Abstract
Cervical cancer is a common gynecological malignancy afflicting women all over the world. Ginsenoside Rh2 (GRh2), especially 20(S)-GRh2, is a biologically active component in the natural plant ginseng, which can exhibit anticancer effects. Here, we aimed to investigate the effect of 20(S)-GRh2 on cervical cancer and elucidate the underlying mechanism through RNA-seq. In this study, the CCK-8 assay showed that 20(S)-GRh2 inhibited HeLa cell viability in a time- and dose-dependent manner. Caspase 3 activity and Annexin V staining results showed that 20(S)-GRh2 induced apoptosis of HeLa cells. Gene function enrichment analysis revealed that the biological process gene ontology (GO) terms were associated with the apoptotic signaling pathway. Biological process GO terms’ similarity network indicated that apoptosis might be from endoplasmic reticulum stress (ERs). Kyoto Encyclopedia of Genes and Genomes enrichment analysis revealed that 20(S)-GRh2 primarily modulates apoptosis pathway genes. Combined protein–protein interaction network, hub gene screening, and qPCR validation data showed that ERs-related genes (ATF4 and DDIT3) and the downstream apoptotic genes (JUN, FOS, BBC3, and PMAIP1) were potential novel targets of 20(S)-GRh2-inducing cervical cancer cell apoptosis. Differential transcript usage analysis indicated that DDIT3 is also a differential transcript and its usage of the isoform (ENST00000552740.5) was reduced by 20(S)-GRh2. Molecular docking suggested that 20(S)-GRh2 binds to the targets (ATF4, DDIT3, JUN, FOS, BBC3, and PMAIP1) with high affinity. In conclusion, our findings indicated that 20(S)-GRh2 might promote ERs-related apoptosis of cervical cancer cells by regulating the DDIT3-based targets’ signal pathway. The role of 20(S)-GRh2 at the transcriptome level provides novel targets and evidence for the treatment of cervical cancer.
1. Introduction
Cervical cancer (CC) is a common gynecological malignant tumor that ranks fourth in morbidity and mortality among all cancers in women worldwide [1]. Surgery is currently the best treatment option for patients at an early stage of CC [2]. However, despite treatment with surgical resection with adjuvant radiotherapy and chemotherapy, the prognosis of patients with advanced CC is still poor [3]. The side effects of traditional chemotherapeutic drugs and the acquired drug resistance of tumors further support the need to identify new efficient antitumor drugs with low toxicity [4].
Natural products have attracted much attention as cancer treatments because of their strong potency and low toxicity, and several products have shown efficacy in cancer control and treatment [5,6]. Ginsenoside Rh2 (GRh2) is the natural extract of Panax ginseng C.A. Meyer (ginseng) and exhibits biological activities in inhibiting cell proliferation and inducing cell apoptosis in oral cancer, colon cancer, and prostate cancer [7,8,9]. GRh2, a major deglycosylated metabolite of ginsenoside Rg3, Rb1, Rb2, and Rc, exhibits stronger tumor-suppressive effects than the parent saponin, along with good biosafety and low molecular weight [10,11]. It is noteworthy that the molecular structure of GRh2 has R-type and S-type, and the 20(S)-GRh2 configuration plays the main anticancer effect [12]. Thus, application of 20(S)-GRh2 may be a promising anticancer therapeutic strategy. However, the underlying targets of 20(S)-GRh2 anticervical cancer remained elusive.
Herein, we performed transcriptome profiling (RNA-Seq) to seek potential targets in 20(S)-GRh2-treated HeLa cells, with the aim of elucidating the underlying mechanisms of 20(S)-GRh2.
2. Results
2.1. 20(S)-GRh2 Induces HeLa Cell Apoptosis
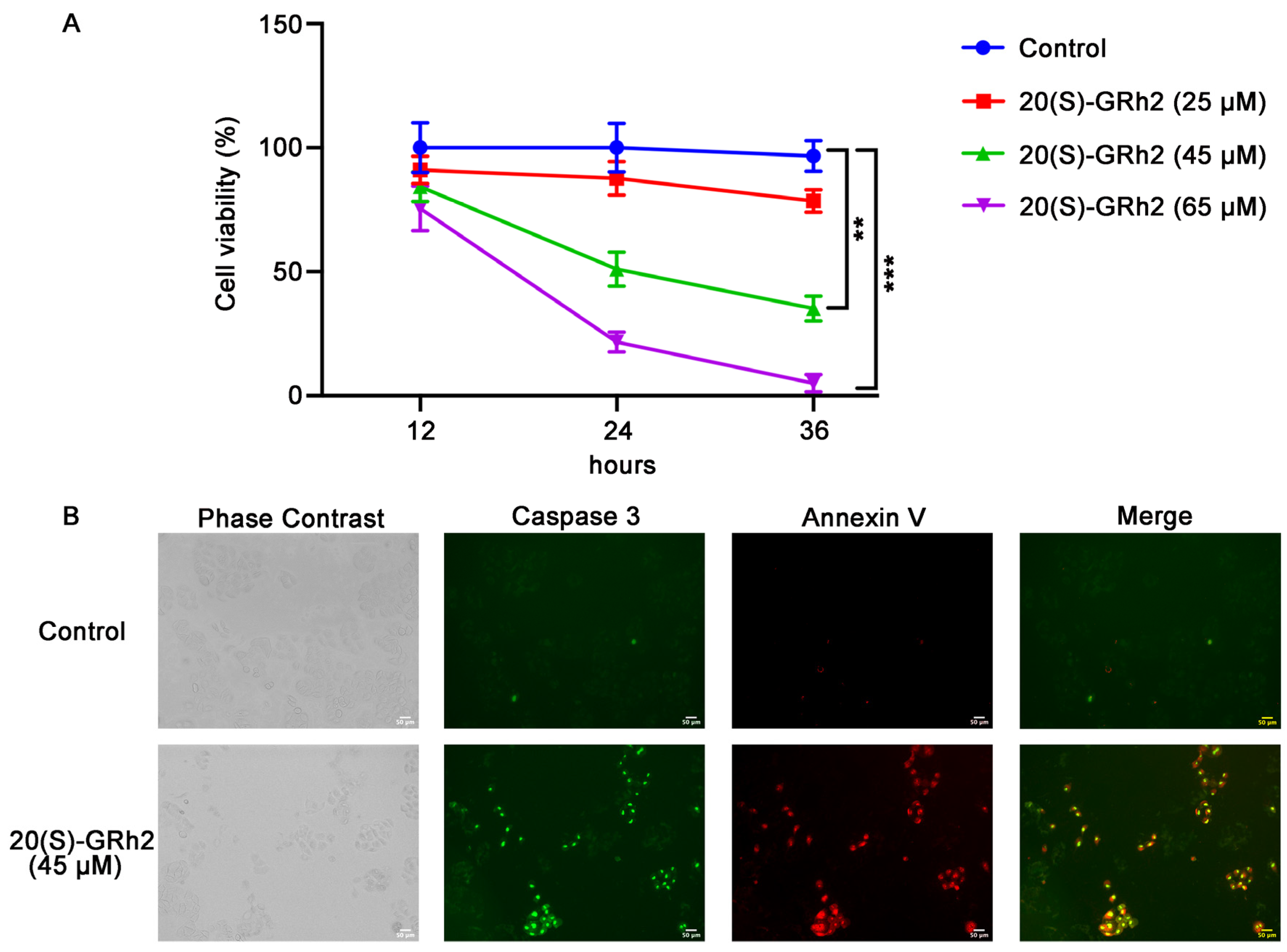
20(S)-GRh2 treatment resulted in marked inhibition (IC50 of approximately 45 μM at 24 h) of HeLa cell proliferation in a dose- and time-dependent manner (Figure 1A). To avoid an excessive response of cells to drug toxicity, we treated with 45 μM 20(S)-GRh2 continuously for 24 h in subsequent experiments.
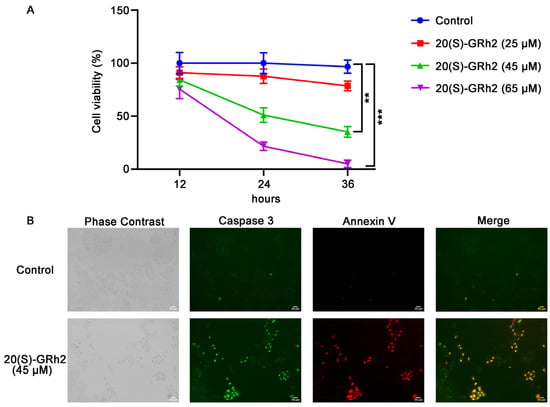
Figure 1.
20(S)-GRh2 inhibited HeLa cell proliferation and induced HeLa cell apoptosis. (A) HeLa cells were treated with control (0.1% v/v DMSO) or 25, 45, 65 µM 20(S)-GRh2 at different time intervals (12 h, 24 h, 36 h), and CCK-8 assay was used to detect cell viability. Data are presented as mean ± standard deviation of three independent experiments (two-way ANOVA, ** p < 0.01, *** p < 0.001 vs. control). (B) Images of HeLa cells treated with 45 µM 20(S)-GRh2 or control (0.1% v/v DMSO) and examined using the Live Cell Caspase-3 Activity and Annexin V Apoptosis Detection reagent. Green fluorescence represents caspase 3 activity, red fluorescence represents apoptotic cells, and phase contrast shows the total cells. The images are representative from three independent experiments. PS, phosphatidylserine; 20(S)-GRh2, 20(S)-ginsenoside Rh2.
To explore whether 20(S)-GRh2-mediated inhibition of proliferation in HeLa cells involved apoptosis, cells were treated with 20(S)-GRh2 and examined using caspase 3 substrate and Annexin V-mCherry. As shown in Figure 1B, a significant increase in apoptosis, determined by increased caspase 3 activity and phosphatidylserine (PS) eversion, was observed in HeLa cells treated with 20(S)-GRh2 compared with the control group. These results demonstrated that 20(S)-GRh2 inhibits cell proliferation of HeLa cells via inducing apoptosis.
