Inhibitory Effects of Myriocin on Non-Enzymatic Glycation of Bovine Serum Albumin
Abstract
1. Introduction
2. Results and Discussion
2.1. Inhibitory Effect of Myr on Non-Enzymatic BSA Glycation
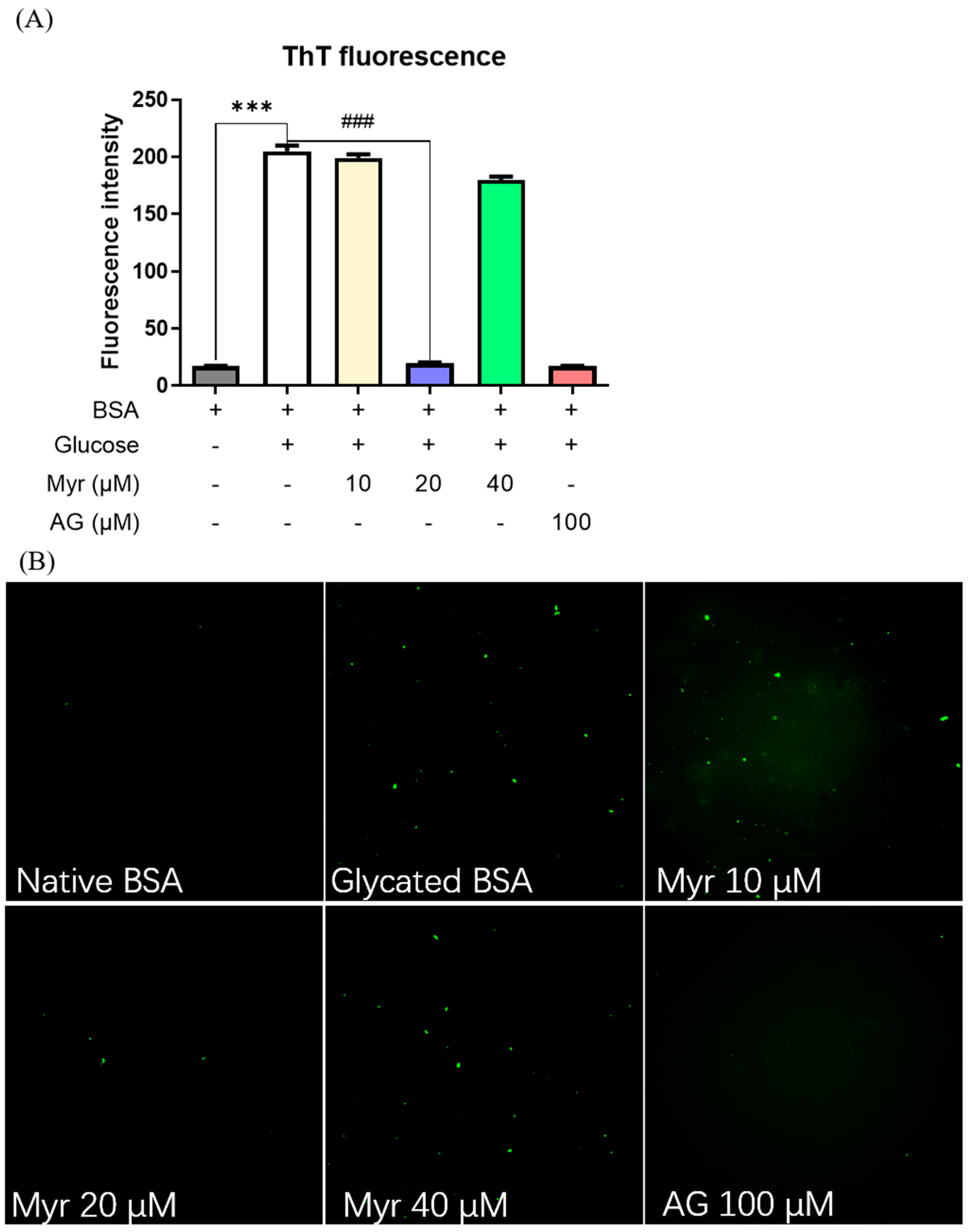
2.2. Anti-Fibrillation Ability of Myr
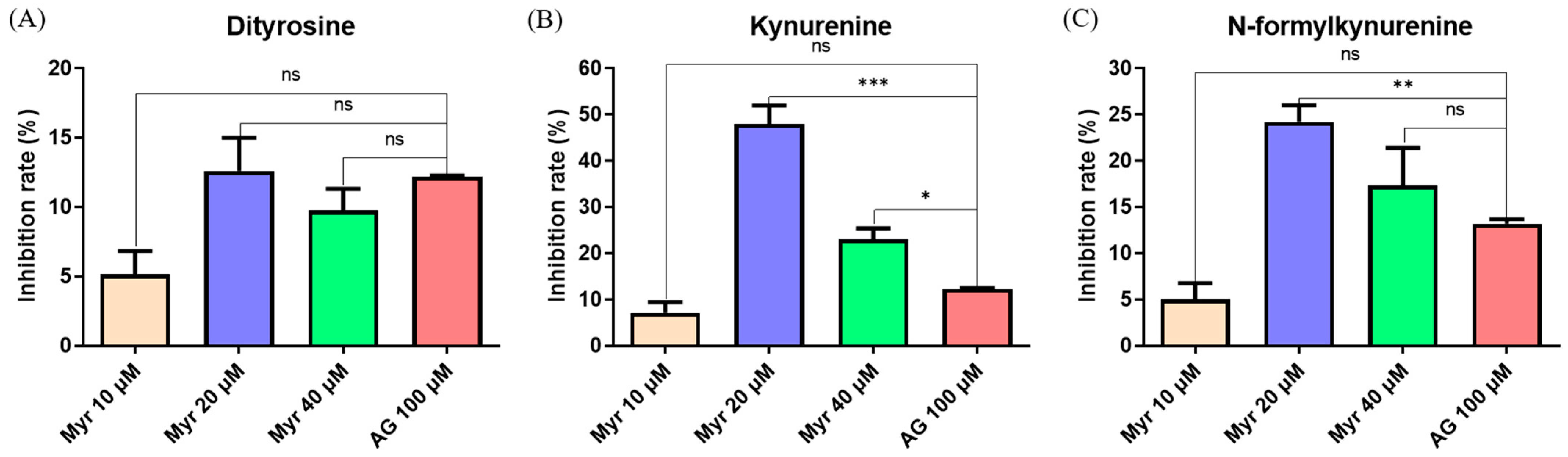
2.3. Inhibitory Effect of Myr on Protein Glycoxidation
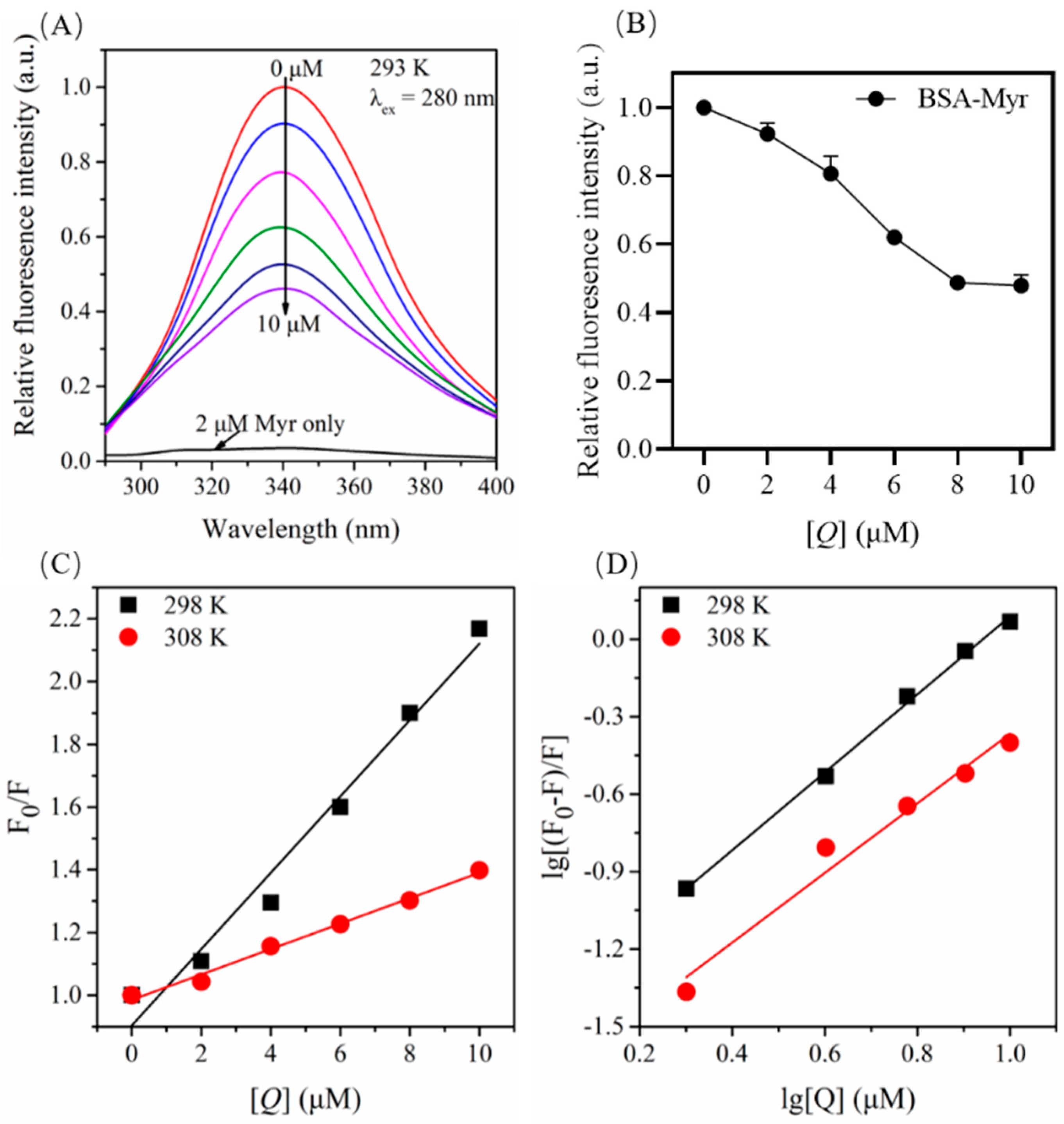
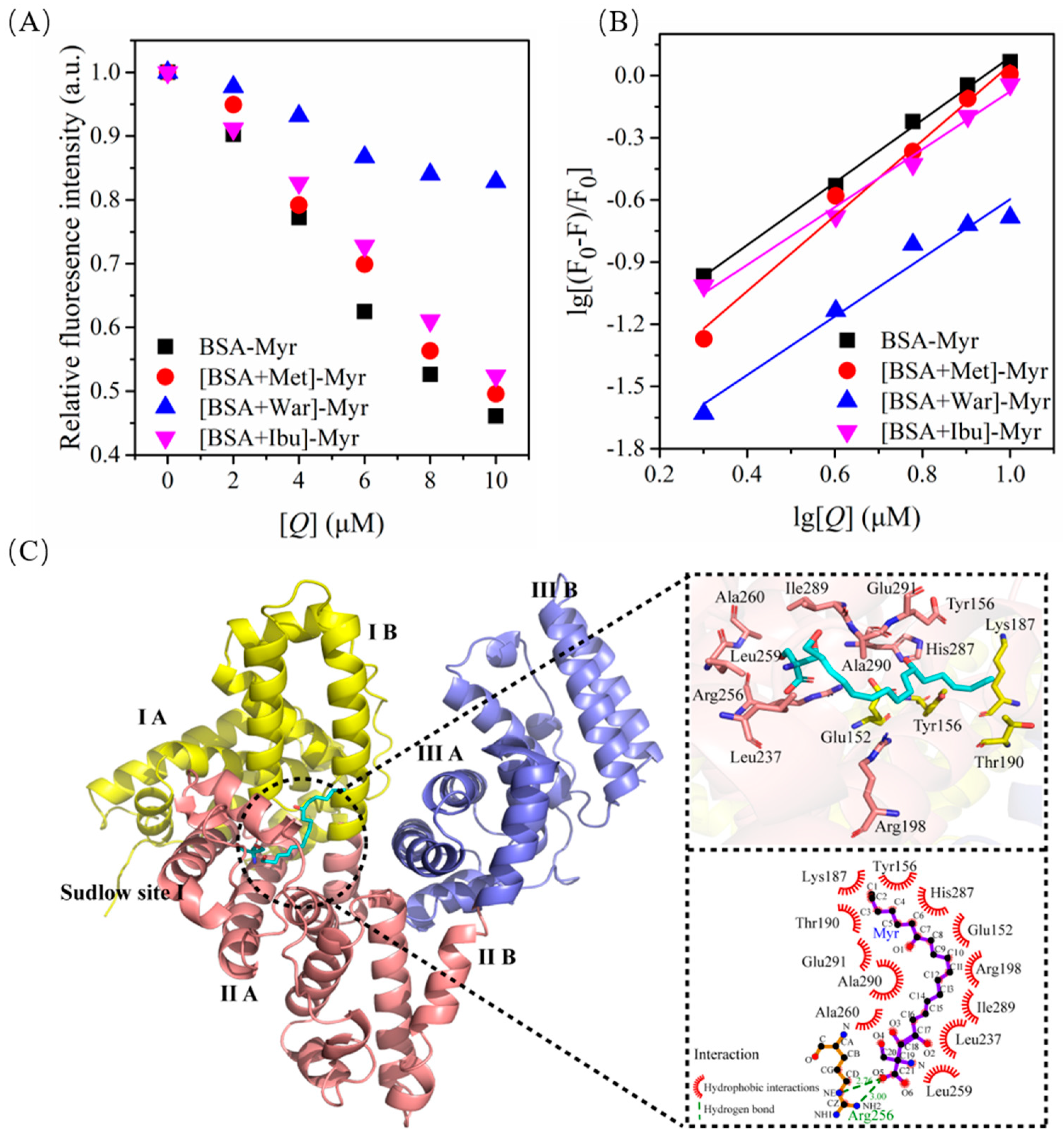
2.4. Quenching Effect of Myr on Endogenous Fluorescence of BSA
2.5. Fluorescence Quenching Mechanism for BSA by Myr
2.6. Binding Constants and Number of Binding Site for the BSA-Myr Complex
2.7. Binding Forces between BSA and Myr
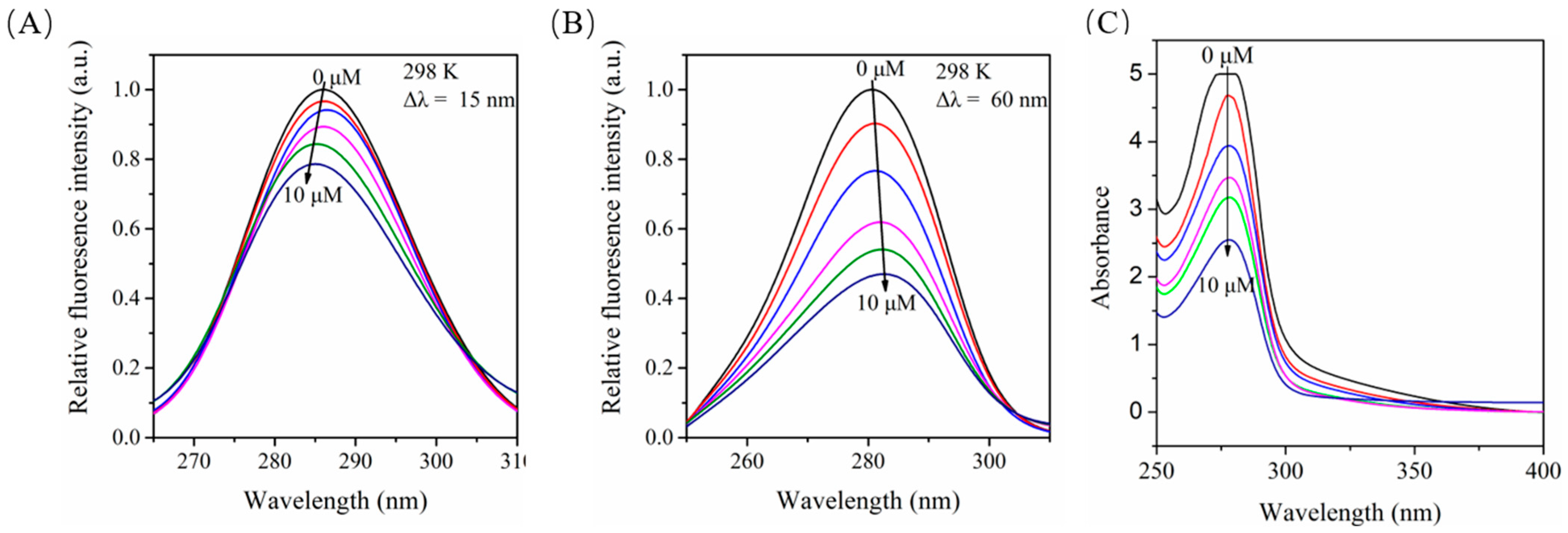
2.8. Influence of Myr on the Structure of BSA as Monitored by Synchronous Fluorescence Spectroscopy
2.9. Effects of Myr on the UV-Vis Absorption of BSA
2.10. Binding Model between BSA and Myr
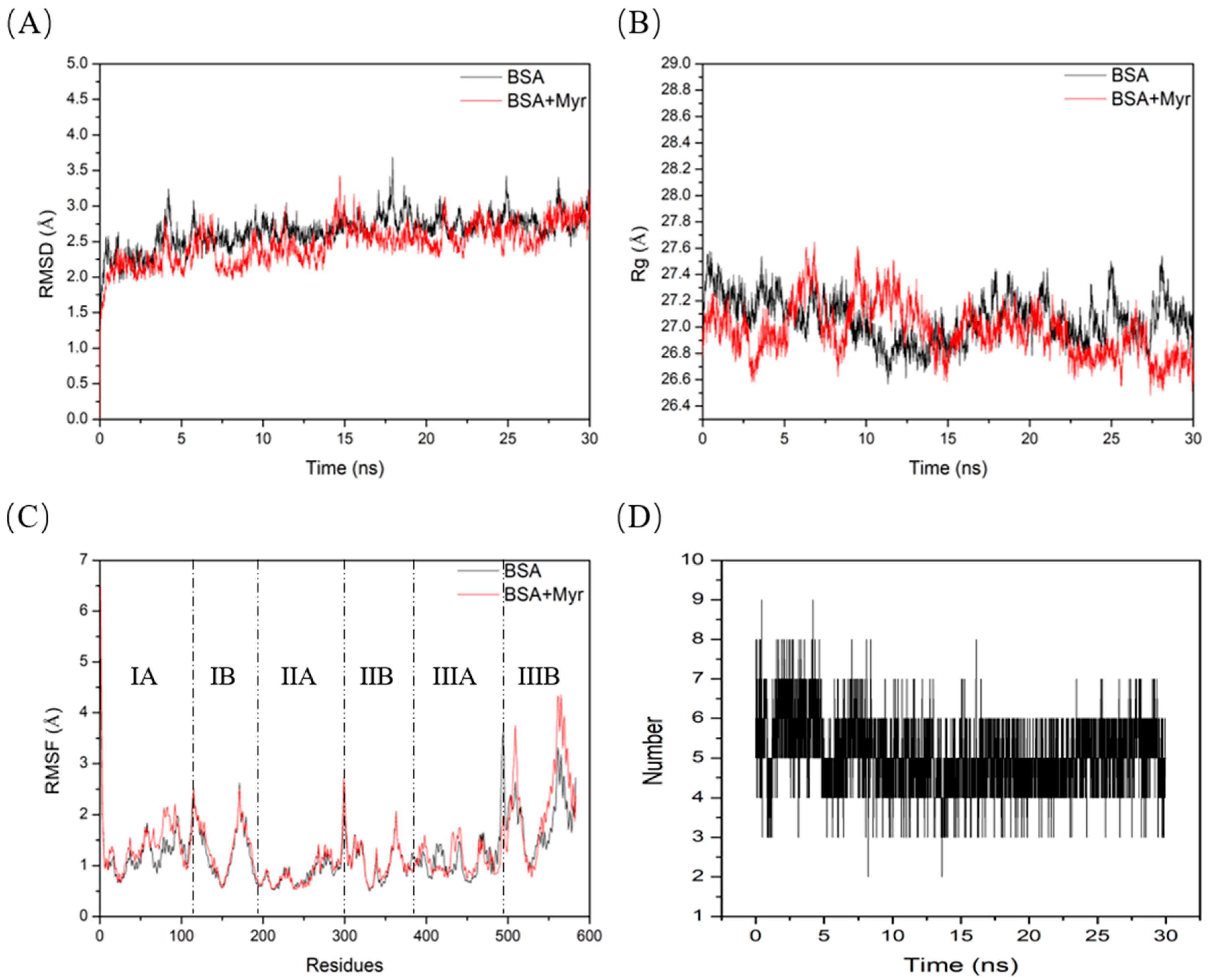
2.11. Molecular Dynamics (MD) Simulation between BSA and Myr
3. Materials and Methods
3.1. Regents
3.2. Non-Enzymatic Glycation of BSA
3.3. Fluorescence Spectroscopy
3.4. Anti-Glycation Activity of Myr
3.4.1. Determination of Fructosamine Content
3.4.2. Determination of Fluorescent AGEs
3.5. Detection of Amyloid-like Fibrils
3.6. Inhibitory Activity of Myr on Glycoxidation Fluorescence Products
3.7. UV-Visible Absorption Spectroscopy
3.8. Probe Displacement Experiment
3.9. Molecular Docking
3.10. Molecular Dynamics (MD) Simulation
3.11. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Myr | Myriocin |
| AG | Aminoguanidine |
| BSA | Bovine serum albumin |
| AGEs | Advanced glycation end products |
| SPT | Serine palmitoyltransferase |
| NBT | Nitroblue tetrazolium |
| ThT | Thioflavin T |
| Trp | Tryptophan |
| Tyr | Tyrosine |
| Phe | Phenylalanine |
| ΔG | Gibbs free energy change |
| ΔH | Enthalpy change |
| ΔS | Entropy change |
| λex | Excitation wavelength |
| λem | Emission wavelength |
| MD | Molecular dynamics |
| RMSD | Root mean square deviation |
| RMSF | Root mean square fluctuation |
| Rg | Radius of gyration |
References
- Twarda-Clapa, A.; Olczak, A.; Białkowska, A.M.; Koziołkiewicz, M. Advanced Glycation End-Products (AGEs): Formation, Chemistry, Classification, Receptors, and Diseases Related to AGEs. Cells 2022, 11, 1312. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.-Y.; Lu, C.-H.; Wu, C.-H.; Li, K.-J.; Kuo, Y.-M.; Hsieh, S.-C.; Yu, C.-L. The Development of Maillard Reaction, and Advanced Glycation End Product (AGE)-Receptor for AGE (RAGE) Signaling Inhibitors as Novel Therapeutic Strategies for Patients with AGE-Related Diseases. Molecules 2020, 25, 5591. [Google Scholar] [CrossRef] [PubMed]
- Peppa, M.; Uribarri, J.; Vlassara, H. Glucose, Advanced Glycation End Products, and Diabetes Complications: What Is New and What Works. Clin. Diabetes 2003, 21, 186–187. [Google Scholar] [CrossRef]
- Singh, V.P.; Bali, A.; Singh, N.; Jaggi, A.S. Advanced Glycation End Products and Diabetic Complications. Korean J. Physiol. Pharmacol. 2014, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Lv, G.-H.; Dai, G.-Y.; Sun, H.-M.; Xu, H.-Q. Food-advanced glycation end products aggravate the diabetic vascular complications via modulating the AGEs/RAGE pathway. Chin. J. Nat. Med. 2016, 14, 844–855. [Google Scholar] [CrossRef]
