Characterization and Classification of Spanish Honey by Non-Targeted LC–HRMS (Orbitrap) Fingerprinting and Multivariate Chemometric Methods
Abstract
:1. Introduction
2. Results and Discussion
2.1. Non-Targeted LC–HRMS Fingerprints
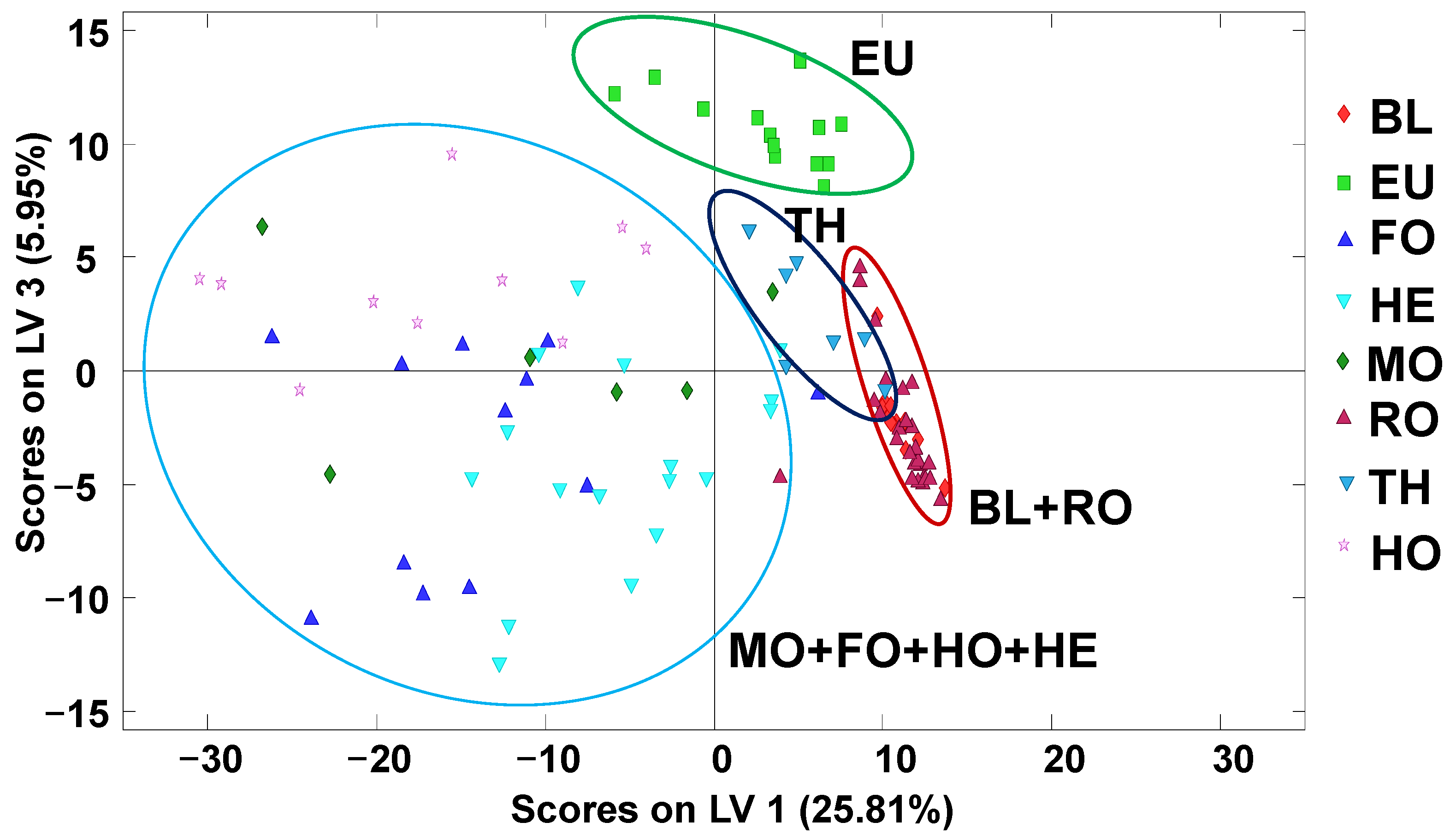
