyclic Lipopeptide Gacamide A by Genome Mining and Repair of the Defective GacA Regulator in Pseudomonas fluorescens Pf0-1. J. Nat. Prod. 2019, 82, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Compeau, G.; Al-Achi, B.J.; Platsouka, E.; Levy, S.B. Survival of rifampin-resistant mutants of Pseudomonas fluorescens and Pseudomonas putida in soil systems. Appl. Environ. Microbiol. 1988, 54, 2432–2438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, D.; Défago, G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Genet. 2005, 3, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Geudens, N.; Kieu, N.P.; Sinnaeve, D.; Ongena, M.; Martins, J.C.; Höfte, M. Biosynthesis, Chemical Structure, and Structure-Activity Relationship of Orfamide Lipopeptides Produced by Pseudomonas protegens and Related Species. Front. Microbiol. 2016, 7, 382. [Google Scholar] [CrossRef]
- Paulsen, I.T.; Press, C.M.; Ravel, J.; Kobayashi, D.Y.; Myers, G.S.A.; Mavrodi, D.V.; DeBoy, R.T.; Seshadri, R.; Ren, Q.; Madupu, R.; et al. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat. Biotechnol. 2005, 23, 873–878. [Google Scholar] [CrossRef]
- Howell, C.R.; Stipanovic, R.D. Control of Rhizoctonia Solani on Cotton Seedlings with Pseudomonas fluorescens and with an Antibiotic Produced by the Bacterium by the Soil Tube Method Described Previously. Phytopathology 1979, 69, 480–482. [Google Scholar] [CrossRef] [Green Version]
- D’Aes, J.; Kieu, N.P.; Leclere, V.; Tokarski, C.; Olorunleke, F.E.; De Maeyer, K.; Jacques, P.; Höfte, M.; Ongena, M. To settle or to move? The interplay between two classes of cyclic lipopeptides in the biocontrol strain Pseudomonas CMR12a. Environ. Microbiol. 2014, 16, 2282–2300. [Google Scholar] [CrossRef] [PubMed]
- Van Der Voort, M.; Meijer, H.J.G.; Schmidt, Y.; Watrous, J.; Dekkers, E.; Mendes, R.; Dorrestein, P.C.; Gross, H.; Raaijmakers, J.M. Genome mining and metabolic profiling of the rhizosphere bacterium Pseudomonas sp. SH-C52 for antimicrobial compounds. Front. Microbiol. 2015, 6, 693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berry, C.L.; Brassinga, A.K.C.; Donald, L.J.; Fernando, W.G.D.; Loewen, P.C.; De Kievit, T.R. Chemical and biological characterization of sclerosin, an antifungal lipopeptide. Can. J. Microbiol. 2012, 58, 1027–1034. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Liu, Y.-P.; Zhang, L.-Q. In silico and Genetic Analyses of Cyclic Lipopeptide Synthetic Gene Clusters in Pseudomonas sp. 11K1. Front. Microbiol. 2019, 10, 544. [Google Scholar] [CrossRef]
- Li, W.; Rokni-Zadeh, H.; De Vleeschouwer, M.; Ghequire, M.G.K.; Sinnaeve, D.; Xie, G.-L.; Rozenski, J.; Madder, A.; Martins, J.C.; De Mot, R. The Antimicrobial Compound Xantholysin Defines a New Group of Pseudomonas Cyclic Lipopeptides. PLoS ONE 2013, 8, e62946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.; Xiao, W.; Chen, G.; Song, D.; Khaskheli, M.A.; Li, P.; Zhang, S.; Feng, G. Identification of Pseudomonas mosselii BS011 gene clusters required for suppression of Rice Blast Fungus Magnaporthe oryzae. J. Biotechnol. 2018, 282, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kruijt, M.; Tran, H.; Raaijmakers, J.M. Functional, genetic and chemical characterization of biosurfactants produced by plant growth-promoting Pseudomonas putida 267. J. Appl. Microbiol. 2009, 107, 546–556. [Google Scholar] [CrossRef] [PubMed]
- Vallet-Gely, I.; Novikov, A.; Augusto, L.; Liehl, P.; Bolbach, G.; Péchy-Tarr, M.; Cosson, P.; Keel, C.; Caroff, M.; Lemaitre, B. Association of Hemolytic Activity of Pseudomonas entomophila, a Versatile Soil Bacterium, with Cyclic Lipopeptide Production. Appl. Environ. Microbiol. 2010, 76, 910–921. [Google Scholar] [CrossRef] [Green Version]
- Rokni-Zadeh, H.; Li, W.; Sánchez-Rodríguez, A.; Sinnaeve, D.; Rozenski, J.; Martins, J.C.; De Mot, R. Genetic and Functional Characterization of Cyclic Lipopeptide White-Line-Inducing Principle (WLIP) Production by Rice Rhizosphere Isolate Pseudomonas putida RW10S2. Appl. Environ. Microbiol. 2012, 78, 4826–4834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omoboye, O.O.; Oni, F.E.; Batool, H.; Yimer, H.Z.; De Mot, R.; Höfte, M. Pseudomonas Cyclic Lipopeptides Suppress the Rice Blast Fungus Magnaporthe oryzae by Induced Resistance and Direct Antagonism. Front. Plant Sci. 2019, 10, 901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehnaz, S.; Saleem, R.S.Z.; Yameen, B.; Pianet, I.; Schnakenburg, G.; Pietraszkiewicz, H.; Valeriote, F.; Josten, M.; Sahl, H.-G.; Franzblau, S.G.; et al. Lahorenoic Acids A–C, ortho-Dialkyl-Substituted Aromatic Acids from the Biocontrol Strain Pseudomonas aurantiaca PB-St2. J. Nat. Prod. 2013, 76, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Thrane, C.; Nielsen, T.H.; Nielsen, M.N.; Sörensen, J.; Olsson, S. Viscosinamide-Producing Pseudomonas fluorescens DR54 Exerts a Biocontrol effect on Pythium ultimum in Sugar Beet Rhizosphere. Design 2000, 33, 139–146. [Google Scholar] [CrossRef]