2.2. Identification of Differentially Expressed Genes (DEGs) in 20(S)-GRh2-Treated HeLa Cells
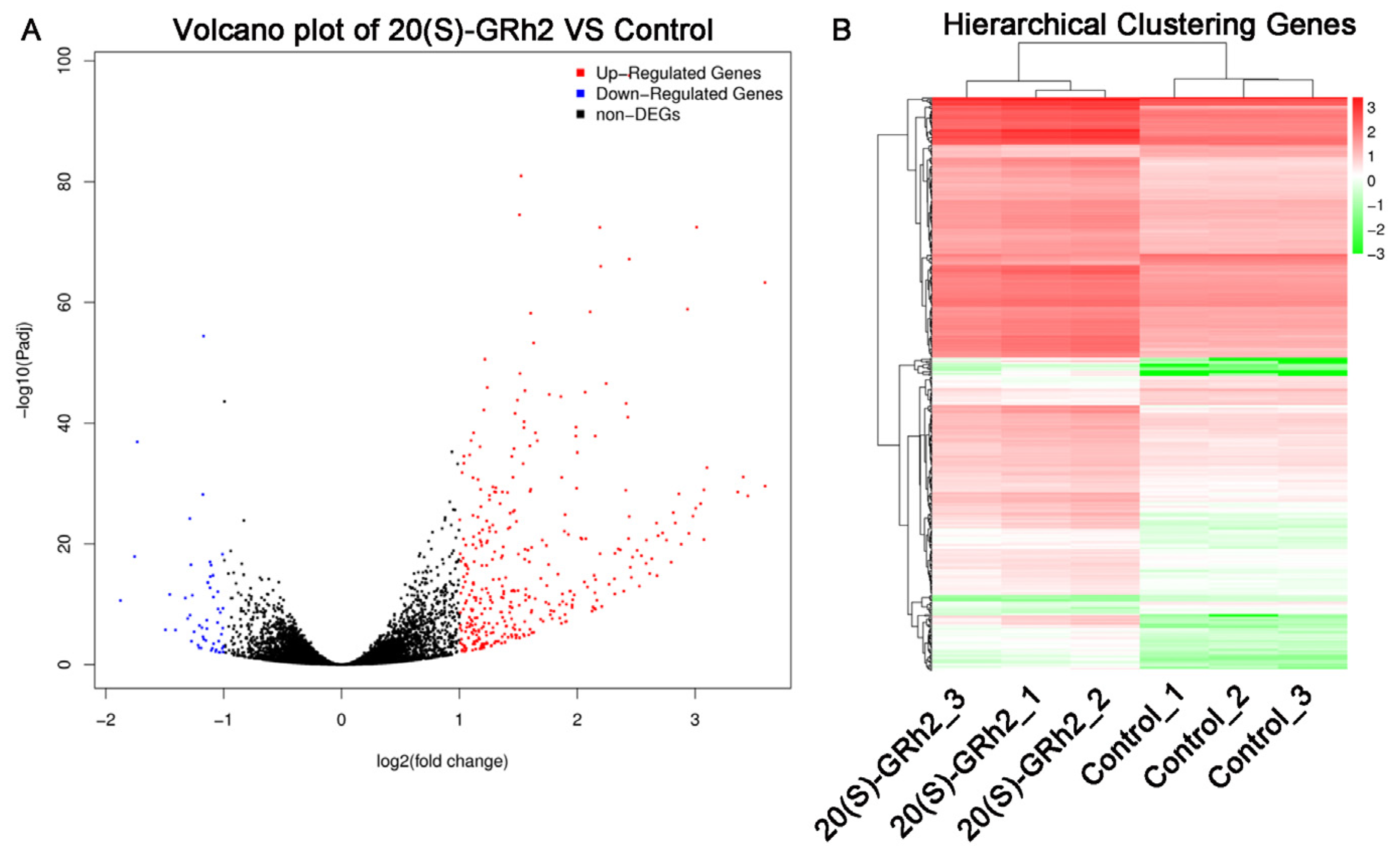
To clarify the potential mechanism by which 20(S)-GRh2 induces HeLa cell apoptosis, we explored DEGs by conducting RNA sequencing on HeLa cells treated with 45 μM 20(S)-GRh2 and control. We screened 361 upregulated DEGs and 53 downregulated DEGs with the criteria of |log2(Fold Change)| > 1 and p < 0.05 (Figure 2A). Cluster analysis of the DEGs, as shown in Figure 2B, demonstrated that biological replicates in each group have great homogeneity.
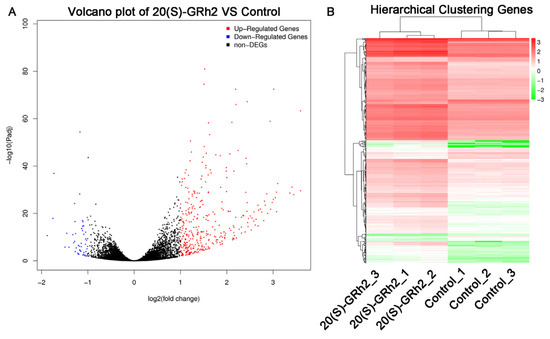
Figure 2.
Identification of DEGs in 20(S)-GRh2-treated HeLa cells. (A) Volcano plots of DEGs are displayed using |log 2(Fold Change)| > 1 and adjusted p-value < 0.05. Red and blue dots mark upregulated genes and downregulated genes, respectively. (B) Cluster heatmap of DEGs with |log 2(Fold Change)| > 1 and adjusted p-value < 0.05. Red, upregulated DEGs; green, downregulated DEGs; DEGs, differentially expressed genes; 20(S)-GRh2, 20(S)-ginsenoside Rh2.
2.3. Enrichment Analysis of DEGs
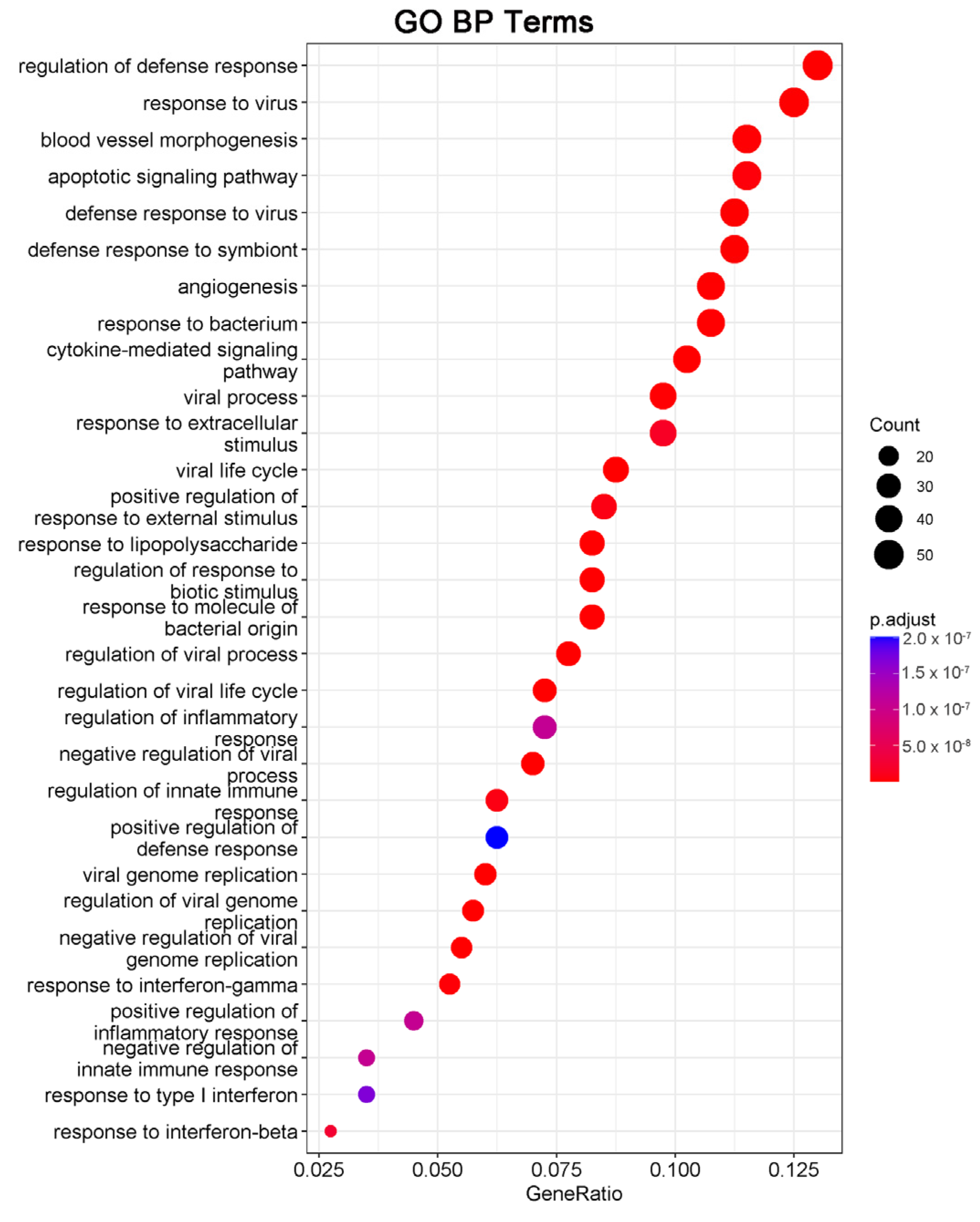
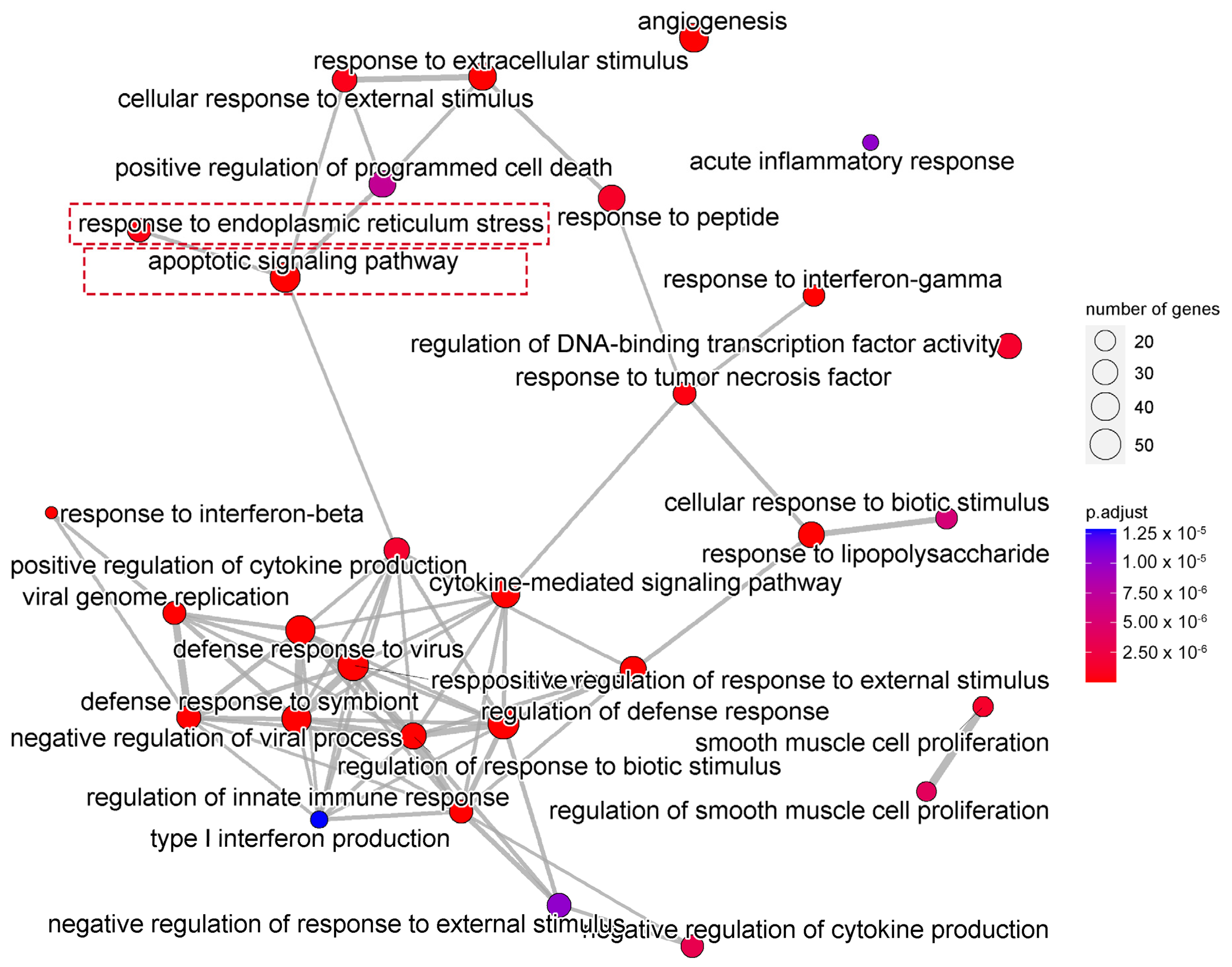
We next performed gene ontology (GO) analysis on the DEGs of 20(S)-GRh2-treated HeLa cells to identify the terms related to biological processes (BP). The top 30 GO BP terms are shown in a bubble plot in Figure 3. The DEGs were mainly enriched in the regulation of defense response, response to virus, blood vessel morphogenesis, and apoptotic signaling pathway. Figure 4 displays the network of the similarity between BP enrichment terms and shows that the apoptotic signaling pathway was closely related to response to endoplasmic reticulum stress (ERs).
