Identification of the New Metabolite of Nebivolol Using Liquid Chromatography Coupled with High-Resolution Mass Spectrometry and Chemometrics
Abstract
:1. Introduction
2. Results and Discussion
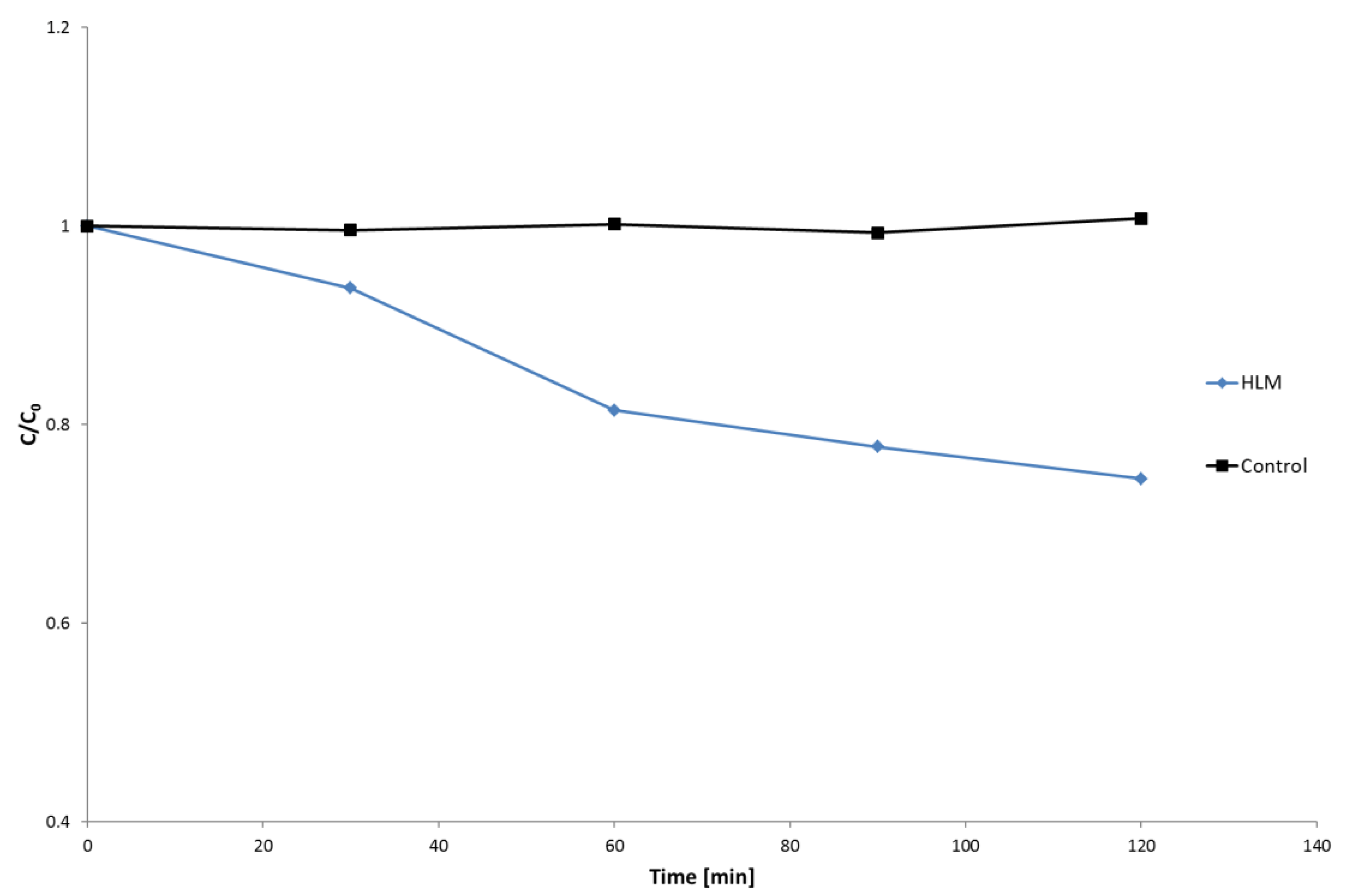
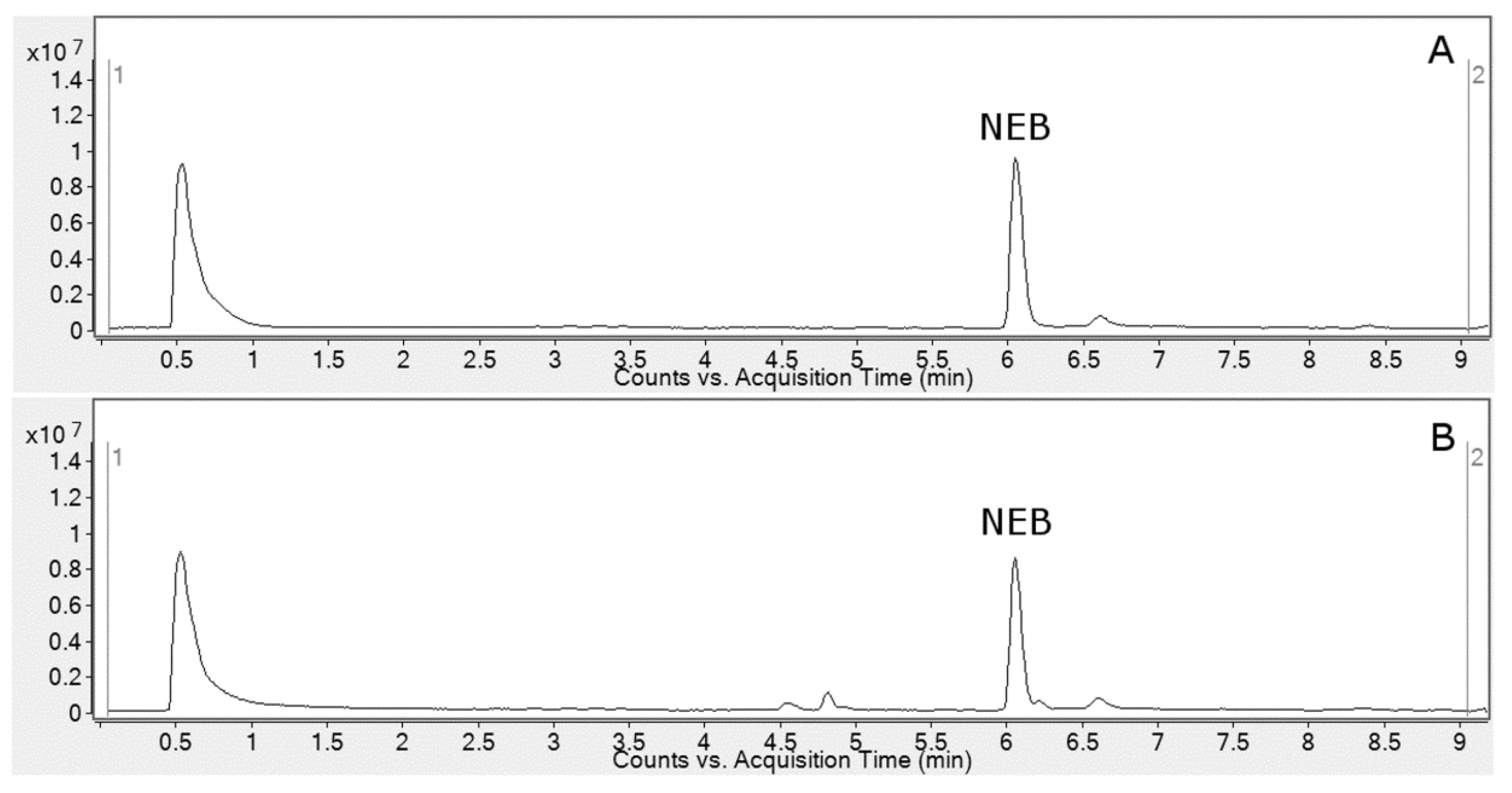
2.1. HLM Biotransformation of Nebivolol
2.2. Chemometric Analysis
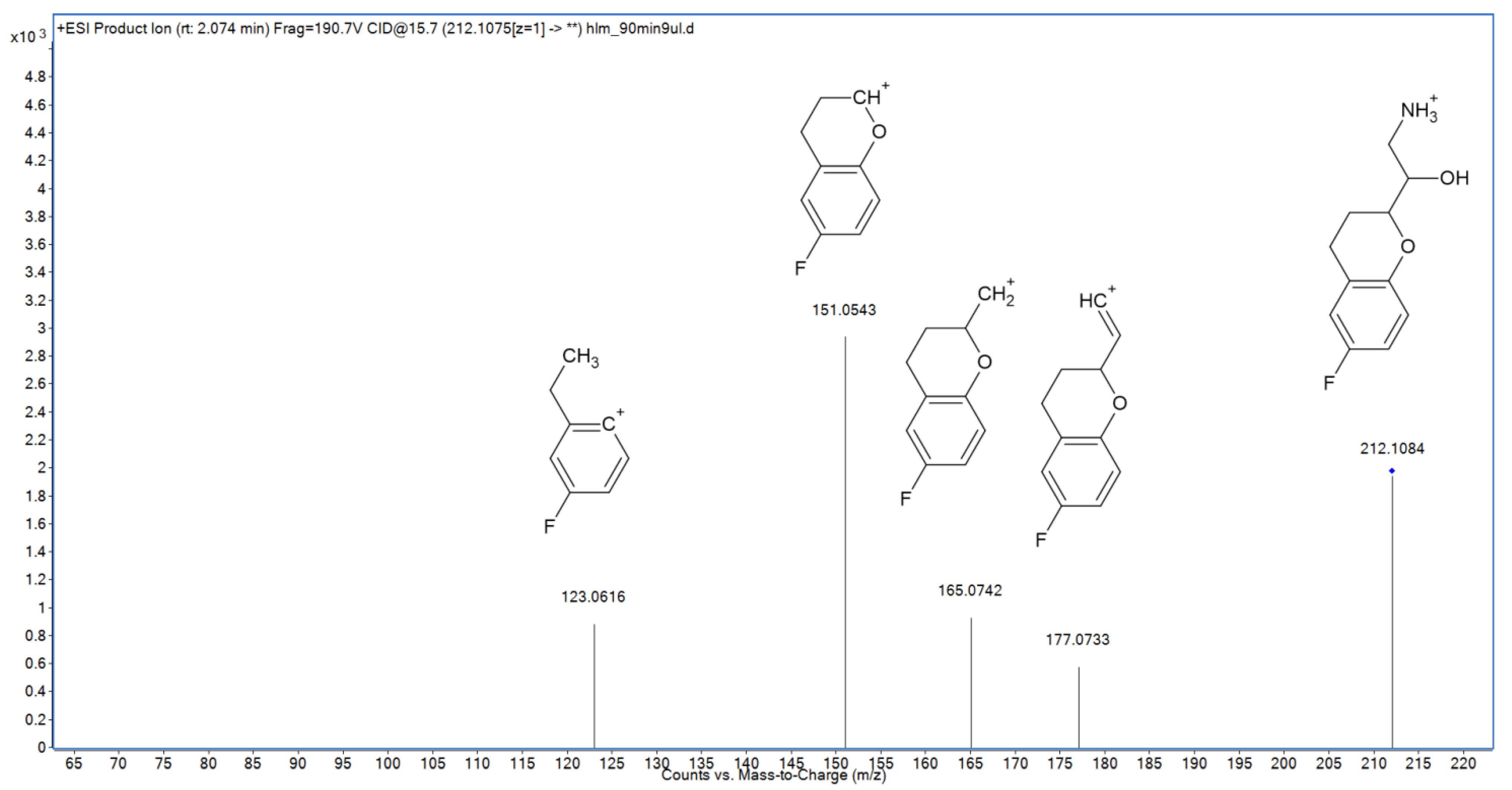
2.3. Metabolite Identification
2.4. Hepatic Biotransformation Pathways
2.5. In Silico Assessment of Toxicity
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. In Vitro Simulation of Metabolism by HLM
3.3. Analytical Procedures
3.4. Chemometric and Toxicity Prediction Software
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Lynch, T.; Price, A. The Effect of Cytochrome P450 Metabolism on Drug Response, Interactions, and Adverse Effects. Am. Fam. Physician 2007, 76, 391–396. [Google Scholar] [PubMed]
- Changjiang, X.; Yong-Tao, L.C.; Kong, A.-N.T. Induction of Phase I, II and III Drug Metabolism/Transport by Xenobiotics. Arch. Pharm. Res. 2005, 28, 249–268. [Google Scholar] [CrossRef]
- Almazroo, O.A.; Miah, M.K.; Venkataramanan, R. Drug Metabolism in the Liver. Clin. Liv. Dis. 2017, 21, 1–20. [Google Scholar] [CrossRef]
- Remmer, H. The role of the liver in drug metabolism. Am. J. Med. 1970, 49, 617–629. [Google Scholar] [CrossRef]