- Xu, J.; Chen, L.-J.; Yu, J.; Wang, H.-J.; Zhang, F.; Liu, Q.; Wu, J. Involvement of Advanced Glycation End Products in the Pathogenesis of Diabetic Retinopathy. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 48, 705–717. [Google Scholar] [CrossRef]
- Rabbani, N.; Godfrey, L.; Xue, M.; Shaheen, F.; Geoffrion, M.; Milne, R.; Thornalley, P.J. Glycation of LDL by Methylglyoxal Increases Arterial Atherogenicity: A Possible Contributor to Increased Risk of Cardiovascular Disease in Diabetes. Diabetes 2011, 60, 1973–1980. [Google Scholar] [CrossRef]
- Baye, E.; Mark, A.B.; Poulsen, M.W.; Andersen, J.M.; Dragsted, L.O.; Bügel, S.G.; de Courten, B. Associations between Urinary Advanced Glycation End Products and Cardiometabolic Parameters in Metabolically Healthy Obese Women. J. Clin. Med. 2019, 8, 1008. [Google Scholar] [CrossRef]
- Davis, K.E.; Prasad, C.; Vijayagopal, P.; Juma, S.; Imrhan, V. Advanced Glycation End Products, Inflammation, and Chronic Metabolic Diseases: Links in a Chain? Crit. Rev. Food Sci. Nutr. 2016, 56, 989–998. [Google Scholar] [CrossRef]
- Shobana, S.; Sreerama, Y.N.; Malleshi, N.G. Composition and enzyme inhibitory properties of finger millet (Eleusine coracana L.) seed coat phenolics: Mode of inhibition of α-glucosidase and pancreatic amylase. Food Chem. 2009, 115, 1268–1273. [Google Scholar] [CrossRef]
- Buckow, R.; Wendorff, J.; Hemar, Y. Conjugation of Bovine Serum Albumin and Glucose under Combined High Pressure and Heat. J. Agric. Food Chem. 2011, 59, 3915–3923. [Google Scholar] [CrossRef] [PubMed]
- Rondeau, P.; Bourdon, E. The glycation of albumin: Structural and functional impacts. Biochimie 2011, 93, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Ledesma-Osuna, A.; Ramos-Clamont, G.; Vazquez-Moreno, L. Characterization of bovine serum albumin glycated with glucose, galactose and lactose. Acta Biochim. Pol. 2008, 55, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Peña, I.; Domínguez, J.M. Thermally Denatured BSA, a Surrogate Additive to Replace BSA in Buffers for High-Throughput Screening. SLAS Discov. 2010, 15, 1281–1286. [Google Scholar] [CrossRef]
- Militello, V.; Vetri, V.; Leone, M. Conformational changes involved in thermal aggregation processes of bovine serum albumin. Biophys. Chem. 2003, 105, 133–141. [Google Scholar] [CrossRef]
- Vetter, S.W. Chapter Five—Glycated Serum Albumin and AGE Receptors. In Advances in Clinical Chemistry; Makowski, G.S., Ed.; Elsevier: Amsterdam, The Netherlands, 2015; Volume 72, pp. 205–275. [Google Scholar]
- Anwar, S.; Khan, S.; Almatroudi, A.; Khan, A.A.; Alsahli, M.A.; Almatroodi, S.A.; Rahmani, A.H. A review on mechanism of inhibition of advanced glycation end products formation by plant derived polyphenolic compounds. Mol. Biol. Rep. 2021, 48, 787–805. [Google Scholar] [CrossRef]
- Zhou, Q.; Cheng, K.-W.; Xiao, J.; Wang, M. The multifunctional roles of flavonoids against the formation of advanced glycation end products (AGEs) and AGEs-induced harmful effects. Trends Food Sci. Technol. 2020, 103, 333–347. [Google Scholar] [CrossRef]
- Liu, J.-l.; He, Y.-l.; Wang, S.; He, Y.; Wang, W.-Y.; Li, Q.-J.; Cao, X.-Y. Ferulic acid inhibits advanced glycation end products (AGEs) formation and mitigates the AGEs-induced inflammatory response in HUVEC cells. J. Funct. Foods 2018, 48, 19–26. [Google Scholar] [CrossRef]