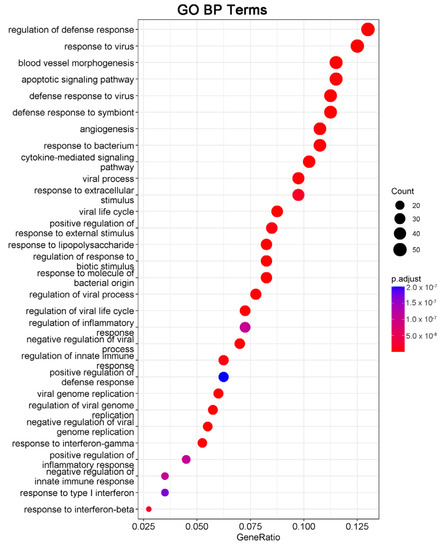
Figure 3.
DEGs in 20(S)-GRh2-treated HeLa cells were analyzed by GO functional enrichment, and the top 30 functionally enriched terms annotated to BP are shown. GO, gene ontology; BP, biological processes; DEGs, differentially expressed genes; 20(S)-GRh2, 20(S)-ginsenoside Rh2.
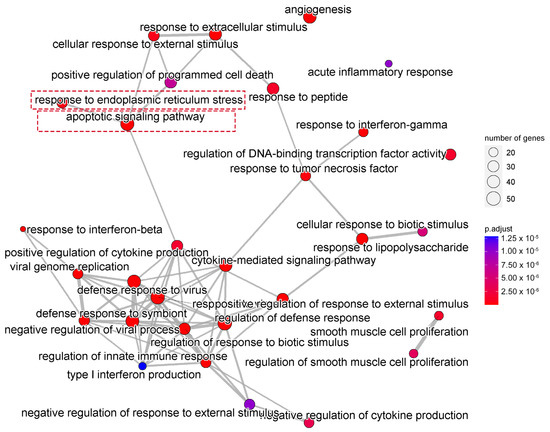
Figure 4.
Network showing the overlapping gene sets among the GO-BP terms after removing redundant genes. GO, gene ontology; BP, biological processes.
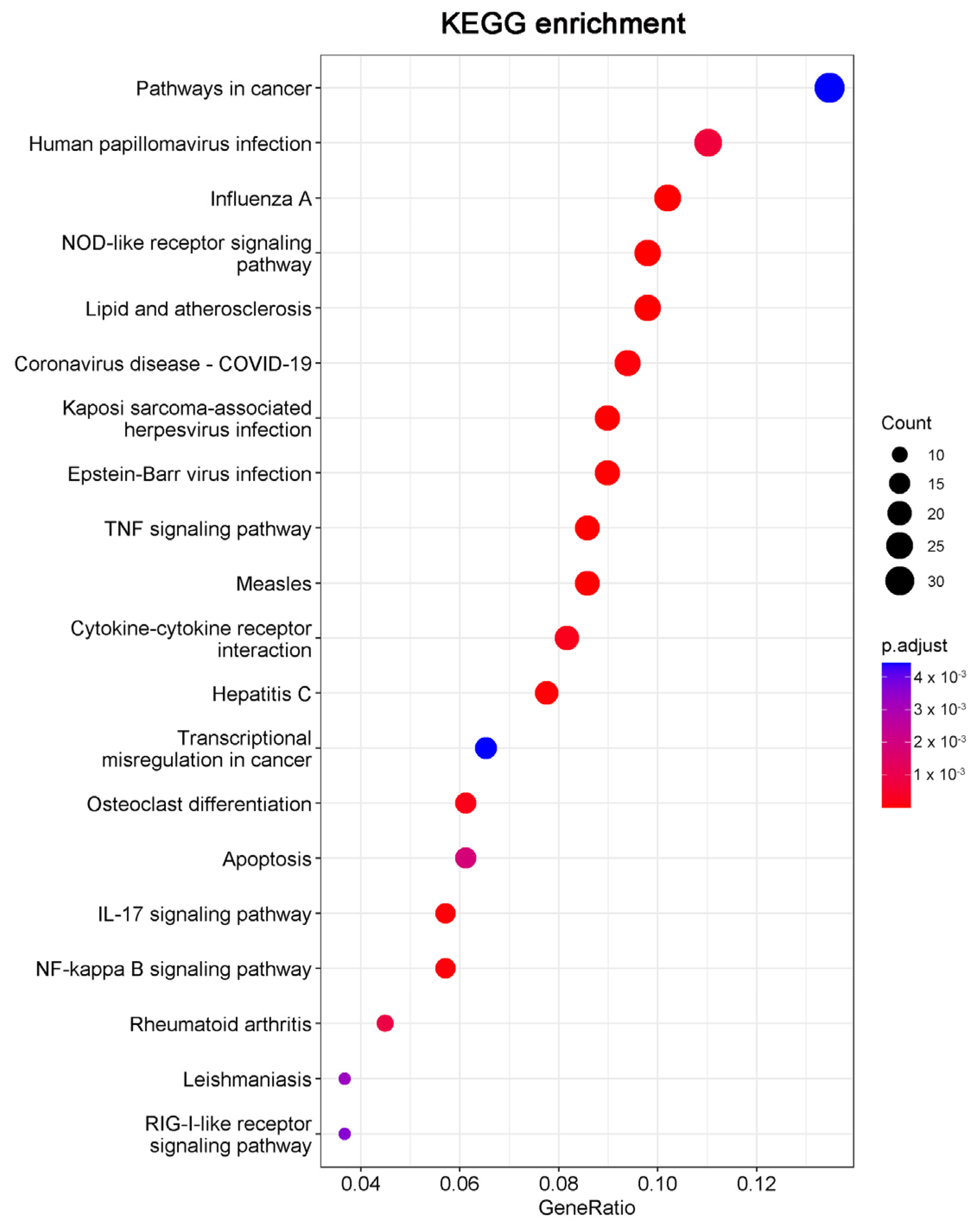
Consistent with the GO results, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis also revealed DEGs were relevant with apoptotic pathways (Figure 5). The 15 DEGs enriched in the apoptosis pathway are listed in Table 1, including 14 significantly upregulated genes (FOS, JUN, ATF4, DDIT3, PMAIP1, BBC3, NFKBIA, TNFSF10, FAS, ERN1, BIRC3, TRAF1, GADD45A, and CTSS genes) and one significantly downregulated gene (TUBA4A gene). These results indicated that 20(S)-GRh2 may exhibit its antitumor activity by regulating these apoptosis-related genes.
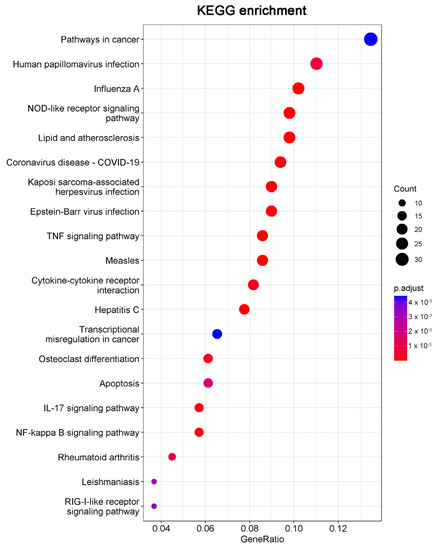
Figure 5.
The top 20 pathways in the KEGG pathway enrichment analysis of the DEGs in 20(S)-GRh2-treated HeLa cells are shown in bubble plots. DEGs, differentially expressed genes; KEGG, Kyoto Encyclopedia of Genes and Genomes; 20(S)-GRh2, 20(S)-ginsenoside Rh2.

Table 1.
DEGs in apoptosis pathways.