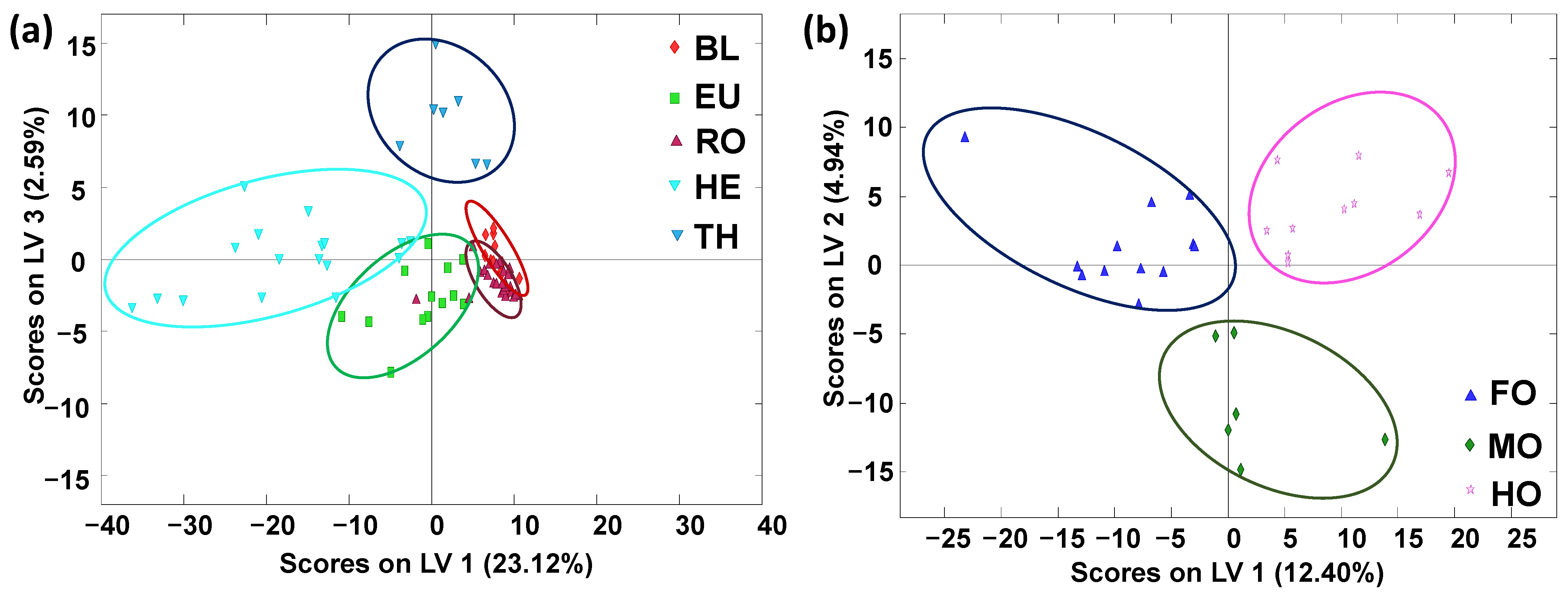
2.2. Honey Exploratory Chemometric Analysis by PCA
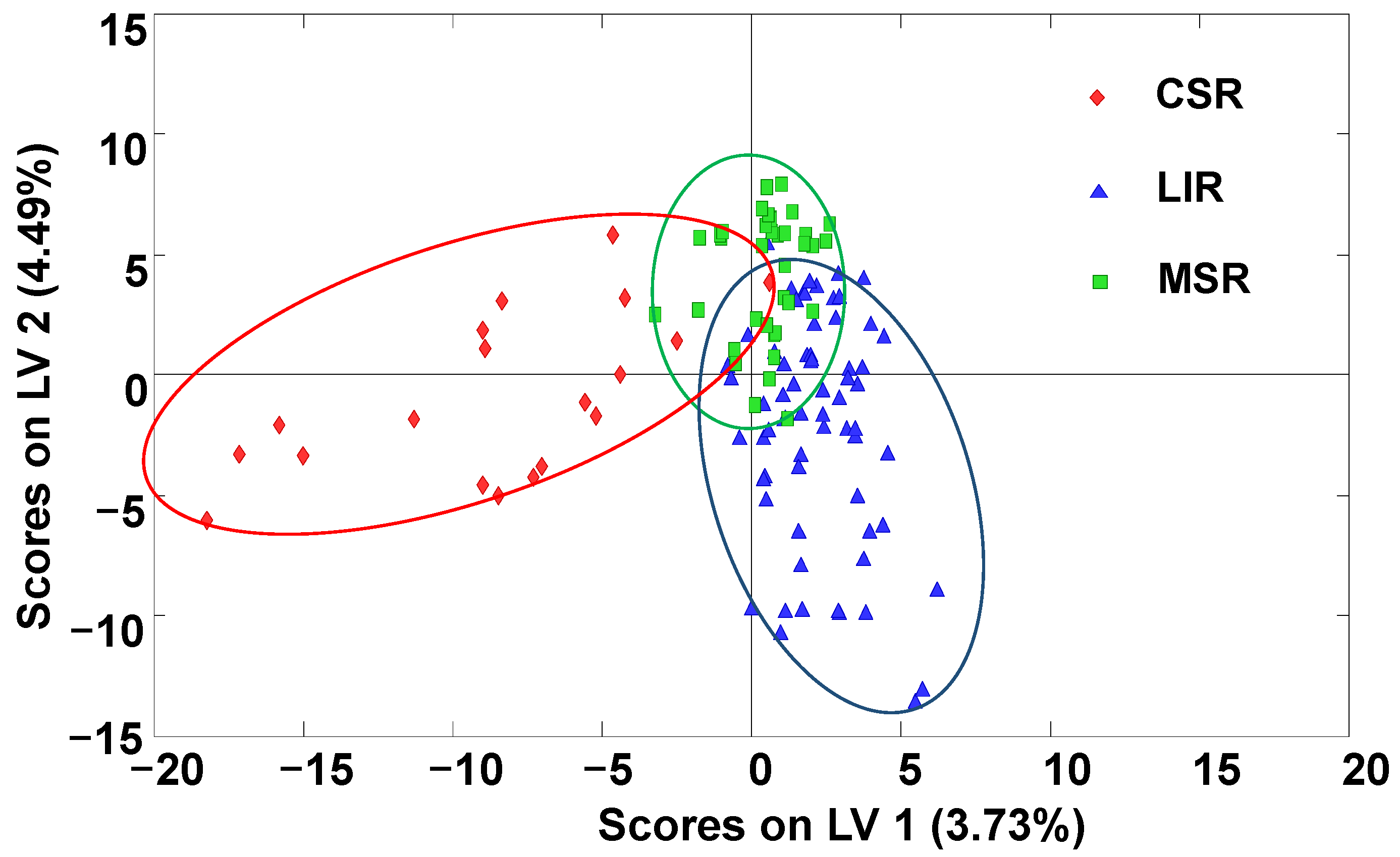
2.3. Classification of Honey Samples by Supervised PLS–DA
3. Materials and Methods
3.1. Reagents and Chemicals
3.2. Samples and Sample Treatment
3.3. Non-Targeted LC–HRMS Chromatographic Fingerprinting Method
3.4. Data Matrix
3.5. Chemometric Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- COUNCIL DIRECTIVE 2001/110/EC of 20 December 2001 relating to honey. Off. J. Eur. Communities 2002, L10, 47–52.