- Shen, Y.; Xu, Z.; Sheng, Z. Ability of resveratrol to inhibit advanced glycation end product formation and carbohydrate-hydrolyzing enzyme activity, and to conjugate methylglyoxal. Food Chem. 2017, 216, 153–160. [Google Scholar] [CrossRef]
- Xue, C.; Deng, P.; Quan, W.; Li, Y.; He, Z.; Qin, F.; Wang, Z.; Chen, J.; Zeng, M. Ginger and curcumin can inhibit heterocyclic amines and advanced glycation end products in roast beef patties by quenching free radicals as revealed by electron paramagnetic resonance. Food Control 2022, 138, 109038. [Google Scholar] [CrossRef]
- Hanada, K. Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2003, 1632, 16–30. [Google Scholar] [CrossRef]
- Lee, Y.S.; Choi, K.M.; Lee, S.; Sin, D.M.; Lim, Y.; Lee, Y.M.; Hong, J.T.; Yun, Y.P.; Yoo, H.S. Myriocin, a serine palmitoyltransferase inhibitor, suppresses tumor growth in a murine melanoma model by inhibiting de novo sphingolipid synthesis. Cancer Biol. 2012, 13, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Park, T.-S.; Panek, R.L.; Mueller, S.B.; Hanselman, J.C.; Rosebury, W.S.; Robertson, A.W.; Kindt, E.K.; Homan, R.; Karathanasis, S.K.; Rekhter, M.D. Inhibition of sphingomyelin synthesis reduces atherogenesis in apolipoprotein E-knockout mice. Circulation 2004, 110, 3465–3471. [Google Scholar] [CrossRef] [PubMed]
- Hojjati, M.R.; Li, Z.; Zhou, H.; Tang, S.; Huan, C.; Ooi, E.; Lu, S.; Jiang, X.C. Effect of myriocin on plasma sphingolipid metabolism and atherosclerosis in apoE-deficient mice. J. Biol. Chem. 2005, 280, 10284–10289. [Google Scholar] [CrossRef]
- Park, T.-S.; Panek, R.L.; Rekhter, M.D.; Mueller, S.B.; Rosebury, W.S.; Robertson, A.; Hanselman, J.C.; Kindt, E.; Homan, R.; Karathanasis, S.K. Modulation of lipoprotein metabolism by inhibition of sphingomyelin synthesis in ApoE knockout mice. Atherosclerosis 2006, 189, 264–272. [Google Scholar] [CrossRef]
- Glaros, E.N.; Kim, W.S.; Wu, B.J.; Suarna, C.; Quinn, C.M.; Rye, K.A.; Stocker, R.; Jessup, W.; Garner, B. Inhibition of atherosclerosis by the serine palmitoyl transferase inhibitor myriocin is associated with reduced plasma glycosphingolipid concentration. Biochem Pharm. 2007, 73, 1340–1346. [Google Scholar] [CrossRef]
- Park, T.S.; Rosebury, W.; Kindt, E.K.; Kowala, M.C.; Panek, R.L. Serine palmitoyltransferase inhibitor myriocin induces the regression of atherosclerotic plaques in hyperlipidemic ApoE-deficient mice. Pharm. Res 2008, 58, 45–51. [Google Scholar] [CrossRef]
- Kurek, K.; Piotrowska, D.M.; Wiesiolek-Kurek, P.; Lukaszuk, B.; Chabowski, A.; Gorski, J.; Zendzian-Piotrowska, M. Inhibition of ceramide de novo synthesis reduces liver lipid accumulation in rats with nonalcoholic fatty liver disease. Liver Int. 2014, 34, 1074–1083. [Google Scholar] [CrossRef]
- Kasumov, T.; Li, L.; Li, M.; Gulshan, K.; Kirwan, J.P.; Liu, X.; Previs, S.; Willard, B.; Smith, J.D.; McCullough, A. Ceramide as a Mediator of Non-Alcoholic Fatty Liver Disease and Associated Atherosclerosis. PLoS ONE 2015, 10, e0126910. [Google Scholar] [CrossRef]
- Raichur, S.; Wang, S.T.; Chan, P.W.; Li, Y.; Ching, J.; Chaurasia, B.; Dogra, S.; Öhman, M.K.; Takeda, K.; Sugii, S.; et al. CerS2 Haploinsufficiency Inhibits β-Oxidation and Confers Susceptibility to Diet-Induced Steatohepatitis and Insulin Resistance. Cell Metab. 2014, 20, 687–695. [Google Scholar] [CrossRef]
