Is Genetic Engineering a Route to Enhance Microalgae-Mediated Bioremediation of Heavy Metal-Containing Effluents?
Abstract
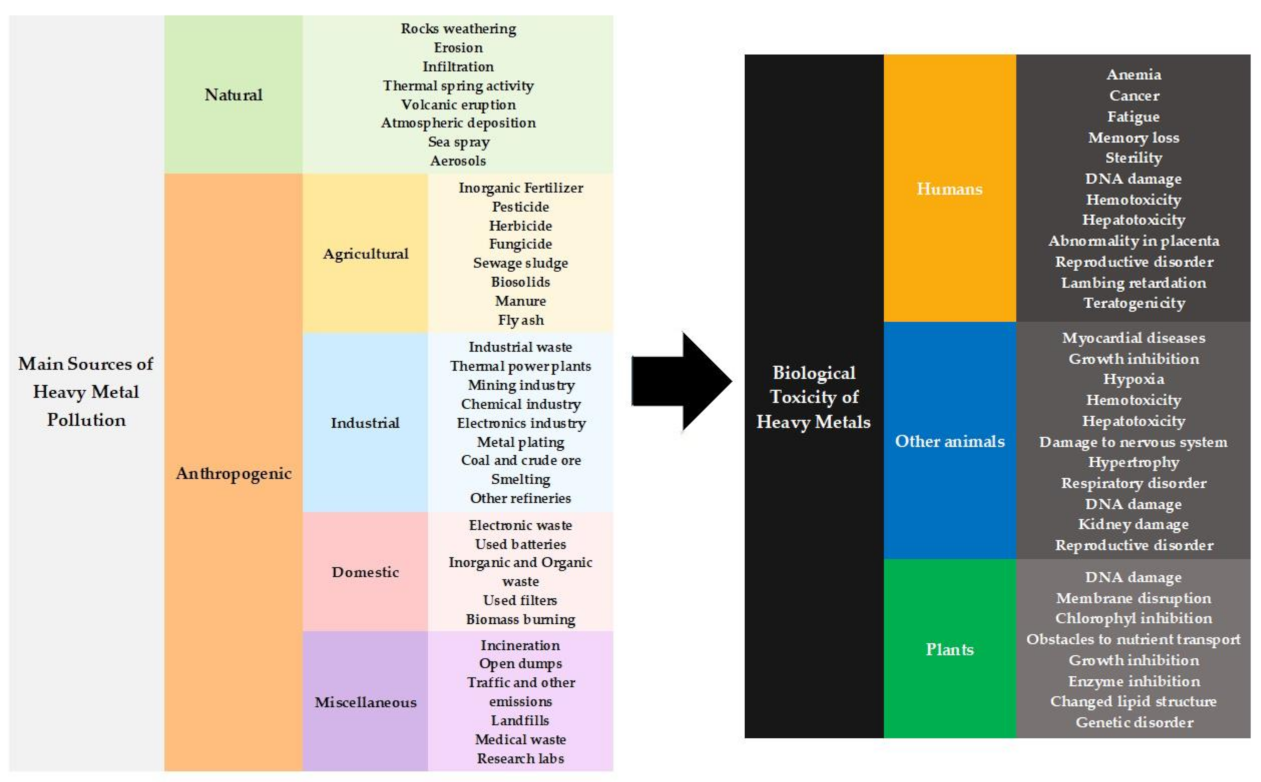
:1. Introduction
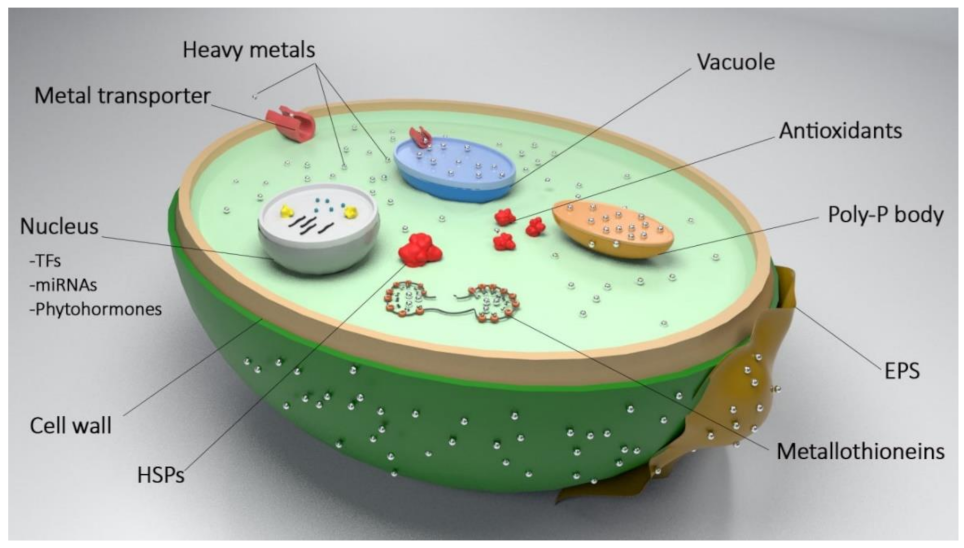
2. Cellular Mechanisms of HM Bioremediation in Microalgae
3. Genetic and Metabolic Engineering Tools for Microalgal Strain Improvement
4. Genetic Engineering Targets to Improve Microalgal HM Bioremediation Capacity
4.1. Metal Transportation
4.2. Metal Chelation
4.3. Metal Biotransformation
4.4. Oxidative Stress Response Regulation
4.5. Metal Stress Response Regulation
4.6. Cell-Surface Bioengineering
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Danouche, M.; el-Ghachtouli, N.; el-Arroussi, H. Phycoremediation mechanisms of heavy metals using living green microalgae: Physicochemical and molecular approaches for enhancing selectivity and removal capacity. Heliyon 2021, 7, e07609. [Google Scholar] [CrossRef]
- Sibi, G. Factors influencing heavy metal removal by microalgae—A review. J. Crit. Rev. 2019, 6, 29–32. [Google Scholar]
- Kumar, V.; Kothari, R.; Kumari, S.; Kumar, P. Sustainable approaches towards wastewater treatment using algal technology along with management of post-harvest biomass. In Advances in Environmental Pollution Management: Wastewater Impacts and Treatment Technologies, 1st ed.; Agro Environ Media: Haridwar, India, 2020; Volume 1, pp. 174–187. [Google Scholar]
- Salam, K.A. Towards sustainable development of microalgal biosorption for treating effluents containing heavy metals. Biofuel Res. J. 2019, 6, 948–961. [Google Scholar] [CrossRef]
- Balzano, S.; Sardo, A.; Blasio, M.; Chahine, T.B.; dell’Anno, F.; Sansone, C.; Brunet, C. Microalgal metallothioneins and phytochelatins and their potential use in bioremediation. Front. Microbiol. 2020, 11, 517. [Google Scholar] [CrossRef] [PubMed]
- Dung, T.T.T.; Cappuyns, V.; Swennen, R.; Phung, N.K. From geochemical background determination to pollution assessment of heavy metals in sediments and soils. Rev. Environ. Sci. Bio/Technol. 2013, 12, 335–353. [Google Scholar] [CrossRef]
- Singh, M.; Kumar, J.; Singh, S.; Singh, V.; Prasad, S.; Singh, M. Adaptation strategies of plants against heavy metal toxicity: A short review. Biochem. Pharmacol. 2015, 4, 161. [Google Scholar]
- Mishra, S.; Bharagava, R.N.; More, N.; Yadav, A.; Zainith, S.; Mani, S.; Chowdhary, P. Heavy metal contamination: An alarming threat to environment and human health. In Environmental Biotechnology for Sustainable Future; Springer: Singapore, 2019; pp. 103–125. [Google Scholar]
- Bruland, K.; Lohan, M. Controls of trace metals in seawater. In Treatise on Geochemistry, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2003; pp. 23–47. [Google Scholar]
- Sunitha, M.S.; Prashant, S.; Kumar, S.A.; Rao, S.; Narasu, M.L.; Kishor, P.K. Cellular and molecular mechanisms of heavy metal tolerance in plants: A brief overview of transgenic plants overexpressing phytochelatin synthase and metallothionein genes. Plant Cell Biotechnol. Mol. Biol. 2013, 14, 33–48. [Google Scholar]