- European Council. European Parliament Directive 2014/63/EU of the European Parliament and of the Council of 15 May 2014 amending Council Directive 2001/110/EC relating to honey. Off. J. Eur. Communities 2014, L164, 1–5. [Google Scholar]
- Committee on Agriculture and Rural Development Prospects and Challenges for the EU Apiculture Sector. European Parliament resolution of 1 March 2018 on prospects and challenges for the EU apiculture sector (2017/2115(INI)). Off. J. Eur. Union 2018, C129, 25–37. [Google Scholar]
- Thrasyvoulou, A.; Tananaki, C.; Goras, G.; Karazafiris, E.; Dimou, M.; Liolios, V.; Kanelis, D.; Gounari, S. Legislation of honey criteria and standards. J. Apic. Res. 2018, 57, 88–96. [Google Scholar] [CrossRef]
- Ahmed, S.; Sulaiman, S.A.; Baig, A.A.; Ibrahim, M.; Liaqat, S.; Fatima, S.; Jabeen, S.; Shamim, N.; Othman, N.H. Honey as a Potential Natural Antioxidant Medicine: An Insight into Its Molecular Mechanisms of Action. Oxid. Med. Cell. Longev. 2018, 2018, 8367846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samarghandian, S.; Farkhondeh, T.; Samini, F. Honey and health: A review of recent clinical research. Pharmacogn. Res. 2017, 9, 121–127. [Google Scholar] [CrossRef]
- Vallianou, N.G. Honey and its Anti-Inflammatory, Anti-Bacterial and Anti-Oxidant Properties. Gen. Med. Open Access 2014, 2, 1000132. [Google Scholar] [CrossRef] [Green Version]
- Al-Qassemi, R.; Robinson, R.K. Some special nutritional properties of honey—A brief review. Nutr. Food Sci. 2003, 33, 254–260. [Google Scholar] [CrossRef]
- Kadri, S.M.; Zaluski, R.; Orsi, R. de O. Nutritional and mineral contents of honey extracted by centrifugation and pressed processes. Food Chem. 2017, 218, 237–241. [Google Scholar] [CrossRef]
- Pyrzynska, K.; Biesaga, M. Analysis of phenolic acids and flavonoids in honey. TrAC Trends Anal. Chem. 2009, 28, 893–902. [Google Scholar] [CrossRef]
- Gašić, U.M.; Milojković-Opsenica, D.M.; Tešić, Ž.L. Polyphenols as possible markers of botanical origin of honey. J. AOAC Int. 2017, 100, 852–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Recklies, K.; Peukert, C.; Kölling-Speer, I.; Speer, K. Differentiation of Honeydew Honeys from Blossom Honeys and According to Their Botanical Origin by Electrical Conductivity and Phenolic and Sugar Spectra. J. Agric. Food Chem. 2021, 69, 1329–1347. [Google Scholar] [CrossRef]
- Pita-Calvo, C.; Vázquez, M. Differences between honeydew and blossom honeys: A review. Trends Food Sci. Technol. 2017, 59, 79–87. [Google Scholar] [CrossRef]
- Wu, L.; Du, B.; Vander Heyden, Y.; Chen, L.; Zhao, L.; Wang, M.; Xue, X. Recent advancements in detecting sugar-based adulterants in honey—A challenge. TrAC Trends Anal. Chem. 2017, 86, 25–38. [Google Scholar] [CrossRef]
- Fakhlaei, R.; Selamat, J.; Khatib, A.; Razi, A.F.A.; Sukor, R.; Ahmad, S.; Babadi, A.A. The toxic impact of honey adulteration: A review. Foods 2020, 9, 1–21. [Google Scholar] [CrossRef]
- Selvaraju, K.; Vikram, P.; Soon, J.M.; Krishnan, K.T.; Mohammed, A. Melissopalynological, physicochemical and antioxidant properties of honey from West Coast of Malaysia. J. Food Sci. Technol. 2019, 56, 2508–2521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thakodee, T.; Deowanish, S.; Duangmal, K. Melissopalynological analysis of stingless bee (Tetragonula pagdeni) honey in Eastern Thailand. J. Asia. Pac. Entomol. 2018, 21, 620–630. [Google Scholar] [CrossRef]