- Correnti, J.; Juskeviciute, E.; Swarup, A.; Hoek, J. Pharmacologic ceramide reduction alleviates alcohol-induced steatosis and hepatomegaly in adiponectin knockout mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 306, G959–G973. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-C.; Lee, R.A.; Tsai, S.L.; Kanamaluru, D.; Gray, N.E.; Yiv, N.; Cheang, R.T.; Tan, J.H.; Lee, J.Y.; Fitch, M.D.; et al. An ANGPTL4–ceramide–protein kinase Cζ axis mediates chronic glucocorticoid exposure–induced hepatic steatosis and hypertriglyceridemia in mice. J. Biol. Chem. 2019, 294, 9213–9224. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-J.; Holland, W.; Wilson, L.; Tanner, J.; Kearns, D.; Cahoon, J.; Pettey, D.; Losee, J.; Duncan, B.; Gale, D.; et al. Ceramide Mediates Vascular Dysfunction in Diet-Induced Obesity by PP2A-Mediated Dephosphorylation of the eNOS-Akt Complex. Diabetes 2012, 61, 1848–1859. [Google Scholar] [CrossRef] [PubMed]
- Park, T.-S.; Hu, Y.; Noh, H.-L.; Drosatos, K.; Okajima, K.; Buchanan, J.; Tuinei, J.; Homma, S.; Jiang, X.-C.; Abel, E.D.; et al. Ceramide is a cardiotoxin in lipotoxic cardiomyopathy. J. Lipid Res. 2008, 49, 2101–2112. [Google Scholar] [CrossRef]
- Ji, R.; Akashi, H.; Drosatos, K.; Liao, X.; Jiang, H.; Kennel, P.; Brunjes, D.; Castillero, E.; Zhang, X.; Deng, L.; et al. Increased de novo ceramide synthesis and accumulation in failing myocardium. JCI Insight 2017, 2, e82922. [Google Scholar] [CrossRef]
- Holland, W.L.; Brozinick, J.T.; Wang, L.P.; Hawkins, E.D.; Sargent, K.M.; Liu, Y.; Narra, K.; Hoehn, K.L.; Knotts, T.A.; Siesky, A.; et al. Inhibition of ceramide synthesis ameliorates glucocorticoid-, saturated-fat-, and obesity-induced insulin resistance. Cell Metab. 2007, 5, 167–179. [Google Scholar] [CrossRef]
- Zabielski, P.; Daniluk, J.; Hady, H.R.; Markowski, A.R.; Imierska, M.; Górski, J.; Blachnio-Zabielska, A.U. The effect of high-fat diet and inhibition of ceramide production on insulin action in liver. J. Cell. Physiol. 2019, 234, 1851–1861. [Google Scholar] [CrossRef]
- Oliveira, L.M.A.; Gomes, R.A.; Yang, D.; Dennison, S.R.; Família, C.; Lages, A.; Coelho, A.V.; Murphy, R.M.; Phoenix, D.A.; Quintas, A. Insights into the molecular mechanism of protein native-like aggregation upon glycation. Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2013, 1834, 1010–1022. [Google Scholar] [CrossRef]
- Ahmad, S.; Khan, M.S.; Akhter, F.; Khan, M.S.; Khan, A.; Ashraf, J.M.; Pandey, R.P.; Shahab, U. Glycoxidation of Biological Macromolecules: A Critical Approach to Halt the Menace of Glycation. Glycobiology 2014, 24, 979–990. [Google Scholar] [CrossRef]
- Thornalley, P.J. Use of aminoguanidine (Pimagedine) to prevent the formation of advanced glycation endproducts. Arch. Biochem. Biophys. 2003, 419, 31–40. [Google Scholar] [CrossRef]
- Boulanger, E.; Wautier, J.-L.; Dequiedt, P.; Schmidt, A.-M. Glycation, glycoxidation and diabetes mellitus. Nephrol. Ther. 2006, 2, S8–S16. [Google Scholar] [PubMed]