- Thrane, C.; Nielsen, M.N.; Rensen, J.S.; Olsson, S. Pseudomonas fluorescens DR54 Reduces Sclerotia Formation, Biomass Development, and Disease Incidence of Rhizoctonia solani Causing Damping-Off in Sugar Beet. Microb. Ecol. 2001, 42, 438–445. [Google Scholar] [CrossRef]
- Tran, H.; Ficke, A.; Asiimwe, T.; Höfte, M.; Raaijmakers, J.M. Role of the cyclic lipopeptide massetolide A in biological control of Phytophthora infestans and in colonization of tomato plants by Pseudomonas fluorescens. New Phytol. 2007, 175, 731–742. [Google Scholar] [CrossRef] [Green Version]
- Van De Mortel, J.E.; De Vos, R.C.; Dekkers, E.; Pineda, A.; Guillod, L.; Bouwmeester, K.; Van Loon, J.J.A.; Dicke, M.; Raaijmakers, J.M. Metabolic and Transcriptomic Changes Induced in Arabidopsis by the Rhizobacterium Pseudomonas fluorescens SS101. Plant Physiol. 2012, 160, 2173–2188. [Google Scholar] [CrossRef] [Green Version]
- D’Aes, J.; Hua, G.K.H.; De Maeyer, K.; Pannecoucque, J.; Forrez, I.; Ongena, M.; Dietrich, L.; Thomashow, L.S.; Mavrodi, D.; Höfte, M. Biological Control of Rhizoctonia Root Rot on Bean by Phenazine- and Cyclic Lipopeptide-Producing Pseudomonas CMR12a. Phytopathology 2011, 101, 996–1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olorunleke, F.E.; Hua, G.K.H.; Kieu, N.P.; Ma, Z.; Höfte, M. Interplay between orfamides, sessilins and phenazines in the control of Rhizoctonia diseases by Pseudomonas sp. CMR12a. Environ. Microbiol. Rep. 2015, 7, 774–781. [Google Scholar] [CrossRef]
- Ma, Z.; Hua, G.K.H.; Ongena, M.; Höfte, M. Role of phenazines and cyclic lipopeptides produced by Pseudomonas sp. CMR12a in induced systemic resistance on rice and bean. Environ. Microbiol. Rep. 2016, 8, 896–904. [Google Scholar] [CrossRef] [PubMed]
- Oni, F.E.; Olorunleke, O.F.; Höfte, M. Phenazines and cyclic lipopeptides produced by Pseudomonas sp. CMR12a are involved in the biological control of Pythium myriotylum on cocoyam (Xanthosoma sagittifolium). Biol. Control. 2018, 129, 109–114. [Google Scholar] [CrossRef]
- Ma, Z.; Ongena, M.; Höfte, M. The cyclic lipopeptide orfamide induces systemic resistance in rice to Cochliobolus miyabeanus but not to Magnaporthe oryzae. Plant Cell Rep. 2017, 36, 1731–1746. [Google Scholar] [CrossRef] [PubMed]
- Gross, H.; Stockwell, V.O.; Henkels, M.D.; Nowak-Thompson, B.; Loper, J.E.; Gerwick, W.H. The Genomisotopic Approach: A Systematic Method to Isolate Products of Orphan Biosynthetic Gene Clusters. Chem. Biol. 2007, 14, 53–63. [Google Scholar] [CrossRef] [Green Version]
- Le, C.; Kruijt, M.; Raaijmakers, J. Involvement of phenazines and lipopeptides in interactions between Pseudomonas species and Sclerotium rolfsii, causal agent of stem rot disease on groundnut. J. Appl. Microbiol. 2011, 112, 390–403. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Kruijt, M.; de Bruijn, I.; Dekkers, E.; Van Der Voort, M.; Schneider, J.H.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A.; et al. Deciphering the Rhizosphere Microbiome for Disease-Suppressive Bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef]
- Lavermicocca, P.; Ballio, A.; Camoni, L.; Di Giorgio, D.; Marchiafava, C. Studies on the Effect of Syringomycin and Syringopeptins on the Functions of Plant Mitochondria. In Pseudomonas Syringae Pathovars and Related Pathogens. Developments in Plant Pathology; Rudolph, K., Burr, T.J., Mansfield, J.W., Stead, D., Vivian, A., von Kietzell, J., Eds.; Springer: Dordrecht, The Netherlands, 1997; pp. 198–201. [Google Scholar]
- Bull, C.T.; Stack, J.P.; Smilanick, J.L. Pseudomonas syringae Strains ESC-10 and ESC-11 Survive in Wounds on Citrus and Control Green and Blue Molds of Citrus. Biol. Control 1997, 8, 81–88. [Google Scholar] [CrossRef]
- Hildebrand, P.D.; Braun, P.G.; McRae, K.B.; Lu, X. Role of the biosurfactant viscosin in broccoli head rot caused by a pectolytic strain of Pseudomonas fluorescens. Can. J. Plant Pathol. 1998, 20, 296–303. [Google Scholar] [CrossRef]
- Gerard, J.; Lloyd, R.; Barsby, T.; Haden, P.; Kelly, M.T.; Andersen, R.J. Massetolides A−H, Antimycobacterial Cyclic Depsipeptides Produced by Two Pseudomonads Isolated from Marine Habitats. J. Nat. Prod. 1997, 60, 223–229. [Google Scholar] [CrossRef]