- Nriagu, J.O.; Pacyna, J.M. Quantitative assessment of worldwide contamination of air, water and soils by trace metals. Nature 1988, 333, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.; Arora, N.; Gupta, P.; Pruthi, P.A.; Poluri, K.M.; Pruthi, V. Microalgae: An emerging source for mitigation of heavy metals and their potential implications for biodiesel production. In Advanced Biofuels; Elsevier: Amsterdam, The Netherlands, 2019; pp. 97–128. [Google Scholar]
- Tchounwou, P.B.; Yedjou, C.G.; Patlolla, A.K.; Sutton, D.J. Heavy metal toxicity and the environment. Mol. Clin. Environ. Toxicol. 2012, 101, 133–164. [Google Scholar]
- Chan, A.; Salsali, H.; McBean, E. Heavy metal removal (copper and zinc) in secondary effluent from wastewater treatment plants by microalgae. ACS Sustain. Chem. Eng. 2014, 2, 130–137. [Google Scholar] [CrossRef]
- Macek, T.; Mackova, M. Potential of biosorption technology. In Microbial Biosorption of Metals; Springer: Berlin/Heidelberg, Germany, 2011; pp. 7–17. [Google Scholar]
- Ahalya, N.; Ramachandra, T.; Kanamadi, R. Biosorption of heavy metals. Reseach J. Chem. Environ. 2003, 7, 71–79. [Google Scholar]
- Gadd, G.M. Biosorption: Critical review of scientific rationale, environmental importance and significance for pollution treatment. J. Chem. Technol. Biotechnol. Int. Res. Process Environ. Clean Technol. 2009, 84, 13–28. [Google Scholar] [CrossRef]
- Eccles, H. Treatment of metal-contaminated wastes: Why select a biological process? Trends Biotechnol. 1999, 17, 462–465. [Google Scholar] [CrossRef]
- Hazen, T.C. Bioremediation. In The Microbiology of the Terrestrial Deep Subsurface; CRC Press: Boca Raton, FL, USA, 2018; pp. 247–266. [Google Scholar]
- Kumar, K.S.; Dahms, H.-U.; Won, E.-J.; Lee, J.-S.; Shin, K.-H. Microalgae—A promising tool for heavy metal remediation. Ecotoxicol. Environ. Saf. 2015, 113, 329–352. [Google Scholar] [CrossRef]
- Fu, F.; Wang, Q. Removal of heavy metal ions from wastewaters: A review. J. Environ. Manag. 2011, 92, 407–418. [Google Scholar] [CrossRef]
- Kaplan, D. Absorption and adsorption of heavy metals by microalgae. Handb. Microalgal Cult. Appl. Phycol. Biotechnol. 2013, 2, 602–611. [Google Scholar]
- John, J. Phycoremediation. In Modern Trends in Applied Aquatic Ecology; Springer: Berlin/Heidelberg, Germany, 2003; pp. 133–147. [Google Scholar]
- Rajamani, S.; Siripornadulsil, S.; Falcão, V.; Torres, M.; Colepicolo, P.; Sayre, R. Phycoremediation of heavy metals using transgenic microalgae. In Transgenic Microalgae Green Cell Factories; Springer: Berlin/Heidelberg, Germany, 2007; pp. 99–109. [Google Scholar]
- Singh, P.; Gupta, S.K.; Guldhe, A.; Rawat, I.; Bux, F. Microalgae isolation and basic culturing techniques. In Handbook of Marine Microalgae; Elsevier: Amsterdam, The Netherlands, 2015; pp. 43–54. [Google Scholar]