- Campmajó, G.; Núñez, O. Chromatographic Fingerprinting Approaches in Food Authentication. In Chromatographic and Related Separation Techniques in Food Integrity and Authenticity. Volume A: Advances in Chromatographic Techniques; Núñez, O., Campmajó, G., Eds.; World Scientific Publishing Europe, Ltd.: London, UK, 2021; pp. 137–165. ISBN 978-1-78634-994-1. [Google Scholar]
- Zzafar, A.; Abbas Bukhari, S.N.; Ahmad, J.; Akhtar, M.S. Authentication and Traceability of Honey. In Fingerprinting Techniques in Food Authentication and Traceability; Siddiqi, K.S., Nollet, L.M.L., Eds.; CRC Press: Boca Raton, FL, USA, 2019; pp. 279–305. [Google Scholar]
- Antignac, J.P.; Courant, F.; Pinel, G.; Bichon, E.; Monteau, F.; Elliott, C.; Le Bizec, B. Mass spectrometry-based metabolomics applied to the chemical safety of food. TrAC Trends Anal. Chem. 2011, 30, 292–301. [Google Scholar] [CrossRef]
- Esslinger, S.; Riedl, J.; Fauhl-Hassek, C. Potential and limitations of non-targeted fingerprinting for authentication of food in official control. Food Res. Int. 2014, 60, 189–204. [Google Scholar] [CrossRef]
- Suhandy, D.; Yulia, M. The use of UV spectroscopy and SIMCA for the authentication of Indonesian honeys according to botanical, entomological and geographical origins. Molecules 2021, 26, 915. [Google Scholar] [CrossRef]
- de Souza, R.R.; Fernandes, D.D.d.S.; Diniz, P.H.G.D. Honey authentication in terms of its adulteration with sugar syrups using UV–Vis spectroscopy and one-class classifiers. Food Chem. 2021, 365, 130467. [Google Scholar] [CrossRef] [PubMed]
- Ruoff, K.; Karoui, R.; Dufour, E.; Luginbbühl, W.; Bosset, J.O.; Bogdanov, S.; Amadò, R. Authentication of the botanical origin of honey by front-face fluorescence spectroscopy. A preliminary study. J. Agric. Food Chem. 2005, 53, 1343–1347. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.; Li, J.; Liu, X.; Yuan, J.; Yuan, W.; Tian, Y.; Xuan, H. Authentication of acacia honey using fluorescence spectroscopy. Food Control 2021, 130, 108327. [Google Scholar] [CrossRef]
- Valinger, D.; Longin, L.; Grbeš, F.; Benković, M.; Jurina, T.; Gajdoš Kljusurić, J.; Jurinjak Tušek, A. Detection of honey adulteration—The potential of UV-VIS and NIR spectroscopy coupled with multivariate analysis. Lwt 2021, 145, 111316. [Google Scholar] [CrossRef]
- Stanek, N.; Teper, D.; Kafarski, P.; Jasicka-Misiak, I. Authentication of phacelia honeys (Phacelia tanacetifolia) based on a combination of HPLC and HPTLC analyses as well as spectrophotometric measurements. Lwt 2019, 107, 199–207. [Google Scholar] [CrossRef]
- Koulis, G.A.; Tsagkaris, A.S.; Aalizadeh, R.; Dasenaki, M.E.; Panagopoulou, E.I.; Drivelos, S.; Halagarda, M.; Georgiou, C.A.; Proestos, C.; Thomaidis, N.S. Honey phenolic compound profiling and authenticity assessment using hrms targeted and untargeted metabolomics. Molecules 2021, 26, 2769. [Google Scholar] [CrossRef]
- Li, Y.; Jin, Y.; Yang, S.; Zhang, W.; Zhang, J.; Zhao, W.; Chen, L.; Wen, Y.; Zhang, Y.; Lu, K.; et al. Strategy for comparative untargeted metabolomics reveals honey markers of different floral and geographic origins using ultrahigh-performance liquid chromatography-hybrid quadrupole-orbitrap mass spectrometry. J. Chromatogr. A 2017, 1499, 78–89. [Google Scholar] [CrossRef]