- De Leij, F.A.A.M.; Sutton, E.J.; Whipps, J.M.; Fenlon, J.S.; Lynch, J.M. Impact of Field Release of Genetically Modified Pseudomonas fluorescens on Indigenous Microbial Populations of Wheat. Appl. Environ. Microbiol. 1995, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, T.H.; Christophersen, C.; Anthoni, U.; Sorensen, J. Viscosinamide, a new cyclic depsipeptide with surfactant and antifungal properties produced by Pseudomonas fluorescens DR54. J. Appl. Microbiol. 1999, 87, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Sinnaeve, D.; Michaux, C.; Van Hemel, J.; Vandenkerckhove, J.; Peys, E.; Borremans, F.A.; Sas, B.; Wouters, J.; Martins, J.C. Structure and X-ray conformation of pseudodesmins A and B, two new cyclic lipodepsipeptides from Pseudomonas bacteria. Tetrahedron 2009, 65, 4173–4181. [Google Scholar] [CrossRef] [Green Version]
- Mazzola, M.; Zhao, X.; Cohen, M.F.; Raaijmakers, J.M. Cyclic Lipopeptide Surfactant Production by Pseudomonas fluorescens SS101 Is Not Required for Suppression of Complex Pythium spp. Populations. Phytopathology 2007, 97, 1348–1355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, F.; Mortishire-Smith, R.J.; Rainey, P.B.; Williams, D.H. Structure of the white-line-inducing principle isolated from Pseudomonas reactans. Acta Crystallogr. Sect. C Cryst. Struct. Commun. 1992, 48, 1965–1968. [Google Scholar] [CrossRef]
- Soler-Rivas, C.; Arpin, N.; Olivier, J.M.; Wichers, H. WLIP, a lipodepsipeptide of Pseudomonas “reactans”, as inhibitor of the symptoms of the brown blotch disease of Agaricus bisporus. J. Appl. Microbiol. 1999, 86, 635–641. [Google Scholar] [CrossRef]
- Zarvandi, S.; Bahrami, T.; Pauwels, B.; Asgharzadeh, A.; Hosseini-Mazinani, M.; Salari, F.; Girard, L.; De Mot, R.; Rokni-Zadeh, H. Draft Genome Sequence of Cyclic Lipopeptide Producer Pseudomonas sp. Strain SWRI103, Isolated from Wheat Rhizosphere. Microbiol. Resour. Announc. 2020, 9, e00538-20. [Google Scholar] [CrossRef]
- Jang, J.Y.; Yang, S.Y.; Kim, Y.C.; Lee, C.W.; Park, M.S.; Kim, J.C.; Kim, I.S. Identification of Orfamide A as an Insecticidal Metabolite Produced by Pseudomonas protegens F6. J. Agric. Food Chem. 2013, 61, 6786–6791. [Google Scholar] [CrossRef]
- Takeuchi, K.; Noda, N.; Someya, N. Complete Genome Sequence of the Biocontrol Strain Pseudomonas protegens Cab57 Discovered in Japan Reveals Strain-Specific Diversity of This Species. PLoS ONE 2014, 9, e93683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loper, J.E.; Henkels, M.D.; Rangel, L.I.; Olcott, M.H.; Walker, F.L.; Bond, K.L.; Kidarsa, T.A.; Hesse, C.N.; Sneh, B.; Stockwell, V.O.; et al. Rhizoxin analogs, orfamide A and chitinase production contribute to the toxicity of Pseudomonas protegens strain Pf-5 to Drosophila melanogaster. Environ. Microbiol. 2016, 18, 3509–3521. [Google Scholar] [CrossRef]
- Biessy, A.; Novinscak, A.; Blom, J.; Thomashow, L.S.; Cazorla, F.M.; Josic, D.; Filion, M. Diversity of Phytobeneficial Traits Revealed by Whole-Genome Analysis of Worldwide-Isolated Phenazine-Producing Pseudomonas spp. Environ. Microbiol. 2019, 21, 437–455. [Google Scholar]
- Geudens, N.; Nasir, M.N.; Crowet, J.-M.; Raaijmakers, J.M.; Fehér, K.; Coenye, T.; Martins, J.C.; Lins, L.; Sinnaeve, D.; Deleu, M. Membrane Interactions of Natural Cyclic Lipodepsipeptides of the Viscosin Group. Biochim. Biophys. Acta 2016, 1859, 331–339. [Google Scholar] [CrossRef]
- Watrous, J.; Roach, P.; Alexandrov, T.; Heath, B.S.; Yang, J.Y.; Kersten, R.D.; van der Voort, M.; Pogliano, K.; Gross, H.; Raaijmakers, J.; et al. Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. USA 2012, 109, E1743–E1752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reder-Christ, K.; Schmidt, Y.; Dörr, M.; Sahl, H.-G.; Josten, M.; Raaijmakers, J.M.; Gross, H.; Bendas, G. Model membrane studies for characterization of different antibiotic activities of lipopeptides from Pseudomonas. Biochim. Biophys. Acta 2011, 1818, 566–573. [Google Scholar] [CrossRef] [Green Version]
- Eschmidt, R.; Eköberl, M.; Emostafa, A.; Ramadan, E.M.; Emonschein, M.; Jensen, K.B.; Ebauer, R.; Eberg, G. Effects of bacterial inoculants on the indigenous microbiome and secondary metabolites of chamomile plants. Front. Microbiol. 2014, 5, 64. [Google Scholar] [CrossRef]
- Vassilev, V.; Lavermicocca, P.; DI Giorgio, D.; Iacobellis, N.S. Production of syringomycins and syringopeptins by Pseudomonas syringae pv. atrofaciens . Plant Pathol. 1996, 45, 316–322. [Google Scholar] [CrossRef]
- Emanuele, M.; Scaloni, A.; Lavermicocca, P.; Jacobellis, N.; Camoni, L.; Di Giorgio, D.; Pucci, P.; Paci, M.; Segre, A.; Ballio, A. Corceptins, new bioactive lipodepsipeptides from cultures of Pseudomonas corrugata. FEBS Lett. 1998, 433, 317–320. [Google Scholar] [CrossRef] [Green Version]
- Quibod, I.L.; Grande, G.; Oreiro, E.G.; Borja, F.N.; Dossa, G.S.; Mauleon, R.; Cruz, C.V.; Oliva, R. Rice-Infecting Pseudomonas Genomes Are Highly Accessorized and Harbor Multiple Putative Virulence Mechanisms to Cause Sheath Brown Rot. PLoS ONE 2015, 10, e0139256. [Google Scholar] [CrossRef] [Green Version]