- Biswas, H.; Bandyopadhyay, D. Physiological responses of coastal phytoplankton (Visakhapatnam, SW Bay of Bengal, India) to experimental copper addition. Mar. Environ. Res. 2017, 131, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Levy, J.L.; Angel, B.M.; Stauber, J.L.; Poon, W.L.; Simpson, S.L.; Cheng, S.H.; Jolley, D.F. Uptake and internalisation of copper by three marine microalgae: Comparison of copper-sensitive and copper-tolerant species. Aquat. Toxicol. 2008, 89, 82–93. [Google Scholar] [CrossRef] [Green Version]
- Letry, K.A.; Castro, E.D.; Gupta, S.K.; Kumar, M. Industrial wastewater-based algal biorefineries: Application constraints and future prospects. In Application of Microalgae in Wastewater Treatment; Springer: Berlin/Heidelberg, Germany, 2019; pp. 371–392. [Google Scholar]
- Tripathi, S.; Poluri, K.M. Heavy metal detoxification mechanisms by microalgae: Insights from transcriptomics analysis. Environ. Pollut. 2021, 285, 117443. [Google Scholar] [CrossRef]
- Cheng, S.Y.; Show, P.-L.; Lau, B.F.; Chang, J.-S.; Ling, T.C. New prospects for modified algae in heavy metal adsorption. Trends Biotechnol. 2019, 37, 1255–1268. [Google Scholar] [CrossRef]
- Samadani, M.; Perreault, F.; Oukarroum, A.; Dewez, D. Effect of cadmium accumulation on green algae Chlamydomonas reinhardtii and acid-tolerant Chlamydomonas CPCC 121. Chemosphere 2018, 191, 174–182. [Google Scholar] [CrossRef]
- Dal Corso, G. Heavy metal toxicity in plants. In Plants and Heavy Metals; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–25. [Google Scholar]
- Spain, O.; Plöhn, M.; Funk, C. The cell wall of green microalgae and its role in heavy metal removal. Physiol. Plant. 2021, 173, 526–535. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, R.; Muñoz, R.; Taboada, M.E.; Vega, M.; Bolado, S. Comparative uptake study of arsenic, boron, copper, manganese and zinc from water by different green microalgae. Bioresour. Technol. 2018, 263, 49–57. [Google Scholar] [CrossRef] [Green Version]
- Leong, Y.K.; Chang, J.-S. Bioremediation of heavy metals using microalgae: Recent advances and mechanisms. Bioresour. Technol. 2020, 303, 122886. [Google Scholar] [CrossRef] [PubMed]
- Javanbakht, V.; Alavi, S.A.; Zilouei, H. Mechanisms of heavy metal removal using microorganisms as biosorbent. Water Sci. Technol. 2014, 69, 1775–1787. [Google Scholar] [CrossRef]
- Singh, D.V.; Bhat, R.A.; Upadhyay, A.K.; Singh, R.; Singh, D. Microalgae in aquatic environments: A sustainable approach for remediation of heavy metals and emerging contaminants. Environ. Technol. Innov. 2021, 21, 101340. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, F.; Liu, Y.; Huang, F.; Zhang, C. Effect of extracellular polymeric substances on arsenic accumulation in Chlorella pyrenoidosa. Sci. Total Environ. 2020, 704, 135368. [Google Scholar] [CrossRef]
- Zeraatkar, A.K.; Ahmadzadeh, H.; Talebi, A.F.; Moheimani, N.R.; McHenry, M.P. Potential use of algae for heavy metal bioremediation, a critical review. J. Environ. Manag. 2016, 181, 817–831. [Google Scholar] [CrossRef] [PubMed]