2.4. Protein–Protein Interaction (PPI) Network Construction and Hub Gene Screening
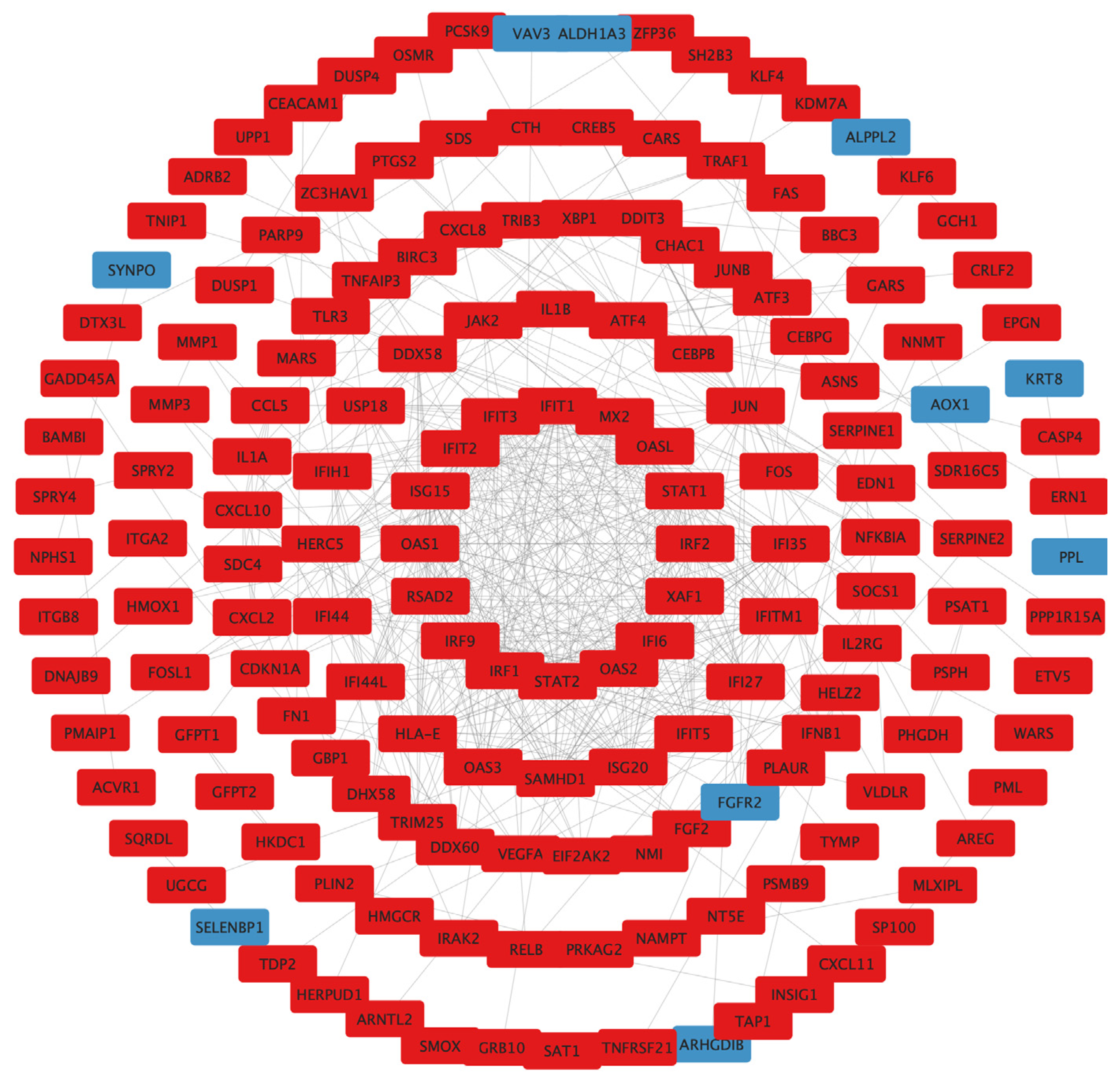
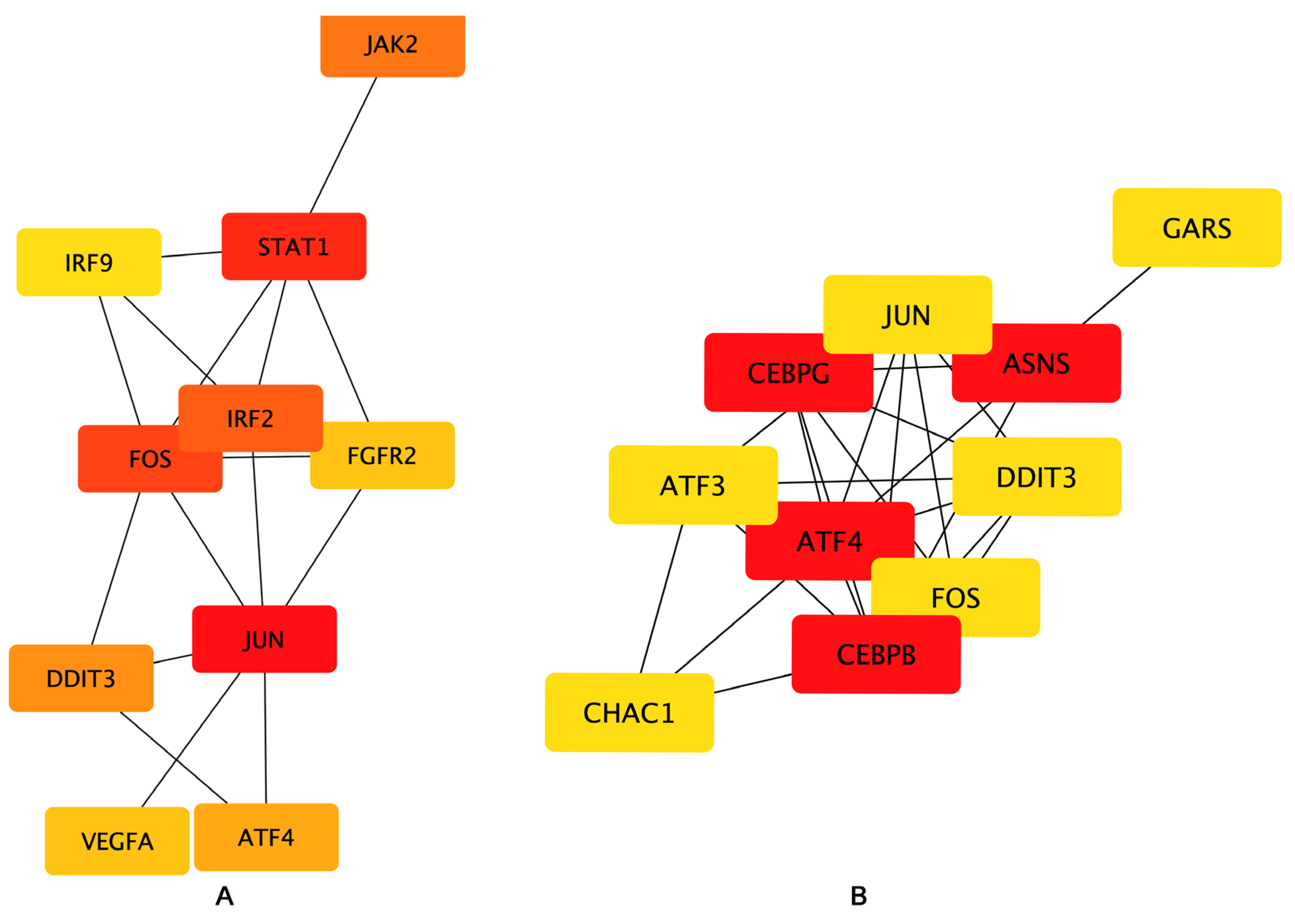
To examine the interaction among the DEGs, a PPI network was constructed using the STRING database, and the results were visualized using Cytoscape software (Figure 6), followed by hub genes-screening in Figure 7. Among the top 10 hub genes, 4 genes (JUN, FOS, ATF4, and DDIT3 genes) belonged to the apoptosis pathway.
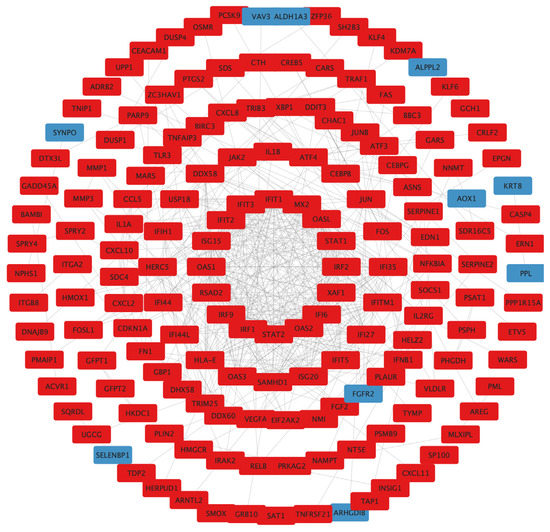
Figure 6.
PPI network construction. Circle size reflects importance in the network (a smaller circle indicates more importance in the network). Red mark, upregulated DEGs; blue mark, downregulated DEGs; PPI, protein–protein interaction; DEGs, differentially expressed genes.
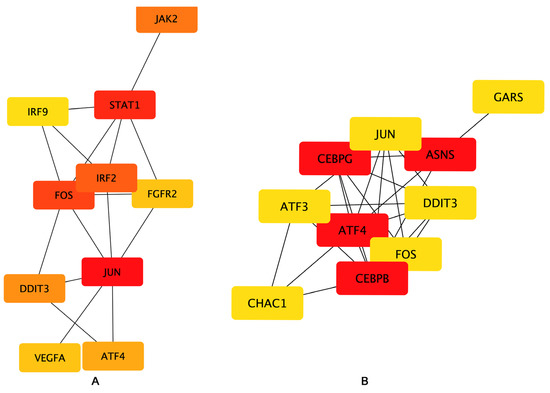
Figure 7.
Hub genes network. The top 10 hub genes of the (A) Bottleneck and (B) EcCentricity algorithm were screened by the CytoHubba plugin. Higher rankings are indicated by deeper red colors.
2.5. qPCR Verification
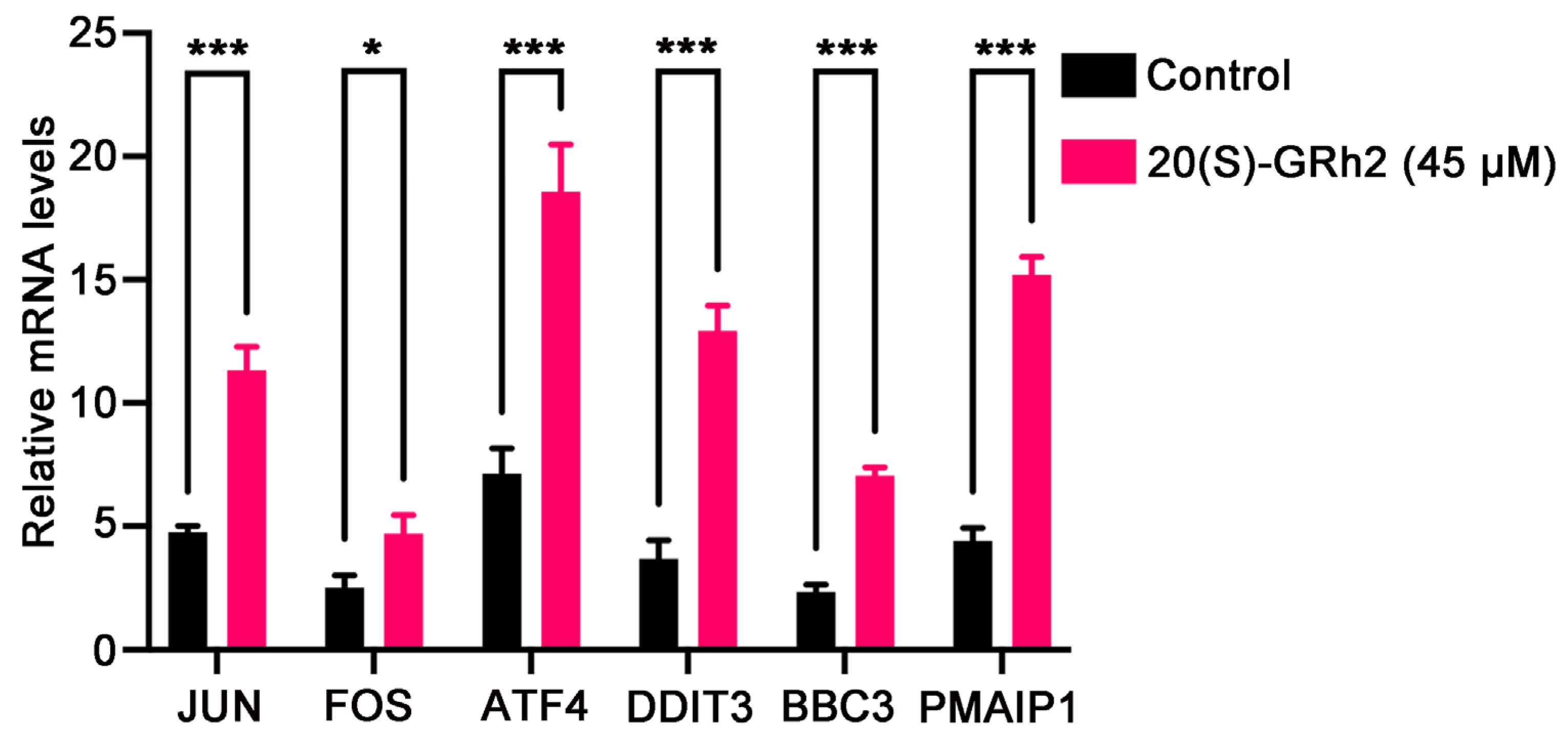
We next examined the mRNA expression of known genes deeply involved in apoptosis including JUN, FOS, ATF4, DDIT3, BBC3, and PMAIP1 in 20(S)-GRh2-treated HeLa cells by qPCR (Figure 8). The results were in line with the RNA-Seq data, in which these apoptosis-relevant genes were upregulated after 20(S)-GRh2 treatment and which indicated that the RNA sequencing data were reliable.