- Ballio, A.; Bossa, F.; Camoni, L.; Di Giorgio, D.; Flamand, M.-C.; Maraite, H.; Nitti, G.; Pucci, P.; Scaloni, A. Structure of fuscopeptins, phytotoxic metabolites of Pseudomonas fuscovaginae. FEBS Lett. 1996, 381, 213–216. [Google Scholar] [CrossRef] [Green Version]
- Flamand, M.-C.; Pelsser, S.; Ewbank, E.; Maraite, H. Production of syringotoxin and other bioactive peptides by Pseudomonas fuscovaginae. Physiol. Mol. Plant Pathol. 1996, 48, 217–231. [Google Scholar] [CrossRef]
- Patel, H.K.; Da Silva, D.P.; Devescovi, G.; Maraite, H.; Paszkiewicz, K.; Studholme, D.J.; Venturi, V. Draft Genome Sequence of Pseudomonas fuscovaginae, a Broad-Host-Range Pathogen of Plants. J. Bacteriol. 2012, 194, 2765–2766. [Google Scholar] [CrossRef] [PubMed]
- Bender, C.L.; Alarcón-Chaidez, F.; Gross, D.C. Pseudomonas syringae Phytotoxins: Mode of Action, Regulation, and Biosynthesis by Peptide and Polyketide Synthetases. Microbiol. Mol. Biol. Rev. 1999, 63, 266–292. [Google Scholar] [CrossRef] [Green Version]
- Lopes, L.D.; Weisberg, A.J.; Davis, E.W., 2nd; Varize, C.d.S.; Silva, M.d.C.P.E.; Chang, J.H.; Loper, J.E.; Andreote, F.D. Genomic and metabolic differences between Pseudomonas putida populations inhabiting sugarcane rhizosphere or bulk soil. PLoS ONE 2019, 14, e0223269. [Google Scholar] [CrossRef] [PubMed]
- Keshavarz-Tohid, V.; Vacheron, J.; Dubost, A.; Prigent-Combaret, C.; Taheri, P.; Tarighi, S.; Taghavi, S.M.; Moënne-Loccoz, Y.; Muller, D. Genomic, phylogenetic and catabolic re-assessment of the Pseudomonas putida clade supports the delineation of Pseudomonas alloputida sp. nov., Pseudomonas inefficax sp. nov., Pseudomonas persica sp. nov., and Pseudomonas shirazica sp. nov. Syst. Appl. Microbiol. 2019, 42, 468–480. [Google Scholar] [CrossRef] [PubMed]
- Girard, L.; Geudens, N.; Pauwels, B.; Höfte, M.; Martins, J.C.; De Mot, R. Transporter Gene-mediated Typing for Detection and Genome Mining of Lipopeptide-producing Pseudomonas. Appl. Environ. Microbiol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Kuiper, I.; Lagendijk, E.L.; Pickford, R.; Derrick, J.; Lamers, G.E.M.; Thomas-Oates, J.E.; Lugtenberg, B.J.J.; Bloemberg, G.V. Characterization of two Pseudomonas putida lipopeptide biosurfactants, putisolvin I and II, which inhibit biofilm formation and break down existing biofilms. Mol. Microbiol. 2003, 51, 97–113. [Google Scholar] [CrossRef] [PubMed]
- Meziane, H.; Van der Sluis, I.; van Loon, L.C.; Höfte, M.; Bakker, P.A.H.M. Determinants of Pseudomonas putida WCS358 Involved in Inducing. Mol. Plant Pathol. 2005, 6, 177–185. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Van Verk, M.C.; Stringlis, I.A.; Zamioudis, C.; Tommassen, J.; Pieterse, C.M.J.; Bakker, P.A.H.M. Unearthing the genomes of plant-beneficial Pseudomonas model strains WCS358, WCS374 and WCS417. BMC Genom. 2015, 16, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Vodovar, N.; Vallenet, D.; Cruveiller, S.; Rouy, Z.; Barbe, V.; Acosta, C.; Cattolico, L.; Jubin, C.; Lajus, A.; Segurens, B.; et al. Complete genome sequence of the entomopathogenic and metabolically versatile soil bacterium Pseudomonas entomophila. Nat. Biotechnol. 2006, 24, 673–679. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.M.; Bakker, P.A.H.M. Induced Systemic Resistance by Beneficial Microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [Green Version]
- Schellenberger, R.; Touchard, M.; Clément, C.; Baillieul, F.; Cordelier, S.; Crouzet, J.; Dorey, S. Apoplastic invasion patterns triggering plant immunity: Plasma membrane sensing at the frontline. Mol. Plant Pathol. 2019, 20, 1602–1616. [Google Scholar] [CrossRef]
- Pel, M.J.C.; Pieterse, C.M.J. Microbial Recognition and Evasion of Host Immunity. J. Exp. Bot. 2012, 64, 1237–1248. [Google Scholar] [CrossRef] [Green Version]
- Behzadi, P.; García-Perdomo, H.A.; Karpiński, T.M. Toll-Like Receptors: General Molecular and Structural Biology. J. Immunol. Res. 2021, 2021, 1–21. [Google Scholar] [CrossRef]
- Medzhitov, R. Toll-like Receptors and Innate Immunity. Nat. Rev. Immunol. 2001, 1, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Grandellis, C.; Garavaglia, B.S.; Gottig, N.; Lonez, C.; Ruysschaert, J.-M.; Ottado, J. DOTAP, a lipidic transfection reagent, triggers Arabidopsis plant defense responses. Planta 2018, 249, 469–480. [Google Scholar] [CrossRef]
- Piasecka, A.; Jedrzejczak-Rey, N.; Bednarek, P. Secondary metabolites in plant innate immunity: Conserved function of divergent chemicals. New Phytol. 2015, 206, 948–964. [Google Scholar] [CrossRef]
- Zipfel, C.; Oldroyd, G.E.D. Plant signalling in symbiosis and immunity. Nature 2017, 543, 328–336. [Google Scholar] [CrossRef]