- Naveed, S.; Li, C.; Lu, X.; Chen, S.; Yin, B.; Zhang, C.; Ge, Y. Microalgal extracellular polymeric substances and their interactions with metal(loid)s: A review. Crit. Rev. Environ. Sci. Technol. 2019, 49, 1769–1802. [Google Scholar] [CrossRef]
- Ubando, A.T.; Africa, A.D.M.; Maniquiz-Redillas, M.C.; Culaba, A.B.; Chen, W.-H.; Chang, J.-S. Microalgal biosorption of heavy metals: A comprehensive bibliometric review. J. Hazard. Mater. 2021, 402, 123431. [Google Scholar] [CrossRef] [PubMed]
- Arunakumara, K.; Zhang, X. Heavy metal bioaccumulation and toxicity with special reference to microalgae. J. Ocean Univ. China 2008, 7, 60–64. [Google Scholar] [CrossRef]
- Mantzorou, A.; Navakoudis, E.; Paschalidis, K.; Ververidis, F. Microalgae: A potential tool for remediating aquatic environments from toxic metals. Int. J. Environ. Sci. Technol. 2018, 15, 1815–1830. [Google Scholar] [CrossRef]
- García-García, J.D.; Sánchez-Thomas, R.; Moreno-Sánchez, R. Bio-recovery of non-essential heavy metals by intra- and extracellular mechanisms in free-living microorganisms. Biotechnol. Adv. 2016, 34, 859–873. [Google Scholar] [CrossRef]
- Monteiro, C.M.; Castro, P.M.; Malcata, F.X. Metal uptake by microalgae: Underlying mechanisms and practical applications. Biotechnol. Prog. 2012, 28, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Worms, I.; Simon, D.F.; Hassler, C.; Wilkinson, K. Bioavailability of trace metals to aquatic microorganisms: Importance of chemical, biological and physical processes on biouptake. Biochimie 2006, 88, 1721–1731. [Google Scholar] [CrossRef]
- Hirata, K.; Tsuji, N.; Miyamoto, K. Biosynthetic regulation of phytochelatins, heavy metal-binding peptides. J. Biosci. Bioeng. 2005, 100, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Penen, F.; Isaure, M.P.; Dobritzsch, D.; Bertalan, I.; Castillo-Michel, H.; Proux, O.; Gontier, E.; le Coustumer, P.; Schaumlöffel, D. Pools of cadmium in Chlamydomonas reinhardtii revealed by chemical imaging and XAS spectroscopy. Metallomics 2017, 9, 910–923. [Google Scholar] [CrossRef]
- Perales-Vela, H.V.; Peña-Castro, J.M.; Cañizares-Villanueva, R.O. Heavy metal detoxification in eukaryotic microalgae. Chemosphere 2006, 64, 1–10. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]
- Tukaj, S.; Tukaj, Z. Distinct chemical contaminants induce the synthesis of Hsp70 proteins in green microalgae Desmodesmus subspicatus: Heat pretreatment increases cadmium resistance. J. Therm. Biol. 2010, 35, 239–244. [Google Scholar] [CrossRef]
- Gielen, H.; Remans, T.; Vangronsveld, J.; Cuypers, A. MicroRNAs in metal stress: Specific roles or secondary responses? Int. J. Mol. Sci. 2012, 13, 15826–15847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.E.; Jin, X.F.; Feng, Y.; Islam, E. Molecular mechanisms and genetic basis of heavy metal tolerance/hyperaccumulation in plants. J. Integr. Plant Biol. 2005, 47, 1025–1035. [Google Scholar] [CrossRef]