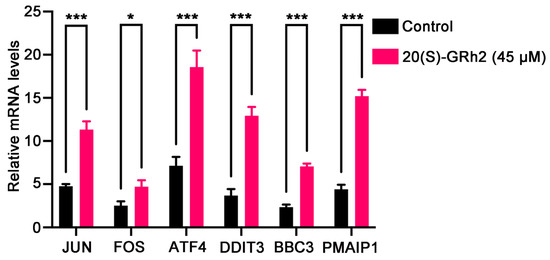
Figure 8.
Validation of selected DEGs by qPCR. The mRNA expression levels of the DEGs were quantified in control and 45 μM 20(S)-GRh2 groups by qPCR. GAPDH mRNA was used as loading control. Data presented as mean ± standard deviation of three independent experiments (two-tailed Student’s t-test). * p < 0.05; *** p < 0.001 vs. control. DEGs, differentially expressed genes; 20(S)-GRh2, 20(S)-ginsenoside Rh2.
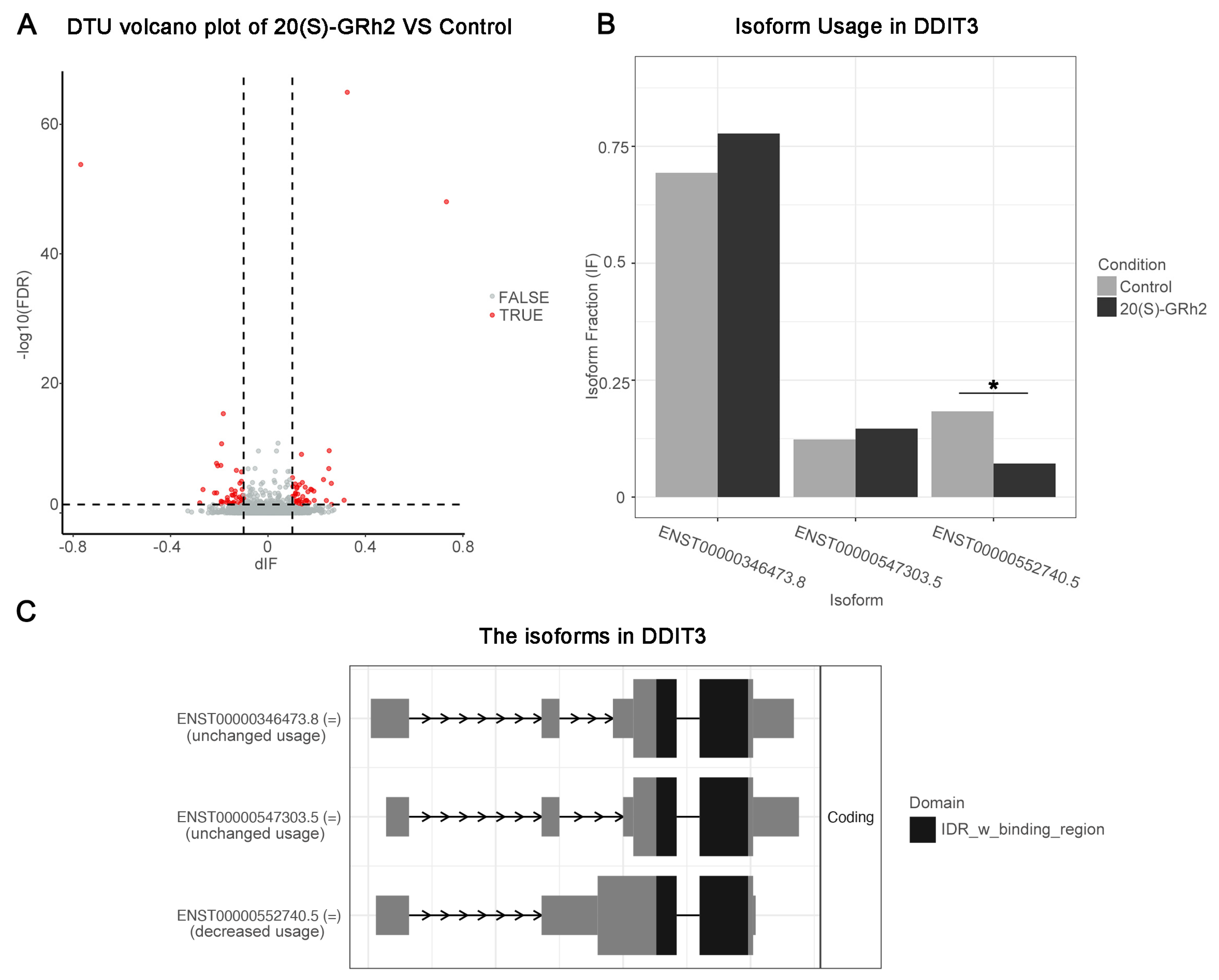
2.6. Differential Transcript Usage (DTU) Analysis
Significant gene expression modifications may occur at the transcriptional level and are ignored in classical DEG analysis [13]. To overcome this limitation, we used DTU analysis to characterize isomeric differences, which in turn measure the relative contribution of transcripts to overall gene expression. The transcript abundance was quantified using Kallisto, and 86 differential transcript usage events were identified by the IsoformSwitch R package (Figure 9A, Table S1). Notably, the usage of short isoform (ENST00000552740.5) in DDIT3, also as a differential transcript, was significantly reduced after 20(S)-GRh2 treatment (Figure 9B). The domain result in isoform landscape of DDIT3 showed that all three isoforms have an IDR _w_ binding region domain, and the domain of the short isoform is closer to the coding end. In addition, the short isoform has a shorter exon length near the end of the coding region (Figure 9C).
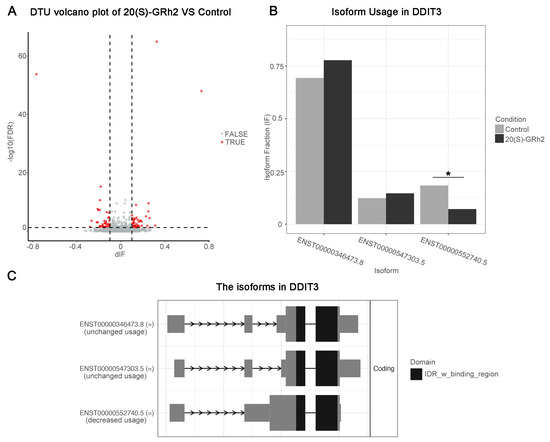
Figure 9.
DTU analysis. (A) Volcano plots showing genes with DTU (red dots mark DTU; differential isoform fraction (dIF) > 0.05 and FDR < 0.05) in the control and G-Rh2-treated groups. (B) Quantitation of the isoform fraction in control and 20(S)-GRh2 treated groups. (C) Isoform landscape of all 3 isoforms of DDIT3 in 20(S)-GRh2 treatment group compared with control. * p < 0.05. 20(S)-GRh2, 20(S)-ginsenoside Rh2.
2.7. Molecular Docking
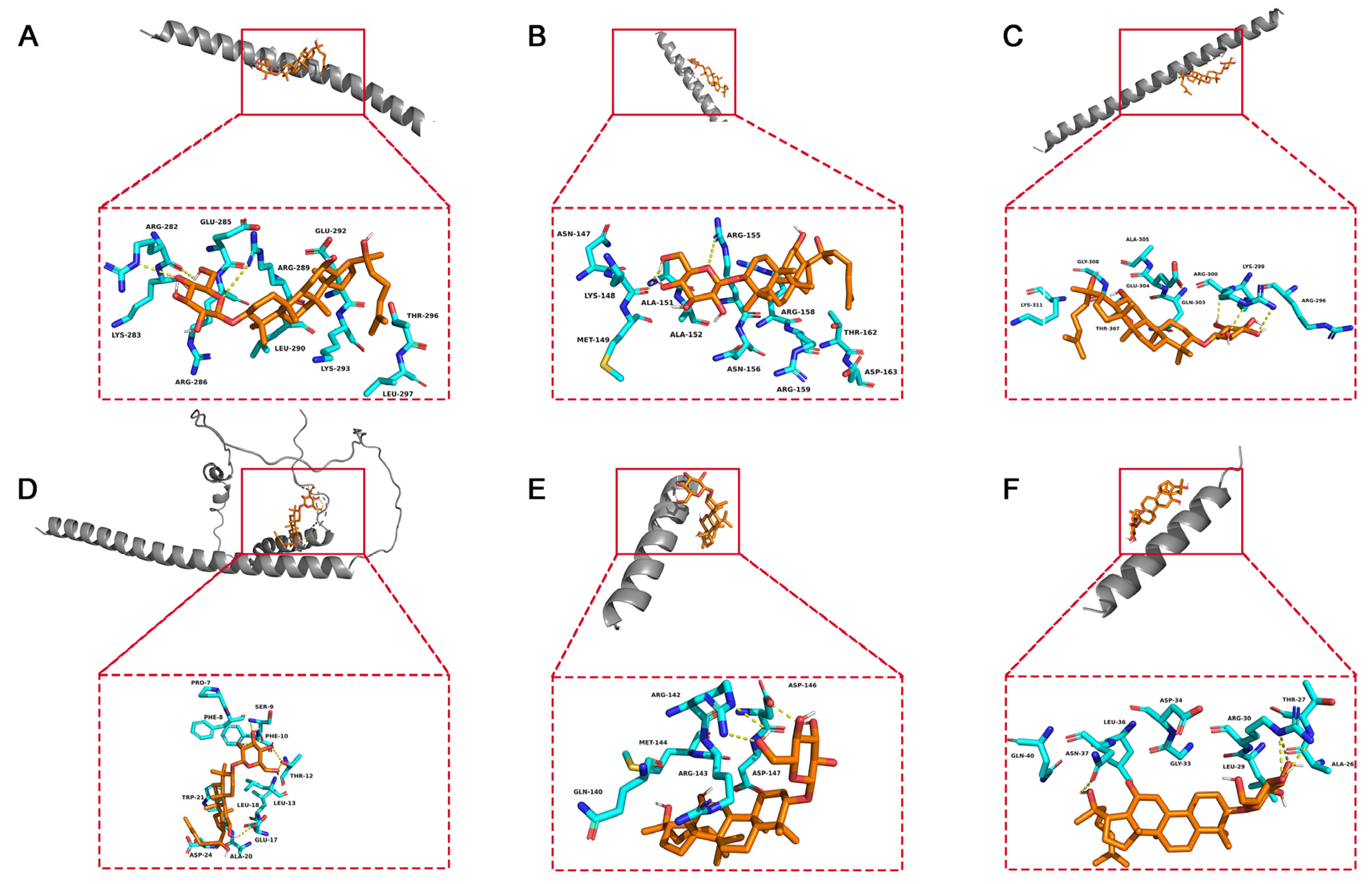
To elucidate the binding mode of 20(S)-GRh2 for their targets (JUN, FOS, ATF4, DDIT3, BBC3, and PMAIP1), molecular docking analysis was performed. The results showed that 20(S)-GRh2 is bound to its residues of targets primarily through hydrogen-bonding interactions and weak interactions (Figure 10 and Table 2). Furthermore, the active sites of each target were occupied successfully by G-Rh2. The binding energy for 20(S)-GRh2-target complexes are lower than −5.0 kcal/mol, indicating highly stable binding.