- McDonnell, A.M.; Dang, C.H. Basic Review of the Cytochrome P450 System. J. Adv. Pract. Oncol. 2013, 4, 263–268. [Google Scholar] [CrossRef]
- Gawlik, M.; Savic, V.; Jovanovic, M.; Skibiński, R. Mimicking of Phase I Metabolism Reactions of Molindone by HLM and Photocatalytic Methods with the Use of UHPLC-MS/MS. Molecules 2020, 25, 1367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dery, A.S.; Hamilton, L.A.; Starr, J.A. Nebivolol for the Treatment of Heart Failure. Am. J. Health Syst. Pharm. 2011, 68, 879–886. [Google Scholar] [CrossRef]
- Van Peer, A.; Snoeck, E.; Woestenborghs, R.; Van De Velde, V.; Mannens, G.; Meuldermans, W.; Heykants, J. Clinical Pharmacokinetics of Nebivolol. Clin. Drug Investig. 1991, 3, 25–30. [Google Scholar] [CrossRef]
- Hu, X.; Lan, T.; Dai, D.; Xu, R.-A.; Yuan, L.; Zhou, Q.; Li, Y.; Cai, J.; Hu, G. Evaluation of 24 CYP2D6 Variants on the Metabolism of Nebivolol In Vitro. Drug Metab. Dispos. 2016, 44, 1828–1831. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.-Y.; Hu, X.-X.; Li, M.-F.; Wang, H.; Zhang, L.-Q.; Hu, G.-X.; Cai, J.-P. Functional Characterization of CYP2C19 Variants in Nebivolol 4-Hydroxlation in Vitro. Drug Test Anal. 2018, 10, 807–813. [Google Scholar] [CrossRef] [PubMed]