- Kebeish, R.; el-Ayouty, Y.; Husain, A. Effect of copper on growth, bioactive metabolites, antioxidant enzymes and photosynthesis-related gene transcription in Chlorella vulgaris. World J. Biol. Biol. Sci. 2014, 2, 34–43. [Google Scholar]
- Li, M.; Hu, C.; Zhu, Q.; Chen, L.; Kong, Z.; Liu, Z. Copper and zinc induction of lipid peroxidation and effects on antioxidant enzyme activities in the microalga Pavlova viridis (Prymnesiophyceae). Chemosphere 2006, 62, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.N.; Kisiala, A.B.; Emery, R.N. The roles of phytohormones in metal stress regulation in microalgae. J. Appl. Phycol. 2020, 32, 3817–3829. [Google Scholar] [CrossRef]
- Kumar, G.; Shekh, A.; Jakhu, S.; Sharma, Y.; Kapoor, R.; Sharma, T.R. Bioengineering of microalgae: Recent advances, perspectives, and regulatory challenges for industrial application. Front. Bioeng. Biotechnol. 2020, 8, 914. [Google Scholar] [CrossRef]
- Guihéneuf, F.; Khan, A.; Tran, L.-S.P. Genetic engineering: A promising tool to engender physiological, biochemical, and molecular stress resilience in green microalgae. Front. Plant Sci. 2016, 7, 400. [Google Scholar] [CrossRef] [Green Version]
- Fajardo, C.; de Donato, M.; Carrasco, R.; Martínez-Rodríguez, G.; Mancera, J.M.; Fernández-Acero, F.J. Advances and challenges in genetic engineering of microalgae. Rev. Aquac. 2020, 12, 365–381. [Google Scholar] [CrossRef]
- Poliner, E.; Clark, E.; Cummings, C.; Benning, C.; Farre, E.M. A high-capacity gene stacking toolkit for the oleaginous microalga, Nannochloropsis oceanica CCMP1779. Algal Res. 2020, 45, 101664. [Google Scholar] [CrossRef]
- Crozet, P.; Navarro, F.J.; Willmund, F.; Mehrshahi, P.; Bakowski, K.; Lauersen, K.J.; Pérez-Pérez, M.-E.; Auroy, P.; Gorchs Rovira, A.; Sauret-Gueto, S. Birth of a photosynthetic chassis: A MoClo toolkit enabling synthetic biology in the microalga Chlamydomonas reinhardtii. ACS Synth. Biol. 2018, 7, 2074–2086. [Google Scholar] [CrossRef] [Green Version]
- Adler-Agnon, Z.; Leu, S.; Zarka, A.; Boussiba, S.; Khozin-Goldberg, I. Novel promoters for constitutive and inducible expression of transgenes in the diatom Phaeodactylum tricornutum under varied nitrate availability. J. Appl. Phycol. 2018, 30, 2763–2772. [Google Scholar] [CrossRef]
- Shin, J.-H.; Choi, J.; Jeon, J.; Kumar, M.; Lee, J.; Jeong, W.-J.; Kim, S.-R. The establishment of new protein expression system using N starvation inducible promoters in Chlorella. Sci. Rep. 2020, 10, 12713. [Google Scholar] [CrossRef] [PubMed]
- Doron, L.; Segal, N.A.; Shapira, M. Transgene expression in microalgae—From tools to applications. Front. Plant Sci. 2016, 7, 505. [Google Scholar] [CrossRef] [PubMed]
- Ng, I.S.; Keskin, B.B.; Tan, S.I. A critical review of genome editing and synthetic biology applications in metabolic engineering of microalgae and cyanobacteria. Biotechnol. J. 2020, 15, 1900228. [Google Scholar] [CrossRef]