- Stringlis, I.A.; Zhang, H.; Pieterse, C.M.J.; Bolton, M.D.; de Jonge, R. Microbial small molecules—weapons of plant subversion. Nat. Prod. Rep. 2018, 35, 410–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, K.; Yu, K.; Pieterse, C.M.; Pieterse, C.M.; Bakker, P.A.; Bakker, P.A.; Berendsen, R.L.; Berendsen, R.L.; Yu, K.; Yu, K.; et al. Beneficial microbes going underground of root immunity. Plant Cell Environ. 2019, 42, 2860–2870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spence, C.; Alff, E.; Johnson, C.; Ramos, C.; Donofrio, N.; Sundaresan, V.; Bais, H. Natural rice rhizospheric microbes suppress rice blast infections. BMC Plant Biol. 2014, 14, 130. [Google Scholar] [CrossRef] [Green Version]
- Brodey, C.L.; Rainey, P.B.; Tester, M.; Johnstone, K. Bacterial Blotch Disease of the Cultivated Mushroom Os Caused By an Iron Channel Forming Lipidesipsipeptide Toxin.Pdf. Mol. Plant. Microbe Interact. 1991, 4, 407–411. [Google Scholar] [CrossRef]
- Kozlova, O.V.; Egorov, S.Y.; Kupriyanova-Ashina, F.G.; Rid, N.; El’-Registan, G.I. Analysis of the Ca2+ response of mycelial fungi to external effects by the recombinant aequorin method. Microbiology 2004, 73, 629–634. [Google Scholar] [CrossRef]
- Aiyar, P.; Schaeme, D.; García-Altares, M.; Flores, D.C.; Dathe, H.; Hertweck, C.; Sasso, S.; Mittag, M. Antagonistic bacteria disrupt calcium homeostasis and immobilize algal cells. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Rainey, P.; Brodey, C.L.; Johnstone, K. Biological properties and spectrum of activity of tolaasin, a lipodepsipeptide toxin produced by the mushroom pathogen Pseudomonas tolaasii. Physiol. Mol. Plant Pathol. 1991, 39, 57–70. [Google Scholar] [CrossRef]
- Steigenberger, J.; Verleysen, Y.; Geudens, N.; Martins, J.C.; Heerklotz, H. The Optimal Lipid Chain Length of a Membrane-Permeabilizing Lipopeptide Results From the Balance of Membrane Partitioning and Local Damage. Front. Microbiol. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- Leveau, J.H.J.; Preston, G. Bacterial mycophagy: Definition and diagnosis of a unique bacterial–fungal interaction. New Phytol. 2008, 177, 859–876. [Google Scholar] [CrossRef]
- Wong, W.C.; Preece, T.F. Identification of Pseudomonas tolaasi: The White Line in Agar and Mushroom Tissue Block Rapid Pitting Tests. J. Appl. Bacteriol. 1979, 47, 401–407. [Google Scholar] [CrossRef]
- De Vleeschouwer, M.; Van Kersavond, T.; Verleysen, Y.; Sinnaeve, D.; Coenye, T.; Martins, J.C.; Madder, A. Identification of the Molecular Determinants Involved in Antimicrobial Activity of Pseudodesmin A, a Cyclic Lipopeptide From the Viscosin Group. Front. Microbiol. 2020, 11, 646. [Google Scholar] [CrossRef] [Green Version]
- De Bruijn, I.; de Kock, M.J.D.; de Waard, P.; van Beek, T.A.; Raaijmakers, J.M. Massetolide A Biosynthesis in Pseudomonas fluorescens. J. Bacteriol. 2008, 190, 2777–2789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saxon, E.B.; Jackson, R.W.; Bhumbra, S.; Smith, T.; Sockett, R.E. Bdellovibrio bacteriovorus HD100 guards against Pseudomonas tolaasii brown-blotch lesions on the surface of post-harvest Agaricus bisporus supermarket mushrooms. BMC Microbiol. 2014, 14, 163. [Google Scholar] [CrossRef] [Green Version]
- Jia, J.; Wang, X.; Deng, P.; Ma, L.; Baird, S.M.; Li, X.; Lu, S. Pseudomonas glycinae sp. nov. isolated from the soybean rhizosphere. MicrobiologyOpen 2020, 9, e1101. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Rha, E.; Kim, H.; Lee, S.-G. Complete Genome Sequence of the Soil Bacterium Pseudomonas kribbensis Strain 46-2 T. Microbiol. Resour. Announc. 2018, 7, e01161-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pascual, J.; García-López, M.; Bills, G.; Genilloud, O. Pseudomonas granadensis sp. nov., a new bacterial species isolated from the Tejeda, Almijara and Alhama Natural Park, Granada, Spain. Int. J. Syst. Evol. Microbiol. 2015, 65, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Cautain, B.; de Pedro, N.; Schulz, C.; Pascual, J.; Sousa, T.D.S.; Martínez, I.G.; Pérez-Victoria, I.; Asensio, F.; González, I.; Bills, G.; et al. Identification of the Lipodepsipeptide MDN-0066, a Novel Inhibitor of VHL/HIF Pathway Produced by a New Pseudomonas Species. PLoS ONE 2015, 10, e0125221. [Google Scholar] [CrossRef] [Green Version]
- Berry, C.L.; Nandi, M.; Manuel, J.; Brassinga, A.K.C.; Fernando, W.D.; Loewen, P.C.; de Kievit, T.R. Characterization of the Pseudomonas sp. DF41 quorum sensing locus and its role in fungal antagonism. Biol. Control. 2014, 69, 82–89. [Google Scholar] [CrossRef]
- Grgurina, I.; Mariotti, F.; Fogliano, V.; Gallo, M.; Scaloni, A.; Iacobellis, N.S.; Cantore, P.L.; Mannina, L.; Castelli, V.V.A.; Greco, M.L.; et al. A new syringopeptin produced by bean strains of Pseudomonas syringae pv. syringae. Biochim. Biophys. Acta 2002, 1597, 81–89. [Google Scholar] [CrossRef]