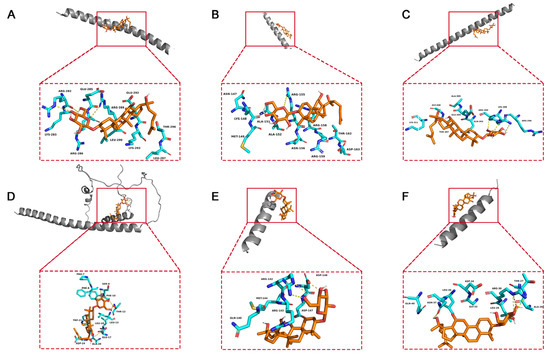
Figure 10.
Molecular docking models of G-Rh2 and (A) JUN, (B) FOS, (C) ATF4, (D) DDIT3, (E) BBC3, and (F) PMAIP1. 20(S)-GRh2, 20(S)-ginsenoside Rh2.

Table 2.
Molecular docking results.
3. Discussion
In this study, a HeLa cell line was used to evaluate the effect of 20(S)-GRh2 antihuman cervical cancer in vitro. Our results showed that 20(S)-GRh2 upregulated ATF4, DDIT3, JUN, FOS, BBC3, and PMAIP1 gene expression, as well as significantly inhibited the isoform (ENST00000552740.5) usage of the transcript DDIT3 to induce endoplasmic reticulum stress-related apoptosis in HeLa cells. Our research provided novel evidence for the mechanism of 20(S)-GRh2 anticervical cancer.
In accordance with previous results [14], we found that 20(S)-GRh2 inhibited the cell viability of HeLa in a dose–time parallel dependent manner. Because rapid proliferation and apoptosis escape are important hallmarks of tumor cells [15], we next mainly focused on the roles and targets of 20(S)-GRh2 in apoptotic events. We found that 20(S)-GRh2 induced apoptosis in HeLa cells with increased caspase 3 activity and phosphatidylserine eversion, which is consistent with its induction of apoptotic events in multiple cancers [16,17].
To understand the molecular mechanism of 20(S)-GRh2 in cervical cancer, we collected HeLa cells administrated with or without 20(S)-GRh2 for RNA-seq and found that upregulated genes after 20(S)-GRh2 treatment were functionally annotated with apoptosis pathways. Additionally, the apoptosis might be mainly related with response to endoplasmic reticulum stress (ERs). In addition, combined with PPI and hub gene network analysis, we found that the upregulation of ATF4, DDIT3, JUN, FOS, BBC3, and PMAIP1 gene expression is closely related to apoptosis. Given that sustained activation of ERs can trigger downstream apoptotic signaling pathways, targeting ERs-related genes has emerged as a potential strategy for cancer therapy [18,19]. Indeed, ATF4 acts as a pivotal mediator of ERs-induced apoptosis, and its translocation into the nucleus promotes ER protein folding and adaptation to stress by inducing the expression of UPR target genes such as the DDIT3 gene [20,21,22]. A similar result was also reported in lung cancer that showed that 20(S)-GRh2 triggers ER stress-related ATF4/DDIT3-induced apoptosis [23]. Although, DDIT3 is not expressed or weakly expressed under normal physiology but rapidly increases after severe or long-term ERs induction [24]. Induction of DDIT3 is strongly associated with the initiation of apoptosis associated with ERs, and silencing DDIT3 expression protects cells from apoptosis in chronic ERs [25]. In addition, DDIT3 transcriptionally regulates major apoptosis mediators of the Bcl-2 protein family, such as BBC3 and PMAIP1 [26,27]. BBC3 and PMAIP1 attach to the mitochondrial membrane through inhibiting the antiapoptotic Bcl-2 family members and activate proapoptotic Bax/Bak to enhance the permeability of the mitochondrial membrane, thereby inducing the apoptotic mitochondrial pathway [28,29]. A previous study stated that ATF4 is an important regulator of PMAIP1 transcription and promotes apoptosis in HeLa cells [30]. A recent report revealed that DDIT3 and the JUN/FOS heterodimer (AP-1 complex) jointly mediate BBC3-induced apoptosis, especially the physical association with JUN [31,32,33]. It implies that ERs-related genes (ATF4 and DDIT3) and downstream apoptosis genes (JUN, FOS, BBC3, and PMAIP1) are the potential targets of 20(S)-GRh2 and might be interacting with each other to be involved in the apoptosis induction in cervical cancer cells.
In addition, DTU analysis was performed to comprehensively analyze RNA-seq datasets at the level of transcripts. Importantly, DDIT3 is also as a differential transcript after 20(S)-GRh2 treatment, and its three isoforms all have the same IDR (intrinsically disordered regions)_w _binding region domain at the N-terminus. Recent studies have shown that IDR induces protein liquid–liquid phase transition and is involved in oncogene activation and carcinogenesis [34,35]. Furthermore, the IDR_w_binding region domain of the differential transcript isoform ENST00000552740.5 in DDIT3 exposed to 20(S)-GRh2 is closer to the end of the coding region; however, whether this phenotype is directly linked to the cervical cancer apoptosis induced by 20(S)-GRh2 remains to be investigated. A recent study showed that the development of cancer is accompanied by aberrant changes in many splicing events, and its exons are biased toward shorter lengths [36]. Notably, we found that ENST00000552740.5 had a shorter exon length near the end of the coding region and its usage was significantly reduced after 20(S)-GRh2 exposure. These results suggested that modulation of the DDIT3 isoform switch by 20(S)-GRh2 treatment may be a potential therapeutic strategy for anticervical cancer.
Considering the established association between apoptosis-related targets and 20(S)-GRh2, we also investigated the binding activity between the two using molecular docking. It has been reported that hydrogen bonds and weak interactions are the major contributions to strengthening the drug–target docking state [37,38]. In this study, G-Rh2 exhibited high-affinity activity with targets (ATF4, DDIT3, JUN, FOS, BBC3, and PMAIP1), binding to the residues of the targets through hydrogen bonds and weak interactions. These results further supported that apoptosis in 20(S)-GRh2-exposed HeLa cells is induced by ER stress-related genes ATF4 and DDIT3 and downstream apoptosis genes JUN, FOS, BBC3, and PMAIP1.
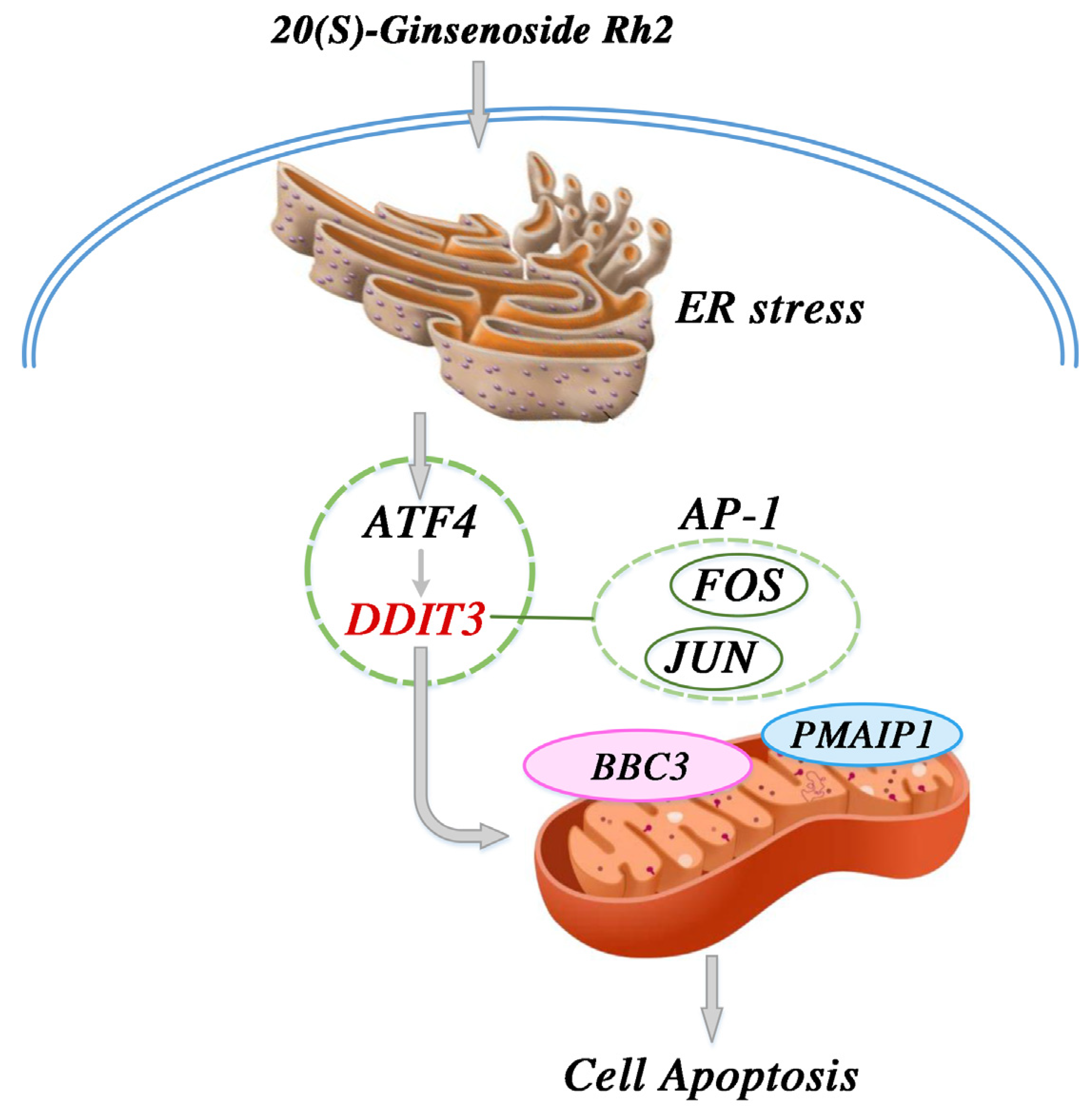
From our results and the above findings, we propose a potential signaling pathway model to help in understanding the mechanism of 20(S)-GRh2-induced apoptosis in HeLa cells. We suggest that increased DDIT3 is the consequence of the induction of ERs and occurs following activation of ATF4 after 20(S)-GRh2 treatment. The activation of DDIT3 is also accompanied with the activation of BBC3 and PMAIP1. Moreover, we proposed that DDIT3 collaborates with JUN and FOS in the AP-1 complex to mediate BBC3-induced apoptosis. In summary, JUN, FOS, BBC3, and PMAIP1 may be key downstream targets of 20(S)-GRh2-induced HeLa apoptosis, as all are regulated by an ERs-mediated cascade of ATF4 and DDIT3 (summarized in Figure 11). Notably, the proposed signaling pathway model is still a hypothesis, and more research is required to determine the specific mechanism of the 20(S)-GRh2-related apoptosis pathway.