- Sinha, R.; Shukla, P. Current trends in protein engineering: Updates and progress. Curr. Protein Pept. Sci. 2019, 20, 398–407. [Google Scholar] [CrossRef]
- Yang, K.K.; Wu, Z.; Arnold, F.H. Machine-learning-guided directed evolution for protein engineering. Nat. Methods 2019, 16, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Pillai, S.; Behra, R.; Nestler, H.; Suter, M.J.-F.; Sigg, L.; Schirmer, K. Linking toxicity and adaptive responses across the transcriptome, proteome, and phenotype of Chlamydomonas reinhardtii exposed to silver. Proc. Natl. Acad. Sci. USA 2014, 111, 3490–3495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, A.; Medhi, K.; Malaviya, P.; Thakur, I.S. Omics approaches for microalgal applications: Prospects and challenges. Bioresour. Technol. 2019, 291, 121890. [Google Scholar] [CrossRef]
- El-Sheekh, M.; el-Dalatony, M.; Thakur, N.; Zheng, Y.; Salama, E.-S. Role of microalgae and cyanobacteria in wastewater treatment: Genetic engineering and omics approaches. Int. J. Environ. Sci. Technol. 2021, 18, 1–22. [Google Scholar] [CrossRef]
- Hainline, B. Algomics for the development of a sustainable microalgae biorefinery. Single Cell Biol. 2016, 5, 2. [Google Scholar]
- May, P.; Wienkoop, S.; Kempa, S.; Usadel, B.; Christian, N.; Rupprecht, J.; Weiss, J.; Recuenco-Munoz, L.; Ebenhöh, O.; Weckwerth, W. Metabolomics- and proteomics-assisted genome annotation and analysis of the draft metabolic network of Chlamydomonas reinhardtii. Genetics 2008, 179, 157–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Lopez, D.; Casero, D.; Cokus, S.J.; Merchant, S.S.; Pellegrini, M. Algal functional annotation tool: A web-based analysis suite to functionally interpret large gene lists using integrated annotation and expression data. BMC Bioinform. 2011, 12, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Dal’Molin, C.G.; Nielsen, L.K. Algae genome-scale reconstruction, modelling and applications. In The Physiology of Microalgae; Springer: Berlin/Heidelberg, Germany, 2016; pp. 591–598. [Google Scholar]
- Boyle, N.R.; Morgan, J.A. Flux balance analysis of primary metabolism in Chlamydomonas reinhardtii. BMC Syst. Biol. 2009, 3, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, N.; Wang, L.; Kang, Y.; Luo, K.; Zeng, D.; Man, Y.B.; Zhang, Q.; Zeng, L.; Luo, J.; Jiang, F. The comparison of transcriptomic response of green microalga Chlorella sorokiniana exposure to environmentally relevant concentration of cadmium (II) and 4-n-nonylphenol. Environ. Geochem. Health 2020, 42, 2881–2894. [Google Scholar] [CrossRef]