- Cominacini, L.; FrattaPasini, A.; Garbin, U.; Nava, C.; Davoli, A.; Criscuoli, M.; Crea, A.; Sawamura, T.; Lo Cascio, V. Nebivolol and Its 4-Keto Derivative Increase Nitric Oxide in Endothelial Cells by Reducing Its Oxidative Inactivation. J. Am. Coll. Cardiol. 2003, 42, 1838–1844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briciu, C.; Neag, M.; Muntean, D.; Bocsan, C.; Buzoianu, A.; Antonescu, O.; Gheldiu, A.-M.; Achim, M.; Popa, A.; Vlase, L. Phenotypic Differences in Nebivolol Metabolism and Bioavailability in Healthy Volunteers. Clujul Med. 2015, 88, 208–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangrella, M.; Rossi, F.; Fici, F.; Rossi, F. Pharmacology of Nebivolol. Pharm. Res. 1998, 38, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Van Rooy, P.; De Crée, J. Haemodynamic Effects of Nebivolol in Poor and Extensive Metabolisers. Clin. Drug Investig. 1991, 3, 161–163. [Google Scholar] [CrossRef]
- Hendrickx, J.; Bockx, M.; Zwijsen, C.; Borgmans, C.; Mannens, G.; Meuldermans, W.; Heykants, J. Location of the Hydroxyl Functions in Hydroxylated Metabolites of Nebivolol in Different Animal Species and Human Subjects as Determined by On-Line High-Performance Liquid Chromatography-Diode-Array Detection. J. Chromatogr. A 1996, 729, 341–354. [Google Scholar] [CrossRef]
- Cheng, J.W.M. Nebivolol: A Third-Generation β-Blocker for Hypertension. Clin. Ther. 2009, 31, 447–462. [Google Scholar] [CrossRef]