Figure 11.
Proposed signal pathway for the anticervical cancer mechanism of 20(S)-GRh2. 20(S)-GRh2, 20(S)-ginsenoside Rh2.
4. Materials and Methods
4.1. Cell Lines
HeLa cells (CC cell line) were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in high glucose DMEM (Hyclone, South Logan, UT, USA) with 10% fetal bovine serum (FBS) (Hyclone) and 1% penicillin-streptomycin (100 U/mL) (Hyclone) in a humidified incubator (Thermo Scientific, Waltham, MA, USA) at 37 °C in 5% CO2.
4.2. Cell Viability Assay
HeLa cells were plated at 2 × 104 cells/well in a six-well plate and treated with 20(S)-GRh2 (25, 45, and 65 μM, Must Bio-Technology Co., Ltd., Chengdu, China) for 12, 24, and 36 h. Cell viability was conducted using CCK-8 kits (Boster, Wuhan, China) on an Infinite M200 PRO plate reader (Tecan, Männedorf, Switzerland) at 450 nm. The 50% inhibitory concentration (IC50) at 24 h was calculated according to the values of cell viability [39].
4.3. Cell Apoptosis Analysis
The Live Cell Caspase-3 Activity and Annexin V Apoptosis Detection Kit (Beyotime, Shanghai, China) were used to evaluate HeLa cell apoptosis. Briefly, HeLa cells were cultured in a 12-well plate for 24 h at 37 °C and then treated with 20(S)-GRh2 (45 μM) for 24 h. HeLa cells were then incubated with Caspase-3 substrate (5 μM) and Annexin V–mCherry (5 μM) for 30 min in the dark at room temperature. The cells were washed twice with PBS and analyzed immediately under an EVOS FL autofluorescence microscope (Life Technologies, Carlsbad, CA, USA).
4.4. RNA-Seq Analysis
HeLa cells were treated with 20(S)-GRh2 (45 μM) or 0.1% v/v DMSO for 24 h. Total RNA was collected with an RNA simple extraction kit (Tiangen, Beijing, China). RNA samples were sent to the BGI Company (https://www.bgi.com/ (accessed on 30 July 2020)) (Shenzhen, China), and samples (5 mg) were used for library construction using the TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, USA). Libraries were quantified by the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and sequenced on an Illumina HiSeq 4000 platform (Illumina, San Diego, CA, USA).
4.5. DEG Screening and Functional Enrichment Analysis
The DEGs between control and 45 μM 20(S)-GRh2 groups were screened using DESeq2. We set the screening condition as |log 2(Fold Change)| > 1 and p adjusted value < 0.05. Gene ontology (GO) categories and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analysis of DEGs were conducted using DAVID database and visualized by the “Cluster Profiler” of R.
4.6. Protein–Protein Interaction (PPI) Network and Hub Gene Analysis
The PPI network of DEGs (connection score > 0.9) was built using the STRING database (https://string-db.org/ (accessed on 7 February 2022)) and visualized using Cytoscape. The top 10 hub genes were screened by the CytoHubba plug-in with the Bottleneck and EcCentricity algorithm [40,41].
4.7. qPCR Analysis
Total RNA was extracted using an RNA simple extraction kit (Tiangen) and then reverse transcribed into cDNA using the Primescript RT kit (Takara) following the manufacturer’s instructions. The TB Green Premix Ex Taq kit (Takara) was used for qPCR quantification. The relative expression of genes was quantified by the 2−ΔΔCT method [42]. GAPDH mRNA was an internal control. Primer sequences were as follows: PMAIP1_forward: 5′-GTGCCCTTGGAAACGGAAGA-3′, reverse: 5′-CCAGCCGCCCAGTCTAATCA-3′; ATF4_forward: 5′-CACTAGGTACCGCCAGAAGA-3′, reverse: 5′-AATCCGCCCTCTCTTTTAGA-3′; BBC3_forward: 5′-CTGTGAATCCTGTGCTCTGC-3′, reverse: 5′-AATGAATGCCAGTGGTCACA-3′; DDIT3_forward: 5′-GGAAACAGAGTGGTCATTCCC-3′, reverse: 5′-CTGCTTGAGCCGTT CATTCTC-3′; JUN_forward: 5′-TCCAAGTGCCGAAAAAGGAAG-3′; reverse: 5′-CGAGTTCTGAGCTTTCAAGGT-3′; FOS_forward: 5′-CCGGGGATAGCCTCTCTTACT-3′; reverse: 5′-CCAGGTCCGTGCAGAAGTC-3′ and GAPDH_forward: 5′-ACAACTTTGGTATCGTGGAAGG-3′, reverse: 5′-GCCATCACGCCACAGTTTC-3′.
4.8. Differential Transcript Usage (DTU) Analysis
Raw fastq files from RNA-seq datasets were processed with Trimmomatic to remove adapters and low-quality reads, followed by quantification at the transcript level by Kallisto with default settings [43]. For differential transcript usage (DTU) analysis, the Kallisto transcript abundance table was applied as input to import Rdata function of R package IsoformSwitchAnalyzeR to create a switchAnalyzeRlist object. Then, we set a cutoff (gene expression > 10 and isoform expression > 3 and differential isoform fraction (dIF) > 0.05) to filter out unqualified genes and isoforms. The single isoform genes were also removed. To test differential transcript usage, the IsoformSwitchTestDEXseq function was employed, and those isoforms with dIF > 0.05 and FDR < 0.05 were considered to be significant. The transcript isoform landscape was visualized with the switchplot function of IsoformSwitchAnalyzeR.
4.9. Molecular Docking
The 3D structure of proteins was downloaded from Protein Data Bank (PDB, https://www.rcsb.org/ (accessed on 28 September 2022)) and AlphaFold protein structure databases (https://alphafold.ebi.ac.uk/ (accessed on 28 September 2022)), including JUN (PDB ID: 1S9K Entity 5), FOS (PDB ID: 1S9K Entity 4), ATF4 (PDB ID: 1CI6 Entity 1), DDIT3 (Alphafold ID: AF-P35638-F1), BBC3 (PDB ID: 6QG8 Entity 2), and PMAIP1(PDB ID: 3MQP Entity 2). The 3D structure of 20(S)-GRh2 was downloaded from PubChem (PubChem CID 119307). Finally, molecular docking for G-Rh2 and proteins were performed with AutoDock 4.0 and visualized with Pymol.
4.10. Statistical Analysis
Statistical analysis was performed using GraphPad Prism 9.0 with experiments that were independently repeated three times. Ordinary two-way ANOVA followed by Tukey’s post-hoc test or two-tailed Student’s t-test were applied to compare 20(S)-GRh2 and control groups. p <0.05 was considered significant.
5. Conclusions
In the present study, we found that 20(S)-GRh2 inhibited cell proliferation and induced cell apoptosis in HeLa cells via upregulating with ERs-related genes (ATF4, DDIT3) and its downstream apoptosis genes (JUN, FOS, BBC3, and PMAIP1). These findings indicated that 20(S)-GRh2 may be a potential drug for CC treatment. A limitation of our study was that the prediction mechanism was not validated in vivo. Further studies are needed to better understand the anticancer mechanism of 20(S)-GRh2 in vivo. Despite the limitation, our study identified potential target genes induced by 20(S)-GRh2 related to the apoptosis of HeLa that may expand sights for the treatment of CC.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules27227865/s1, Table S1: DTU_result.
Author Contributions
Conceptualization, M.F. and M.L.; methodology, Y.L. and C.W.; software, L.J.; validation, Y.L. and S.Y.; formal analysis, X.W.; investigation, X.W. and L.J.; resources, X.W. and C.W.; data curation, Y.L.; writing—original draft preparation, Y.L.; writing—review and editing, Y.L. and M.L.; visualization, X.W. and J.Q.; supervision, P.Z. and D.Z.; project administration, J.W. and M.L.; funding acquisition, P.Z., D.Z. and M.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Scientific and Technological Development Planning Foundation of Jilin Province (YDZJ202201ZYTS683), the Scientific and Technological Developing Scheme of Ji Lin Province (20210204188YY), and by the National Natural Science Foundation of China (U19A2013).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated for this study can be found in the NCBI SRA accession (BioProject ID: PRJNA846264, https://www.ncbi.nlm.nih.gov/sra/PRJNA846264 (accessed on 6 June 2022)).
Acknowledgments
We thank Gabrielle White Wolf from Liwen Bianji (Edanz) (www.liwenbianji.cn (accessed on 30 September 2022)) for editing the English text of a draft of this manuscript.
Conflicts of Interest
The authors declare that they have no conflict interest.
Sample Availability
Samples of some reported nanomaterials are available from the authors upon request.