- Jamers, A.; Blust, R.; de Coen, W.; Griffin, J.L.; Jones, O.A. Copper toxicity in the microalga Chlamydomonas reinhardtii: An integrated approach. Biometals 2013, 26, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Jamers, A.; Blust, R.; de Coen, W.; Griffin, J.L.; Jones, O.A. An omics based assessment of cadmium toxicity in the green alga Chlamydomonas reinhardtii. Aquat. Toxicol. 2013, 126, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Olsson, S.; Puente-Sanchez, F.; Gómez, M.J.; Aguilera, A. Transcriptional response to copper excess and identification of genes involved in heavy metal tolerance in the extremophilic microalga Chlamydomonas acidophila. Extremophiles 2015, 19, 657–672. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, L.R.S.; Cao, E.P. Metagenomic analysis reveals the presence of heavy metal response genes from cyanobacteria thriving in Balatoc Mines, Benguet Province, Philippines. Philipp. J. Sci. 2019, 148, 71–82. [Google Scholar]
- Ye, J.; Song, Z.; Wang, L.; Zhu, J. Metagenomic analysis of microbiota structure evolution in phytoremediation of a swine lagoon wastewater. Bioresour. Technol. 2016, 219, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Kumari, P.; Kumar, Y. Bioinformatics and computational tools in bioremediation and biodegradation of environmental pollutants. In Bioremediation for Environmental Sustainability; Elsevier: Amsterdam, The Netherlands, 2021; pp. 421–444. [Google Scholar]
- Hanikenne, M.; Krämer, U.; Demoulin, V.; Baurain, D. A comparative inventory of metal transporters in the green alga Chlamydomonas reinhardtii and the red alga Cyanidioschizon merolae. Plant Physiol. 2005, 137, 428–446. [Google Scholar] [CrossRef] [Green Version]
- Rosakis, A.; Köster, W. Transition metal transport in the green microalga Chlamydomonas reinhardtii—Genomic sequence analysis. Res. Microbiol. 2004, 155, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Blaby-Haas, C.E.; Merchant, S.S. The ins and outs of algal metal transport. Biochim. Biophys. Acta Mol. Cell Res. 2012, 1823, 1531–1552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nevo, Y.; Nelson, N. The NRAMP family of metal-ion transporters. Biochim. Biophys. Acta Mol. Cell Res. 2006, 1763, 609–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Ma, Y.; Xing, G.; Li, W.; Kong, X.; Li, J.; Wang, L.; Yuan, H.; Yang, J. Revelation of microalgae’s lipid production and resistance mechanism to ultra-high Cd stress by integrated transcriptome and physiochemical analyses. Environ. Pollut. 2019, 250, 186–195. [Google Scholar] [CrossRef]
- Puente-Sánchez, F.; Díaz, S.; Penacho, V.; Aguilera, A.; Olsson, S. Basis of genetic adaptation to heavy metal stress in the acidophilic green alga Chlamydomonas acidophila. Aquat. Toxicol. 2018, 200, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Rosakis, A.; Köster, W. Divalent metal transport in the green microalga Chlamydomonas reinhardtii is mediated by a protein similar to prokaryotic Nramp homologues. Biometals 2005, 18, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Beauvais-Flück, R.; Slaveykova, V.I.; Cosio, C. Cellular toxicity pathways of inorganic and methyl mercury in the green microalga Chlamydomonas reinhardtii. Sci. Rep. 2017, 7, 8034. [Google Scholar] [CrossRef] [Green Version]