- Skibiński, R.; Trawiński, J.; Gawlik, M. Characterization of Phase I Hepatic Metabolites of Anti-Premature Ejaculation Drug Dapoxetine by UHPLC-ESI-Q-TOF. Molecules 2021, 26, 3794. [Google Scholar] [CrossRef] [PubMed]



| Name | Reaction Type | Retention Time (min) | Measured Mass (m/z) | Theoretical Mass (m/z) | Mass Error (ppm) | Molecular Formula [M + H]+ |
|---|---|---|---|---|---|---|
| NEB | – | 6.05 | 406.1831 | 406.1824 | 1.72 | C22H26F2NO4 |
| M1 | Al-OH | 4.84 | 422.1761 | 422.1774 | 3.07 | C22H26F2NO5 |
| M2 | Al-OH | 4.57 | 422.1785 | 422.1774 | 2.61 | C22H26F2NO5 |
| M3 | Ar-OH | 5.58 | 422.1761 | 422.1774 | 3.08 | C22H26F2NO5 |
| M4 | Al-OH, Deh | 5.30 | 420.1618 | 420.1617 | 0.24 | C22H24F2NO5 |
| M5 | Al-OH | 3.45 | 438.1726 | 438.1723 | 0.68 | C22H26F2NO6 |
| M6 | N-dealk | 2.08 | 212.1083 | 212.1081 | 0.94 | C11H15FNO2 |
| Toxicity Model | NEB | M6 | M1 | M2 | M3 | M4 | M5 |
|---|---|---|---|---|---|---|---|
| Mouse IP | 2.34 | 2.88 | 2.45 | 2.42 | 2.4 | 2.36 | 2.45 |
| Mouse OR | 2.59 | 2.68 | 2.68 | 2.65 | 2.67 | 2.81 | 2.87 |
| Mouse IV | 1.72 | 2.04 | 1.73 | 1.83 | 1.78 | 1.8 | 1.75 |
| Mouse SC | 3.32 | 2.84 | 3.45 | 3.38 | 3.36 | 3.3 | 3.53 |
| Rat IP | 1.70 | 2.54 | 1.76 | 1.85 | 1.84 | 1.93 | 1.81 |
| Rat OR | 2.83 | 2.76 | 2.88 | 2.86 | 3.01 | 2.8 | 2.96 |
| Rat OR T.E.S.T. | 3.15 | 2.64 | 3.06 | 3.4 | 2.67 | 3.01 | 2.77 |