Abbreviations
| Ginsenoside Rh2 | GRh2 |
| DEGs | Differentially expressed genes |
| ERs | Endoplasmic reticulum stress |
| GO | Gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| CC | Cervical cancer |
| FBS | Fetal bovine serum |
| PS | Phosphatidylserine |
| BP | Biological process |
References
- Arbyn, M.; Weiderpass, E.; Bruni, L.; de Sanjosé, S.; Saraiya, M.; Ferlay, J.; Bray, F. Estimates of incidence and mortality of cervical cancer in 2018: A worldwide analysis. Lancet Glob. Health 2020, 8, e191–e203. [Google Scholar] [CrossRef]
- Basaran, D.; Leitao, M.M., Jr. The Landmark Series: Minimally Invasive Surgery for Cervical Cancer. Ann. Surg. Oncol. 2021, 28, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Vasan, N.; Baselga, J.; Hyman, D.M. A view on drug resistance in cancer. Nature 2019, 575, 299–309. [Google Scholar] [CrossRef]
- Utlu, Z.; Bilen, H. Evaluation of cutaneous side-effects associated with chemotherapeutic use in oncological patients. Adv. Dermatol. Allergol./Postępy Dermatol. Alergol. 2021, 38, 1078–1085. [Google Scholar] [CrossRef]
- Lou, J.S.; Yao, P.; Tsim KW, K. Cancer Treatment by Using Traditional Chinese Medicine: Probing Active Compounds in Anti-multidrug Resistance During Drug Therapy. Curr. Med. Chem. 2018, 25, 5128–5141. [Google Scholar] [CrossRef]
- He, M.; Xia, L.; Li, J. Potential Mechanisms of Plant-Derived Natural Products in the Treatment of Cervical Cancer. Biomolecules 2021, 11, 1539. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yi, J.; Kim, E.; Choo, Y.; Hai, H.; Kim, K.; Kim, E.-K.; Ryoo, Z.; Kim, M. 20(S)-Ginsenoside Rh2 Suppresses Oral Cancer Cell Growth by Inhibiting the Src-Raf-ERK Signaling Pathway. Anticancer Res. 2021, 41, 227–235. [Google Scholar] [CrossRef]
- Ma, J.; Gao, G.; Lu, H.; Fang, D.; Li, L.; Wei, G.; Chen, A.; Yang, Y.; Zhang, H.; Huo, J. Reversal effect of ginsenoside Rh2 on oxaliplatin-resistant colon cancer cells and its mechanism. Exp. Ther. Med. 2019, 18, 630–636. [Google Scholar] [CrossRef]
- Ben-Eltriki, M.; Deb, S.; Guns, E.S.T. 1α,25-Dihydroxyvitamin D3 synergistically enhances anticancer effects of ginsenoside Rh2 in human prostate cancer cells. J. Steroid Biochem. Mol. Biol. 2021, 209, 105828. [Google Scholar] [CrossRef]
- Mathiyalagan, R.; Wang, C.; Kim, Y.J.; Castro-Aceituno, V.; Ahn, S.; Subramaniyam, S.; Simu, S.Y.; Jiménez-Pérez, Z.E.; Yang, D.C.; Jung, S.-K. Preparation of Polyethylene Glycol-Ginsenoside Rh1 and Rh2 Conjugates and Their Efficacy against Lung Cancer and Inflammation. Molecules 2019, 24, 4367. [Google Scholar] [CrossRef]
- Luo, H.; Vong, C.T.; Chen, H.; Gao, Y.; Lyu, P.; Qiu, L.; Zhao, M.; Liu, Q.; Cheng, Z.; Zou, J.; et al. Naturally occurring anti-cancer compounds: Shining from Chinese herbal medicine. Chin. Med. 2019, 14, 48. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Park, S.; Huang, H.; Kim, E.; Yi, J.; Choi, S.-K.; Ryoo, Z.; Kim, M. Anticancer effects and potential mechanisms of ginsenoside Rh2 in various cancer types (Review). Oncol. Rep. 2021, 45, 33. [Google Scholar] [CrossRef] [PubMed]
- Marques-Coelho, D.; Iohan, L.D.; Melo de Farias, A.R.; Flaig, A.; Lambert, J.C.; Costa, M.R. Differential transcript usage unravels gene expression alterations in Alzheimer’s disease human brains. npj Aging Mech. Dis. 2021, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, J.; Qiao, J.; Liu, S.; Wang, S.; Zhao, D.; Bai, X.; Liu, M. Ginsenoside Rh2 inhibits HeLa cell energy metabolism and induces apoptosis by upregulating voltage-dependent anion channel 1. Int. J. Mol. Med. 2020, 46, 1695–1706. [Google Scholar] [CrossRef]
- Mironova, N.; Vlassov, V. Surveillance of Tumour Development: The Relationship Between Tumour-Associated RNAs and Ribonucleases. Front. Pharmacol. 2019, 10, 1019. [Google Scholar] [CrossRef]
- Liu, Z.-H.; Li, J.; Xia, J.; Jiang, R.; Zuo, G.-W.; Li, X.-P.; Chen, Y.; Xiong, W.; Chen, D.-L. Ginsenoside 20(s)-Rh2 as potent natural histone deacetylase inhibitors suppressing the growth of human leukemia cells. Chem. Biol. Interact. 2015, 242, 227–234. [Google Scholar] [CrossRef]
- Shi, Q.; Shi, X.; Zuo, G.; Xiong, W.; Li, H.; Guo, P.; Wang, F.; Chen, Y.; Li, J.; Chen, D.-L. Anticancer effect of 20(S)-ginsenoside Rh2 on HepG2 liver carcinoma cells: Activating GSK-3β and degrading β-catenin. Oncol. Rep. 2016, 36, 2059–2070. [Google Scholar] [CrossRef]
- Kim, C.; Kim, B. Anti-Cancer Natural Products and Their Bioactive Compounds Inducing ER Stress-Mediated Apoptosis: A Review. Nutrients 2018, 10, 1021. [Google Scholar] [CrossRef]
- Zhao, T.; Du, J.; Zeng, H. Interplay between endoplasmic reticulum stress and non-coding RNAs in cancer. J. Hematol. Oncol. 2020, 13, 163. [Google Scholar] [CrossRef]
- Tian, F.; Zhao, J.; Bu, S.; Teng, H.; Yang, J.; Zhang, X.; Li, X.; Dong, L. KLF6 Induces Apoptosis in Human Lens Epithelial Cells Through the ATF4-ATF3-CHOP Axis. Drug Des. Dev. Ther. 2020, 14, 1041–1055. [Google Scholar] [CrossRef]
- Park, K.-M.; Park, J.-Y.; Pyo, J.; Lee, S.-Y.; Kim, H.-S. Induction of DR5-Dependent Apoptosis by PGA2 through ATF4-CHOP Pathway. Molecules 2022, 27, 3804. [Google Scholar] [CrossRef] [PubMed]
- Ying, Y.; Xue, R.; Yang, Y.; Zhang, S.X.; Xiao, H.; Zhu, H.; Li, J.; Chen, G.; Ye, Y.; Yu, M.; et al. Activation of ATF4 triggers trabecular meshwork cell dysfunction and apoptosis in POAG. Aging 2021, 13, 8628–8642. [Google Scholar] [CrossRef] [PubMed]
- Ge, G.; Yan, Y.; Cai, H. Ginsenoside Rh2 Inhibited Proliferation by Inducing ROS Mediated ER Stress Dependent Apoptosis in Lung Cancer Cells. Biol. Pharm. Bull. 2017, 40, 2117–2124. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.-Y.; Di, Z.-M.; Cao, Q.; Xu, W.-S.; Bi, S.-X.; Yu, J.-S.; Shen, Y.-X.; Yu, Y.-Q.; Feng, L.-J. Xanthatin induces glioma cell apoptosis and inhibits tumor growth via activating endoplasmic reticulum stress-dependent CHOP pathway. Acta Pharmacol. Sin. 2020, 41, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Tian, Z.; Guo, Y.; Liu, X.; Ma, Y.; Du, X.; Wang, R.; Zhang, S.; Shi, L.; Guo, H.; et al. Microcystin-leucine arginine exposure contributes to apoptosis and follicular atresia in mice ovaries by endoplasmic reticulum stress-upregulated Ddit3. Sci. Total Environ. 2021, 756, 144070. [Google Scholar] [CrossRef]
- Jung, K.-J.; Min, K.-J.; Bae, J.H.; Kwon, T.K. Carnosic acid sensitized TRAIL-mediated apoptosis through down-regulation of c-FLIP and Bcl-2 expression at the post translational levels and CHOP-dependent up-regulation of DR5, Bim, and PUMA expression in human carcinoma caki cells. Oncotarget 2015, 6, 1556–1568. [Google Scholar] [CrossRef]
- Ma, B.; Li, J.; Yang, W.-K.; Zhang, M.-G.; Xie, X.-D.; Bai, Z.-T. N-trans-Feruloyloctopamine Wakes Up BBC3, DDIT3, CDKN1A, and NOXA Signals to Accelerate HCC Cell Apoptosis. Anal. Cell. Pathol. 2021, 2021, 1560307. [Google Scholar] [CrossRef]
- Manne, R.K.; Agrawal, Y.; Malonia, S.K.; Banday, S.; Edachery, S.; Patel, A.; Kumar, A.; Shetty, P.; Santra, M.K. FBXL20 promotes breast cancer malignancy by inhibiting apoptosis through degradation of PUMA and BAX. J. Biol. Chem. 2021, 297, 101253. [Google Scholar] [CrossRef]
- Nakajima, W.; Tanaka, N. Noxa induces apoptosis in oncogene-expressing cells through catch-and-release mechanism operating between Puma and Mcl-1. Biochem. Biophys. Res. Commun. 2011, 413, 643–648. [Google Scholar] [CrossRef]
- Núñez-Vázquez, S.; Sánchez-Vera, I.; Saura-Esteller, J.; Cosialls, A.M.; Noisier, A.F.; Albericio, F.; Lavilla, R.; Pons, G.; Iglesias-Serret, D.; Gil, J. NOXA upregulation by the prohibitin-binding compound fluorizoline is transcriptionally regulated by integrated stress response-induced ATF3 and ATF4. FEBS J. 2021, 288, 1271–1285. [Google Scholar] [CrossRef]
- Santos, M.M.; Tannuri, A.C.A.; Coelho, M.C.M.; Gonçalves, J.D.O.; Serafini, S.; da Silva, L.F.F.; Tannuri, U. Immediate expression of c-fos and c-jun mRNA in a model of intestinal autotransplantation and ischemia-reperfusion in situ. Clinics 2015, 70, 373–379. [Google Scholar] [CrossRef]
- Marotta, P.; Salatiello, F.; Ambrosino, L.; Berruto, F.; Chiusano, M.L.; Locascio, A. The Ascidia Ciona robusta Provides Novel Insights on the Evolution of the AP-1 Transcriptional Complex. Front. Cell Dev. Biol. 2021, 9, 709696. [Google Scholar] [CrossRef] [PubMed]
- Ryder, C.B.; McColl, K.; Distelhorst, C.W. Acidosis blocks CCAAT/enhancer-binding protein homologous protein (CHOP)- and c-Jun-mediated induction of p53-upregulated mediator of apoptosis (PUMA) during amino acid starvation. Biochem. Biophys. Res. Commun. 2013, 430, 1283–1288. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhang, M.; Li, Y.; Mei, S.; Qin, J.; Yan, J. c-Jun/Ap-1 is upregulated in an Ang II-induced abdominal aortic aneurysm formation model and mediates Chop expression in mouse aortic smooth muscle cells. Mol. Med. Rep. 2019, 19, 3459–3468. [Google Scholar] [CrossRef]
- Mehta, S.; Zhang, J. Liquid–liquid phase separation drives cellular function and dysfunction in cancer. Nat. Rev. Cancer 2022, 22, 239–252. [Google Scholar] [CrossRef]
- Zhang, Y.; Qian, J.; Gu, C.; Yang, Y. Alternative splicing and cancer: A systematic review. Signal Transduct. Target. Ther. 2021, 6, 78. [Google Scholar] [CrossRef]
- Newberry, R.W.; Raines, R.T. Secondary Forces in Protein Folding. ACS Chem. Biol. 2019, 14, 1677–1686. [Google Scholar] [CrossRef]
- Patil, R.; Das, S.; Stanley, A.; Yadav, L.; Sudhakar, A.; Varma, A.K. Optimized Hydrophobic Interactions and Hydrogen Bonding at the Target-Ligand Interface Leads the Pathways of Drug-Designing. PLoS ONE 2010, 5, e12029. [Google Scholar] [CrossRef]
- Malik, C.; Ghosh, S. Regulation of Single-Channel Conductance of Voltage-Dependent Anion Channel by Mercuric Chloride in a Planar Lipid Bilayer. J. Membr. Biol. 2020, 253, 357–371. [Google Scholar] [CrossRef]
- Fang, Y.; Wang, Y.; Zeng, D.; Zhi, S.; Shu, T.; Huang, N.; Zheng, S.; Wu, J.; Liu, Y.; Huang, G.; et al. Comprehensive analyses reveal TKI-induced remodeling of the tumor immune microenvironment in EGFR/ALK-positive non-small-cell lung cancer. OncoImmunology 2021, 10, 1951019. [Google Scholar] [CrossRef]
- Willis, C.R.; Ames, R.; Deane, C.S.; Phillips, B.E.; Boereboom, C.L.; Abdulla, H.; Bukhari, S.S.; Lund, J.; Williams, J.P.; Wilkinson, D.J.; et al. Network analysis of human muscle adaptation to aging and contraction. Aging 2020, 12, 740–755. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar]
- Hamilton, M.J.; Girke, T.; Martinez, E. Global isoform-specific transcript alterations and deregulated networks in clear cell renal cell carcinoma. Oncotarget 2018, 9, 23670–23680. [Google Scholar] [CrossRef] [PubMed][Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).