- Prasanna, G.; Saraswathi, N.T. Linolenic acid prevents early and advanced glycation end-products (AGEs) modification of albumin. Int. J. Biol. Macromol. 2017, 95, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, F.; Lu, Q.; Liu, R.; Tian, J.; Huang, Y. Study on interaction between human salivary α-amylase and sorghum procyanidin tetramer: Binding characteristics and structural analysis. Int. J. Biol. Macromol. 2018, 118, 1136–1141. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. (Ed.) Quenching of Fluorescence. In Principles of Fluorescence Spectroscopy; Springer: Boston, MA, USA, 2006; pp. 277–330. [Google Scholar]
- Yue, Y.; Sun, Y.; Dong, Q.; Liu, R.; Yan, X.; Zhang, Y.; Liu, J. Interaction of human serum albumin with novel imidazole derivatives studied by spectroscopy and molecular docking. Luminescence 2016, 31, 671–681. [Google Scholar] [CrossRef]
- Xu, H.; Lu, Y.; Zhang, T.; Liu, K.; Liu, L.; He, Z.; Xu, B.; Wu, X. Characterization of binding interactions of anthraquinones and bovine β-lactoglobulin. Food Chem. 2019, 281, 28–35. [Google Scholar] [CrossRef]
- Shi, J.H.; Pan, D.-Q.; Jiang, M.; Liu, T.-T.; Wang, Q. In vitro study on binding interaction of quinapril with bovine serum albumin (BSA) using multi-spectroscopic and molecular docking methods. J. Biomol. Struct. Dyn. 2017, 35, 2211–2223. [Google Scholar] [CrossRef]
- Wang, Q. Forward and adjoint sensitivity computation of chaotic dynamical systems. J. Comput. Phys. 2013, 235, 1–13. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, P.; Yang, Z.; Du, F.; Li, Z.; Wu, C.; Fang, A.; Xu, X.; Zhou, G. Molecular dynamics simulation exploration of the interaction between curcumin and myosin combined with the results of spectroscopy techniques. Food Hydrocoll. 2020, 101, 105455. [Google Scholar] [CrossRef]
- Rabbani, N.; Xue, M.; Thornalley, P. Dicarbonyls and glyoxalase in disease mechanisms and clinical therapeutics. Glycoconj. J. 2016, 33, 513–525. [Google Scholar] [CrossRef]
- Lv, C.; Zhao, G.; Ning, Y. Interactions between plant proteins/enzymes and other food components, and their effects on food quality. Crit. Rev. Food Sci. Nutr. 2017, 57, 1718–1728. [Google Scholar] [CrossRef]
- Zhu, G.-F.; Wang, Y.; Liu, J.; Wang, H.; Xi, L.; Du, L.-F. Interaction Between Ginkgolic Acid and Human Serum Albumin by Spectroscopy and Molecular Modeling Methods. J. Solut. Chem. 2014, 43, 1232–1249. [Google Scholar] [CrossRef]
- Olsson, T.S.G.; Williams, M.A.; Pitt, W.R.; Ladbury, J.E. The Thermodynamics of Protein–Ligand Interaction and Solvation: Insights for Ligand Design. J. Mol. Biol. 2008, 384, 1002–1017. [Google Scholar] [CrossRef] [PubMed]
- Al-Shabib, N.A.; Khan, J.M.; Malik, A.; Alsenaidy, M.A.; Rehman, M.T.; AlAjmi, M.F.; Alsenaidy, A.M.; Husain, F.M.; Khan, R.H. Molecular insight into binding behavior of polyphenol (rutin) with beta lactoglobulin: Spectroscopic, molecular docking and MD simulation studies. J. Mol. Liq. 2018, 269, 511–520. [Google Scholar] [CrossRef]
- Gao, W.; Li, N.; Chen, Y.; Xu, Y.; Lin, Y.; Yin, Y.; Hu, Z. Study of interaction between syringin and human serum albumin by multi-spectroscopic method and atomic force microscopy. J. Mol. Struct. 2010, 983, 133–140. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, Z.; Xing, Y.; Hou, C.; Zhou, Q.; Sun, Y.; Sun, Y.; Xu, H.; Gao, J. Mechanism of the interaction between benthiavalicarb-isopropyl and human serum albumin. Spectrosc. Lett. 2020, 53, 360–371. [Google Scholar] [CrossRef]
- Singha Roy, A.; Tripathy, D.R.; Ghosh, A.K.; Dasgupta, S. An alternate mode of binding of the polyphenol quercetin with serum albumins when complexed with Cu(II). J. Lumin. 2012, 132, 2943–2951. [Google Scholar] [CrossRef]