| Ames Percepta | 0.06 | 0.61 | 0.08 | 0.06 | 0.05 | 0.07 | 0.08 |
| Ames T.E.S.T. | 0.10 | 0.45 | 0.46 | 0.21 | 0.5 | 0.56 | 0.46 |
| Developmental Toxicity | 0.62 | 0.76 | 0.71 | 0.57 | 0.81 | 0.75 | 0.65 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trawiński, J.; Wroński, M.; Gawlik, M.; Skibiński, R. Identification of the New Metabolite of Nebivolol Using Liquid Chromatography Coupled with High-Resolution Mass Spectrometry and Chemometrics. Molecules 2022, 27, 763. https://doi.org/10.3390/molecules27030763
Trawiński J, Wroński M, Gawlik M, Skibiński R. Identification of the New Metabolite of Nebivolol Using Liquid Chromatography Coupled with High-Resolution Mass Spectrometry and Chemometrics. Molecules. 2022; 27(3):763. https://doi.org/10.3390/molecules27030763
Chicago/Turabian StyleTrawiński, Jakub, Michał Wroński, Maciej Gawlik, and Robert Skibiński. 2022. "Identification of the New Metabolite of Nebivolol Using Liquid Chromatography Coupled with High-Resolution Mass Spectrometry and Chemometrics" Molecules 27, no. 3: 763. https://doi.org/10.3390/molecules27030763
APA StyleTrawiński, J., Wroński, M., Gawlik, M., & Skibiński, R. (2022). Identification of the New Metabolite of Nebivolol Using Liquid Chromatography Coupled with High-Resolution Mass Spectrometry and Chemometrics. Molecules, 27(3), 763. https://doi.org/10.3390/molecules27030763