- Ballio, A.; Barra, D.; Bossa, F.; Collina, A.; Grgurina, I.; Marino, G.; Moneti, G.; Paci, M.; Pucci, P.; Segre, A.; et al. Syringopeptins, new phytotoxic lipodepsipeptides of Pseudomonas syringae pv. syringae. FEBS Lett. 1991, 291, 109–112. [Google Scholar] [CrossRef] [Green Version]
- Ravindran, A.; Jalan, N.; Yuan, J.S.; Wang, N.; Gross, D.C. Comparative genomics of Pseudomonas syringae pv. syringae strains B301D and HS 191 and insights into intrapathovar traits associated with plant pathogenesis. MicrobiologyOpen 2015, 4, 553–573. [Google Scholar] [CrossRef]
- Berti, A.D.; Greve, N.J.; Christensen, Q.H.; Thomas, M.G. Identification of a Biosynthetic Gene Cluster and the Six Associated Lipopeptides Involved in Swarming Motility of Pseudomonas syringae pv. tomato DC3000. J. Bacteriol. 2007, 189, 6312–6323. [Google Scholar] [CrossRef] [Green Version]
- Ramkumar, G.; Lee, S.W.; Weon, H.-Y.; Kim, B.-Y.; Lee, Y.H. First report on the whole genome sequence of Pseudomonas cichoriistrain JBC1 and comparison with other Pseudomonas species. Plant Pathol. 2015, 64, 63–70. [Google Scholar] [CrossRef]
- Alam, K.; Islam, M.; Li, C.; Sultana, S.; Zhong, L.; Shen, Q.; Yu, G.; Hao, J.; Zhang, Y.; Li, R.; et al. Genome Mining of Pseudomonas Species: Diversity and Evolution of Metabolic and Biosynthetic Potential. Molecules 2021, 26, 7524. [Google Scholar] [CrossRef] [PubMed]






| Taxonomy | Biocontrol Strains | Host/Origin | Country | LP Family | LP | Reference |
|---|---|---|---|---|---|---|
| P. fluorescens SG | SS101 | Wheat rhizosphere | Netherlands | Viscosin | Massetolide | [34] |
| SBW25 | Sugarbeet phyllosphere | UK | Viscosin | [35] | ||
| DR54 | Sugarbeet rhizosphere | Denmark | Viscosinamide | [36,37] | ||
| A2W4.9, U2W1.5 | White cocoyam rhizosphere | Nigeria | Viscosinamide | [38] | ||
| BRG100 | Green foxtail rhizosphere | Canada | Pseudophomin | [39] | ||
| RE*1-1-14 | Internal part of soybean roots | Germany | Poaemide | [40,41] | ||
| NCPPB1311 | Cultivated mushrooms | UK | WLIP | [42] | ||
| P. koreensis SG | DSS73 | Sugarbeet rhizosphere | Denmark | Amphisin | Amphisin | [14,43] |
| HKI0770 | Forest soil | Forest soil | Anikasin | [44,45] | ||
| CTS17 | Sugarbeet rhizosphere | Denmark | Hodersin | [14,46] | ||
| DSS41 | Sugarbeet rhizosphere | Denmark | Lokisin | [47] | ||
| 2.74 | Tomato hydroponics | Sweden | Lokisin | [48] | ||
| S150 | Tobacco rhizosphere | China | Lokisin | [49] | ||
| COR10 | Red cocoyam rhizosphere | Cameroon | Lokisin | [10] | ||
| UCMA 17988 | Raw bulk tank milk | France | Milkisin | [50] | ||
| COW8 | White cocoyam rhizosphere | Cameroon | Rhizoamide (N2—11:7) † | [11] | ||
| 96.578 | Sugarbeet rhizosphere | Denmark | Tensin | [37,51] | ||
| BW11P2 | Banana rhizoplane | Sri Lanka | Bananamide | Bananamide I, II, III | [12,52] | |
| COW3, COW65 | White cocoyam rhizosphere | Cameroon | Bananamide D, E, F, G | [10,29] | ||
| COW5 | White cocoyam rhizosphere | Cameroon | Cocoyamide | Cocoyamide A | [10] | |
| Pf0-1 | Loam soil | USA | Gacamide A | [53,54] | ||
| P. protegens SG | CHA0 | Tobacco roots | Switzerland | Orfamide | Orfamide | [55,56] |
| Pf-5 | Cotton rhizosphere | USA | Orfamide | [57,58] | ||
| CMR5c | Red cocoyam rhizosphere | Cameroon | Orfamide | [56] | ||
| CMR12a | Red cocoyam rhizosphere | Cameroon | Orfamide, Sessilin | [59] | ||
| P. chlororaphis SG | COR52 | Red cocoyam rhizosphere | Cameroon | Viscosin | Pseudodesmin | [38] |
| P. mandelii SG | In5 | Suppressive potato soil | Greenland | Syringomycin | Nunamycin | [18] |
| In5 | Suppressive potato soil | Greenland | Syringopeptin | Nunapeptin | [18] | |
| P. corrugata SG | SH-C52 | Sugarbeet rhizosphere | Netherlands | Syringomycin | Thanamycin | [60] |
| DF41 | Canola root | Canada | Thanamycin -var1 | [28,61] | ||
| 11K1 | Bean rhizosphere | China | Brasmycin | [62] | ||
| SH-C52 | Sugarbeet rhizosphere | Netherlands | Syringopeptin | Thanapeptin | [60] | |
| DF41 | Canola root | Canada | Sclerosin | [61] | ||
| 11K1 | Bean rhizosphere | China | Braspeptin | [62] | ||
| P. putida G | BW11M1 | Banana rhizoplane | Sri Lanka | Xantholysin | Xantholysin | [12,63] |
| COR51 | Red cocoyam rhizosphere | Cameroon | Xantholysin | [10] | ||
| BS011 | Rice rhizosphere | China | Xantholysin | [64] | ||
| 267 | Black pepper | Vietnam | Putisolvin | Putisolvin I, II | [65] | |
| COR55 | Red cocoyam rhizosphere | Cameroon | Putisolvin III, IV, V | [10,11] | ||
| L48 | Fly | Guadeloupe | Entolysin | Entolysin A, B | [66] | |
| COR5 | Red cocoyam rhizosphere | Cameroon | Entolysin B | [10] | ||
| RW10S2 | Rice rhizosphere | Sri Lanka | Viscosin | WLIP | [67] | |
| COW10 | White cocoyam rhizosphere | Cameroon | WLIP | [10] | ||
| NSE1 | White cocoyam rhizosphere | Nigeria | WLIP | [68] | ||
| COR35 | Red cocoyam rhizosphere | Cameroon | Unclassified | N8 (17:8) † | [11] | |