- Caruso, Í.P.; Vilegas, W.; Cristante de Oliveira, L.; Cornélio, M.L. Fluorescence spectroscopic and dynamics simulation studies on isoorientin binding with human serum albumin. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 228, 117738. [Google Scholar] [CrossRef]
- Jisha, V.; Arun, K.; Hariharan, M.; Ramaiah, D. Site–Selective Binding and Dual Mode Recognition of Serum Albumin by a Squaraine Dye. J. Am. Chem. Soc. 2006, 128, 6024–6025. [Google Scholar] [CrossRef]
- Zhao, M.-L.; Wang, W.; Nie, H.; Cao, S.-S.; Du, L.-F. In silico structure prediction and inhibition mechanism studies of AtHDA14 as revealed by homology modeling, docking, molecular dynamics simulation. Comput. Biol. Chem. 2018, 75, 120–130. [Google Scholar] [CrossRef]
- Xi, L.; Xiong, X.; Li, X.; Zhang, Q.; Yang, W.; Du, L. Insight into the structural stability of wild-type and histidine mutants in Pin1 by experimental and computational methods. Sci. Rep. 2019, 9, 8413. [Google Scholar] [CrossRef]
- Nasiri, R.; Bahrami, H.; Zahedi, M.; Moosavi-Movahedi, A.; Sattarahmady, N. A Theoretical Elucidation of Glucose Interaction with HSA’s Domains. J. Biomol. Struct. Dyn. 2010, 28, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Benvidi, A.; Rezaeinasab, M.; Gharaghani, S.; Abbasi, S. Monitoring the protective ability of thymoquinone mixture with p-cymene against bovine serum albumin (BSA) glycation: MCR-ALS analysis based on combined spectroscopic and electrochemical methods. Int. J. Biol. Macromol. 2018, 107, 2465–2474. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, S.; Preethy, R.; Saraswathi, N.T. Nordihydroguaiaretic acid prevents glycation induced structural alterations and aggregation of albumin. Int. J. Biol. Macromol. 2019, 122, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Bao, M.; Yang, C.; Chen, J.; Zhou, S.; Sun, R.; Wu, C.; Li, X.; Bao, J. Computer-aided identification of a novel pyruvate kinase M2 activator compound. Cell Prolif. 2018, 51, e12509. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Sang, F.; Qian, J.; Lyu, S.; Wang, Y.; Li, Q.; Du, L. Identification of novel potential PI3Kα inhibitors for cancer therapy. J. Biomol. Struct. Dyn. 2020, 39, 3721–3732. [Google Scholar] [CrossRef]
- Wang, Y.; Xi, L.; Yao, J.; Yang, J.; Du, L.-F. Role of pH in structural changes for Pin1 protein: An insight from molecular dynamics study. J. Mol. Model. 2014, 20, 2376. [Google Scholar] [CrossRef]
- Lu, K.; Wang, X.; Chen, Y.; Liang, D.; Luo, H.; Long, L.; Hu, Z.; Bao, J. Identification of two potential glycogen synthase kinase 3β inhibitors for the treatment of osteosarcoma. Acta Biochim. Et Biophys. Sin. 2018, 50, 456–464. [Google Scholar] [CrossRef]
- Zeng, L.; Ding, H.; Hu, X.; Zhang, G.; Gong, D. Galangin inhibits α-glucosidase activity and formation of non-enzymatic glycation products. Food Chem. 2019, 271, 70–79. [Google Scholar] [CrossRef]
- Ding, H.; Ni, M.; Zhang, G.; Liao, Y.; Hu, X.; Zhang, Y.; Gong, D. The inhibition of oleanolic acid on protein non-enzymatic glycation. LWT 2020, 125, 109253. [Google Scholar] [CrossRef]
- Awasthi, S.; Nambiappan Thangavel, S. Silybin, a flavonolignan from milk thistle seeds, restrains the early and advanced glycation end product modification of albumin. RSC Adv. 2015, 5, 87660–87666. [Google Scholar] [CrossRef]