- García-Seval, V.; Martínez-Alfaro, C.; Saurina, J.; Núñez, O.; Sentellas, S. Characterization, Classification and Authentication of Spanish Blossom and Honeydew Honeys by Non-Targeted HPLC-UV and Off-Line SPE HPLC-UV Polyphenolic Fingerprinting Strategies. Foods 2022, 11, 2345. [Google Scholar] [CrossRef]
- Lucci, P.; Saurina, J.; Núñez, O. Trends in LC-MS and LC-HRMS analysis and characterization of polyphenols in food. TrAC-Trends Anal. Chem. 2017, 88, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Martos, I.; Cossentini, M.; Ferreres, F.; Tomás-Barberán, F.A. Flavonoid Composition of Tunisian Honeys and Propolis. J. Agric. Food Chem. 1997, 45, 2824–2829. [Google Scholar] [CrossRef]
- Chambers, M.C.; MacLean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Holman, J.D.; Tabb, D.L.; Mallick, P. Employing ProteoWizard to Convert Raw Mass Spectrometry Data. Curr. Protoc. Bioinform. 2014, 46, 13.24.1–13.24.9. [Google Scholar] [CrossRef] [Green Version]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Massart, D.L.; Vandeginste, B.G.M.; Buydens, L.M.C.; de Jong, S.; Lewi, P.J.; Smeyers-Verbeke, J. Handbook of Chemometrics and Qualimetrics. J. Chem. Inf. Comput. Sci. 1997, 38, 1254. [Google Scholar] [CrossRef]


| Sample Class Variety | Sensitivity (%) | Specificity (%) | Classification Error (%) |
|---|---|---|---|
| BL | 100 | 57.1 | 21.4 |
| RO | 100 | 98.4 | 0.8 |
| EU | 100 | 96.6 | 1.7 |
| TH | 96.2 | 79.6 | 12.7 |
| HE | 100 | 100 | 0 |
| Sample Class Variety | Sensitivity (%) | Specificity (%) | Classification Error (%) |
|---|---|---|---|
| CSR | 94.7 | 99.0 | 3.1 |
| LIR | 92.4 | 98.2 | 4.7 |
| MSR | 91.9 | 87.1 | 10.5 |
| Time (min) | Elution | % Acetonitrile | Flow Rate (µL/min) |
|---|---|---|---|
| 0–5 | Isocratic | 3% | 400 |
| 5–13 | Linear gradient | 3–95% | 400 |
| 13–15 | Isocratic | 95% | 400 |
| 15–15.5 | Linear gradient | 95–3% | 400 |
| 15.5–19 | Isocratic | 3% | 400 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Seval, V.; Saurina, J.; Sentellas, S.; Núñez, O. Characterization and Classification of Spanish Honey by Non-Targeted LC–HRMS (Orbitrap) Fingerprinting and Multivariate Chemometric Methods. Molecules 2022, 27, 8357. https://doi.org/10.3390/molecules27238357
García-Seval V, Saurina J, Sentellas S, Núñez O. Characterization and Classification of Spanish Honey by Non-Targeted LC–HRMS (Orbitrap) Fingerprinting and Multivariate Chemometric Methods. Molecules. 2022; 27(23):8357. https://doi.org/10.3390/molecules27238357
Chicago/Turabian StyleGarcía-Seval, Víctor, Javier Saurina, Sònia Sentellas, and Oscar Núñez. 2022. "Characterization and Classification of Spanish Honey by Non-Targeted LC–HRMS (Orbitrap) Fingerprinting and Multivariate Chemometric Methods" Molecules 27, no. 23: 8357. https://doi.org/10.3390/molecules27238357
APA StyleGarcía-Seval, V., Saurina, J., Sentellas, S., & Núñez, O. (2022). Characterization and Classification of Spanish Honey by Non-Targeted LC–HRMS (Orbitrap) Fingerprinting and Multivariate Chemometric Methods. Molecules, 27(23), 8357. https://doi.org/10.3390/molecules27238357