| P. asplenii SG | COR33 | Red cocoyam rhizosphere | Cameroon | Unclassified | N5 (13:8) † | [11] |
| COR18 | Red cocoyam rhizosphere | Cameroon | N5 (13:8), N7 †, Mycin LP † | [11] | ||
| Novel U2 SG | COR58 | Red cocoyam rhizosphere | Cameroon | Unclassified | N4 (12:10) † | [10,11] |
| Strain and Taxonomy | Plant | Pathogen | Lipopeptide | Experimental Setup | Method * | Reference |
|---|---|---|---|---|---|---|
| P. fluorescens SG | ||||||
| P. fluorescens DR54 | Sugar beet | Pythium ultimum | Viscosinamide | soil, in vitro | Pure | [14,36,70] |
| Sugar beet | Rhizoctonia solani | Viscosinamide | soil, in vitro | Pure | [14,36,71] | |
| Pseudomonas sp. A2W4.9 | - | Pythium myriotylum | Viscosinamide | in vitro | Pure | [38] |
| - | Rhizoctonia solani AG2-2 | Viscosinamide | in vitro | Pure | [38] | |
| P. lactis SS101 | Tomato | Phytophthora infestans | Massetolide A | soil assay, foliar | Mutant, pure | [72] |
| Arabidopsis | Pseudomonas syringae pv. tomato | Massetolide A | soil assay (ISR), in vitro | Mutant | [73] | |
| Hyacinth bulbs | Pythium intermedium, Pythium spp., Phytophthora infestans, Albugo candida | Massetolide A | in vitro | Mutant | [34] | |
| P. fluorescens SBW25 | - | Phytophthora infestans | Viscosin | in vitro | Mutant | [35] |
| P. fluorescens BRG100 | - | Leptosphaeria maculans, Sclerotinia sclerotiorum | Pseudophomin A and B | in vitro | Pure | [39] |
| Pseudomonas sp. COR52 | - | Pythium myriotylum | Pseudodesmin | in vitro | Pure | [38] |
| - | Rhizoctonia solani AG2-2 | Pseudodesmin | in vitro | Pure | [38] | |
| P. poae RE *1-1-14 | - | Phytophthora capsici, Phytophthora infestans | Poaeamide | in vitro | Pure | [41] |
| Pythium ultimum, Rhizoctonia solani | in vitro | Pure | [41] | |||
| P. reactans NCPPB1311 | - | Erwinia carotovora subsp. carotovora, Agaricus bisporus | WLIP | in vitro | Pure | [42] |
| P. reactans | - | Pseudomonas tolaasii | WLIP | in vitro, mushroom cap | Pure | [42] |
| P. tolaasii NCPPB2192 | - | Escherichia coli, Erwinia, Agrobacterium, Pseudomonas, Xanthomonas, Pleurotus spp., Agaricus bisporus | Tolaasin 1 | in vitro | Pure | [42] |
| P. protegens SG | ||||||
| P. sessiligenes CMR12a | Bean | Rhizoctonia solani AG2-2, AG4 | Sessilin | soil assay | Mutant | [74,75] |
| Rhizoctonia solani AG2-2 (web blight) | Sessilin | soil assay (ISR) | Mutant, crude extract | [76] | ||
| Rhizoctonia solani AG2-1, AG4 | Sessilin | in vitro | Crude extract | [75] | ||
| Rice | Pyricularia oryzae | Sessilin | soil assay (ISR) | Mutants | [76] | |
| Chinese cabbage | Rhizoctonia solani AG2-1 | Sessilin | soil assay | Mutant | [75] | |
| Cocoyam | Pythium myriotylum | Sessilin | soil assay | Mutant | [77] | |
| Pythium myriotylum | Sessilin | in vitro | Crude extract | [77] | ||
| Bean | Rhizoctonia solani AG4 | Orfamide | soil assay | Mutant | [75] | |
| Bean | Rhizoctonia solani AG2-2 (web blight) | Orfamide B | soil assay (ISR) | Mutant, pure | [76] | |
| - | Rhizoctonia solani AG2-1, AG4 | Orfamide B | in vitro | Mutant, pure | [75] | |
| Chinese cabbage | Rhizoctonia solani AG4 | Orfamide | soil assay | Mutant | [75] | |
| Cocoyam | Pythium myriotylum | Orfamide | soil assay | Mutant | [77] | |
| - | Pythium myriotylum | Orfamide B | in vitro | Pure | [77] | |
| Rice | Pyricularia oryzae | Orfamide | soil assay (ISR) | Mutants | [76] | |
| Rice | Pyricularia oryzae | Orfamide A | soil assay (ISR) | Pure | [78] | |
| P. protegens CHA0 | Rice | Cochliobolus miyabeanus | Orfamide A | soil assay (ISR) | Mutant | [78] |
| - | Phytophthora porri, Pythium ultimum | in vitro | Pure | [56] | ||
| - | Rhizoctonia solani AG4 | in vitro | Pure | [56] | ||
| Rice | Cochliobolus miyabeanus | soil drench (ISR) | Pure | [78] | ||
| - | Phytophthora ramorum | in vitro | Pure | [79] | ||
| P. aestus CMR5c | - | Rhizoctonia solani AG4 | Orfamide B | in vitro | Pure | [56] |
| - | Pyricularia oryzae | in vitro | Pure | [56] | ||
| - | Phytophthora porri, Pythium ultimum | in vitro | Pure | [56] | ||
| - | Pyricularia oryzae | Orfamide G | in vitro | Pure | [56] | |
| - | Rhizoctonia solani AG4 | in vitro | Pure | [56] | ||
| - | Phytophthora porri, Pythium ultimum | in vitro | Pure | [56] | ||
| P. chlororaphis SG Pseudomonas sp. COR52 | - | Pythium myriotylum | Pseudodesmin | in vitro | Pure | [38] |
| - | Rhizoctonia solani | Pseudodesmin | in vitro | Pure | [38] | |
| P. koreensis SG | ||||||
| P. botevensis COW3 | - | Pythium myriotylum | Bananamide D, E, F, G | in vitro | Pure | [29] |
| Pyricularia oryzae | Bananamide D, E, F, G | in vitro | Pure | [29] | ||
| Rice | Pyricularia oryzae | Bananamide D, E, F, G | soil assay (ISR) | Crude extract | [68] | |
| Pseudomonas sp. COW5 | - | Pythium myriotylum | Cocoyamide | in vitro | Pure | [10] |