- Anguizola, J.; Matsuda, R.; Barnaby, O.S.; Hoy, K.S.; Wa, C.; DeBolt, E.; Koke, M.; Hage, D.S. Review: Glycation of human serum albumin. Clin. Chim. Acta 2013, 425, 64–76. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.M.; Davidson, W.S.; Urbina, E.M.; Dolan, L.M.; Heink, A.; Zang, H.; Lu, L.J.; Shah, A.S. The Effects of Type 2 Diabetes on Lipoprotein Composition and Arterial Stiffness in Male Youth. Diabetes 2013, 62, 2958–2967. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.-M.; Ma, W.-Y.; Wu, C.-C.; Lu, K.-C. Glycated albumin in diabetic patients with chronic kidney disease. Clin. Chim. Acta 2012, 413, 1555–1561. [Google Scholar] [CrossRef] [PubMed]
- Michelis, R.; Kristal, B.; Zeitun, T.; Shapiro, G.; Fridman, Y.; Geron, R.; Sela, S. Albumin oxidation leads to neutrophil activation in vitro and inaccurate measurement of serum albumin in patients with diabetic nephropathy. Free Radic. Biol. Med. 2013, 60, 49–55. [Google Scholar] [CrossRef]
- Ahmad, S.; Siddiqui, Z.; Rehman, S.; Khan, M.; Khan, H.; Khanum, S.; Alouffi, S.; Saeed, M. A Glycation Angle to Look into the Diabetic Vasculopathy: Cause and Cure. Curr. Vasc. Pharmacol. 2017, 15, 352–364. [Google Scholar] [CrossRef]
- Fu, Y.; Jia, Y.; Sun, Y.; Liu, X.; Yi, J.; Cai, S. Dietary Flavonoids Alleviate Inflammation and Vascular Endothelial Barrier Dysfunction Induced by Advanced Glycation End Products In Vitro. Nutrients 2022, 14, 1026. [Google Scholar] [CrossRef]
- Zhang, C.; Guan, J.; Zhang, J.; Yang, J.; Wang, X.; Peng, X. Protective effects of three structurally similar polyphenolic compounds against oxidative damage and their binding properties to human serum albumin. Food Chem. 2021, 349, 129118. [Google Scholar] [CrossRef]
- Khan, M.S.; Qais, F.A.; Rehman, M.T.; Ismail, M.H.; Alokail, M.S.; Altwaijry, N.; Alafaleq, N.O.; AlAjmi, M.F.; Salem, N.; Alqhatani, R. Mechanistic inhibition of non-enzymatic glycation and aldose reductase activity by naringenin: Binding, enzyme kinetics and molecular docking analysis. Int. J. Biol. Macromol. 2020, 159, 87–97. [Google Scholar] [CrossRef]
- Qais, F.; Ahmad, I. Mechanism of non-enzymatic antiglycation action by coumarin: A biophysical study. New J. Chem. 2019, 43. [Google Scholar] [CrossRef]
- Awasthi, S.; Nambiappan Thangavel, S. Carbonyl scavenging and chemical chaperon like function of essential amino acids attenuates non-enzymatic glycation of albumin. RSC Adv. 2016, 6, 24557–24564. [Google Scholar] [CrossRef]
- Bitencourt-Ferreira, G.; Pintro, V.; De Azevedo, W., Jr. Docking with AutoDock4. Methods Mol. Biol. 2019, 2053, 125–148. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.; Shoichet, B. ZINC—A Free Database of Commercially Available Compounds for Virtual Screening. J. Chem. Inf. Model. 2005, 45, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Rauf, M.A. Ligand docking and binding site analysis with pymol and autodock/vina. Int. J. Basic Appl. Sci. 2015, 4, 168–177. [Google Scholar] [CrossRef]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef]
- Asgharzadeh, S.; Shareghi, B.; Farhadian, S.; Tirgir, F. Effect of free L-cysteine on the structure and function of α-chymotrypsin. J. Mol. Liq. 2019, 280, 79–86. [Google Scholar] [CrossRef]