| P. fluorescens Pf0-1 | - | Pseudomonas syringae, Erwinia amylovora | Gacamide | in vitro | Pure | [53] |
| Pseudomonas sp. DSS73 | - | Rhizoctonia solani, Pythium ultimum | Amphisin | in vitro | Mutant, pure | [14,43] |
| P. fluorescens HKI0770 | Polysphondylium violaceum | Anikasin | in vitro | Pure | [44] | |
| Pseudomonas sp. COR10 | - | Pythium myriotylum | Lokisin | in vitro | Pure | [10] |
| Rice | Pyricularia oryzae | Lokisin | soil assay (ISR) | Crude extract | [68] | |
| Pseudomonas sp. UCMA 17988 | Penicillium expansum | Milkisin | in vitro | Pure | [50] | |
| Pseudomonas sp. COW8 | - | Pythium myriotylum | N2 (Rhizoamide (11:7)) | in vitro | Pure | [10,11] |
| Pseudomonas sp. DSS41 | - | Rhizoctonia solani, Pythium ultimum | Lokisin | in vitro | Pure | [14] |
| Pseudomonas sp. 2.74 | Tomato | Pythium ultimum | Lokisin | hydroponic assay | Crude extract | [48] |
| Pseudomonas sp. 96.578 | - | Rhizoctonia solani | Tensin | in vitro | Pure | [46,51] |
| Pseudomonas sp. | - | Rhizoctonia solani, Pythium ultimum | Hodersin | in vitro | Pure | [14] |
| P. corrugata SG | ||||||
| Pseudomonas sp. SH-C52 | Groundnut | Sclerotium rolfsii | Thanamycin | nethouse and field | Mutant | [80] |
| - | Botrytis cinerea, Geotrichum sp., Rhizoctonia solani | Thanamycin | in vitro | Mutant | [60] | |
| Sugar beet | Rhizoctonia solani | Thanamycin | soil assay | Mutant | [81] | |
| - | Rhizoctonia solani | Thanamycin | in vitro | Mutant | [81] | |
| Phytophthora infestans, Pythium ultimum | Thanapeptin | in vitro | Mutant | [60] | ||
| P. brassicacearum DF41 | Canola | Sclerotinia sclerotiorum | Sclerosin | soil assay, foliar spray | Mutant | [61,80] |
| P. brassicacearum 11K1 | Botryosphaeria dothidea | Brasmycin | in vitro | [62] | ||
| Botryosphaeria dothidea | Braspeptin | in vitro | [62] | |||
| P. mandelii SG | ||||||
| P. fluorescens In5 | Rhizoctonia solani | Nunamycin | in vitro | Mutant | [18] | |
| Pythium aphanidermatum | Nunapeptin | in vitro | Mutant | [18] | ||
| P. syringae G | ||||||
| P. syringae pv. syringae B359 (B427) | - | Botrytis cinerea, Rhodotorula pilimanae | Syringotoxin | in vitro | Pure | [82] |
| P. syringae pv. syringae B301 | - | Botrytis cinerea, Geotrichum candidum | Syringomycin E | in vitro | Pure | [82] |
| P. syringae ESC-10 and ESC-11 | Lemon | Penicillium digitatum | in vitro, in planta | Pure | [83] | |
| P. syringae pv. syringae B359 (B427) | Botrytis cinerea, Geotrichum candidum | Syringopeptin (SP22-A, SP25-A) | in vitro | Pure | [82] | |
| P. putida G | ||||||
| P. entomophilia L48 | Cucumber | Pythium ultimum | Entolysin | soil assay | Mutant | [66] |
| Pseudomonas sp. COR5 | - | Pythium myriotylum | Entolysin | in vitro | Pure | [10] |
| Rice | Pyricularia oryzae | Entolysin | soil assay (ISR) | Crude extract | [68] | |
| P. putida 267 | Phytophthora capsici | Putisolvin | in vitro | Mutant | [13] | |
| Pseudomonas sp. COR55 | - | Pythium myriotylum | Putisolvin | in vitro | Pure | [10] |
| Pseudomononas sp. NSE1 | - | Pythium myriotylum | WLIP | in vitro | Pure | [11] |
| - | Rhizoctonia solani AG2-2 | WLIP | in vitro | Pure | [11] | |
| P. promisalinigenes RW10S2 | - | Xanthomonas sp. | WLIP | in vitro | Mutant | [67] |
| Rice | Pyricularia oryzae | WLIP | soil assay (ISR) | Mutant analysis, Crude extract | [68] | |
| P. mosselii BW11M1 | - | Xanthomonas spp., Rhizoctonia solani, Botrytis cinerea | Xantholysin | in vitro | Mutant | [63] |
| P. mosselii BS011 | Rice | Pyricularia oryzae | Xantholysin | soil assay (ISR) | Crude extract | [64] |
| in vitro | Crude extract | [64] | ||||
| - | Pythium myriotylum | Xantholysin A | in vitro | Pure | ||
| Pseudomonas sp. COR51 | Rice | Pyricularia oryzae | Xantholysin | soil assay (ISR) | Crude extract | [68] |
| Pseudomonas sp. COR35 | - | Pythium myriotylum | N8 (17:8) | in vitro | Pure | [10] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oni, F.E.; Esmaeel, Q.; Onyeka, J.T.; Adeleke, R.; Jacquard, C.; Clement, C.; Gross, H.; Ait Barka, E.; Höfte, M. Pseudomonas Lipopeptide-Mediated Biocontrol: Chemotaxonomy and Biological Activity. Molecules 2022, 27, 372. https://doi.org/10.3390/molecules27020372
Oni FE, Esmaeel Q, Onyeka JT, Adeleke R, Jacquard C, Clement C, Gross H, Ait Barka E, Höfte M. Pseudomonas Lipopeptide-Mediated Biocontrol: Chemotaxonomy and Biological Activity. Molecules. 2022; 27(2):372. https://doi.org/10.3390/molecules27020372
Chicago/Turabian StyleOni, Feyisara Eyiwumi, Qassim Esmaeel, Joseph Tobias Onyeka, Rasheed Adeleke, Cedric Jacquard, Christophe Clement, Harald Gross, Essaid Ait Barka, and Monica Höfte. 2022. "Pseudomonas Lipopeptide-Mediated Biocontrol: Chemotaxonomy and Biological Activity" Molecules 27, no. 2: 372. https://doi.org/10.3390/molecules27020372