- Hess, B.; Kutzer, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef]
- Wang, X.; Lu, K.; Luo, H.; Liang, D.; Long, X.; Yuan, Y.; Wu, C.; Bao, J. In silico identification of small molecules as novel LXR agonists for the treatment of cardiovascular disease and cancer. J. Mol. Model. 2018, 24, 57. [Google Scholar] [CrossRef]
- Li, J.; Zhou, N.; Luo, K.; Zhang, W.; Li, X.; Wu, C.; Bao, J. In Silico Discovery of Potential VEGFR-2 Inhibitors from Natural Derivatives for Anti-Angiogenesis Therapy. Int. J. Mol. Sci. 2014, 15, 15994–16011. [Google Scholar] [CrossRef]
- Xi, L.; Wang, Y.; He, Q.; Zhang, Q.; Du, L. Interaction between Pin1 and its natural product inhibitor epigallocatechin-3-gallate by spectroscopy and molecular dynamics simulations. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2016, 169, 134–143. [Google Scholar] [CrossRef]
- Wang, W.; Li, X.; Wang, Q.; Zhu, X.; Zhang, Q.; Du, L. The acidic pH-induced structural changes in apo-CP43 by spectral methodologies and molecular dynamics simulations. J. Mol. Struct. 2018, 1152, 177–188. [Google Scholar] [CrossRef]
- Vermaas, J.V.; Hardy, D.J.; Stone, J.E.; Tajkhorshid, E.; Kohlmeyer, A. TopoGromacs: Automated Topology Conversion from CHARMM to GROMACS within VMD. J. Chem. Inf. Model. 2016, 56, 1112–1116. [Google Scholar] [CrossRef] [PubMed]

| Parameters | 298 K | 308 K |
|---|---|---|
| Ksv (105 mol/L) | 1.21 ± 0.01 | 0.41 ± 0.01 |
| Kq (1013 L/mol/s) | 1.21 ± 0.01 | 0.41 ± 0.01 |
| R2a | 0.989 | 0.996 |
| n | 1.50 ± 0.04 | 1.35 ± 0.12 |
| Ka (104 mol/L) | 3.79 ± 0.18 | 1.93 ± 0.27 |
| R2b | 0.999 | 0.987 |
| ΔG (kJ/mol) | −26.12 ± 0.01 | −24.45 ± 0.01 |
| ΔH (kJ/mol) | −51.53 ± 0.58 | −51.53 ± 0.58 |
| ΔS (J/mol/K) | −85.25 ± 2.38 | −87.91 ± 2.36 |
| System | K′a (104 mol/L) | K′a/Ka (%) | R2 |
|---|---|---|---|
| BSA-Myr | 3.79 ± 0.18 | - | 0.999 |
| [BSA + Met]-Myr | 1.79 ± 0.22 | 45.00 ± 4.12 | 0.994 |
| [BSA + War]-Myr | 0.98 ± 0.15 | 25.76 ± 3.06 | 0.984 |
| [BSA + Ibu]-Myr | 3.38 ± 0.33 | 89.23 ± 4.81 | 0.994 |
| Different AGEs | Excitation Wavelength (nm) | Emission Wavelength (nm) |
|---|---|---|
| Total AGEs | 350 | 440 |
| Vesperlysine | 350 | 405 |
| Crossline | 380 | 440 |
| Argpyrimidine | 320 | 380 |
| Pentosidine | 335 | 385 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, L.; Liu, Y.; Xu, J.; Li, J.; Cheng, G.; Cai, J.; Dang, J.; Yu, M.; Wang, W.; Duan, W.; et al. Inhibitory Effects of Myriocin on Non-Enzymatic Glycation of Bovine Serum Albumin. Molecules 2022, 27, 6995. https://doi.org/10.3390/molecules27206995
He L, Liu Y, Xu J, Li J, Cheng G, Cai J, Dang J, Yu M, Wang W, Duan W, et al. Inhibitory Effects of Myriocin on Non-Enzymatic Glycation of Bovine Serum Albumin. Molecules. 2022; 27(20):6995. https://doi.org/10.3390/molecules27206995
Chicago/Turabian StyleHe, Libo, Yang Liu, Junling Xu, Jingjing Li, Guohua Cheng, Jiaxiu Cai, Jinye Dang, Meng Yu, Weiyan Wang, Wei Duan, and et al. 2022. "Inhibitory Effects of Myriocin on Non-Enzymatic Glycation of Bovine Serum Albumin" Molecules 27, no. 20: 6995. https://doi.org/10.3390/molecules27206995
APA StyleHe, L., Liu, Y., Xu, J., Li, J., Cheng, G., Cai, J., Dang, J., Yu, M., Wang, W., Duan, W., & Liu, K. (2022). Inhibitory Effects of Myriocin on Non-Enzymatic Glycation of Bovine Serum Albumin. Molecules, 27(20), 6995. https://doi.org/10.3390/molecules27206995