- Hirooka, S.; Hirose, Y.; Kanesaki, Y.; Higuchi, S.; Fujiwara, T.; Onuma, R.; Era, A.; Ohbayashi, R.; Uzuka, A.; Nozaki, H. Acidophilic green algal genome provides insights into adaptation to an acidic environment. Proc. Natl. Acad. Sci. USA 2017, 114, E8304–E8313. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Gui, H.; Luo, Z.; Zhen, Z.; Yan, C.; Xing, B. Dissolved organic phosphorus enhances arsenate bioaccumulation and biotransformation in Microcystis aeruginosa. Environ. Pollut. 2019, 252, 1755–1763. [Google Scholar] [CrossRef] [PubMed]
- Ibuot, A.; Dean, A.P.; McIntosh, O.A.; Pittman, J.K. Metal bioremediation by CrMTP4 over-expressing Chlamydomonas reinhardtii in comparison to natural wastewater-tolerant microalgae strains. Algal Res. 2017, 24, 89–96. [Google Scholar] [CrossRef]
- Osundeko, O.; Dean, A.P.; Davies, H.; Pittman, J.K. Acclimation of microalgae to wastewater environments involves increased oxidative stress tolerance activity. Plant Cell Physiol. 2014, 55, 1848–1857. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, A.C.; Argüello, J.M. Toward a molecular understanding of metal transport by P1B-Type ATPases. Curr. Top. Membr. 2012, 69, 113–136. [Google Scholar] [PubMed] [Green Version]
- Williams, L.E.; Mills, R.F. P1B-ATPases—An ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci. 2005, 10, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Ibuot, A.; Webster, R.E.; Williams, L.E.; Pittman, J.K. Increased metal tolerance and bioaccumulation of zinc and cadmium in Chlamydomonas reinhardtii expressing a AtHMA4 C-terminal domain protein. Biotechnol. Bioeng. 2020, 117, 2996–3005. [Google Scholar] [CrossRef]
- Bækgaard, L.; Mikkelsen, M.D.; Sørensen, D.M.; Hegelund, J.N.; Persson, D.P.; Mills, R.F.; Yang, Z.; Husted, S.; Andersen, J.P.; Buch-Pedersen, M.J. A combined zinc/cadmium sensor and zinc/cadmium export regulator in a heavy metal pump. J. Biol. Chem. 2010, 285, 31243–31252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verret, F.; Gravot, A.; Auroy, P.; Preveral, S.; Forestier, C.; Vavasseur, A.; Richaud, P. Heavy metal transport by AtHMA4 involves the N-terminal degenerated metal binding domain and the C-terminal His11 stretch. FEBS Lett. 2005, 579, 1515–1522. [Google Scholar] [CrossRef] [Green Version]
- Ceasar, S.A.; Lekeux, G.; Motte, P.; Xiao, Z.; Galleni, M.; Hanikenne, M. Di-cysteine residues of the Arabidopsis thaliana HMA4 C-terminus are only partially required for cadmium transport. Front. Plant Sci. 2020, 11, 560. [Google Scholar] [CrossRef]
- Lekeux, G.; Laurent, C.; Joris, M.; Jadoul, A.; Jiang, D.; Bosman, B.; Carnol, M.; Motte, P.; Xiao, Z.; Galleni, M. Di-cysteine motifs in the C-terminus of plant HMA4 proteins confer nanomolar affinity for zinc and are essential for HMA4 function in vivo. J. Exp. Bot. 2018, 69, 5547–5560. [Google Scholar] [CrossRef] [Green Version]
- Ramírez-Rodríguez, A.E.; Bañuelos-Hernández, B.; García-Soto, M.J.; Govea-Alonso, D.G.; Rosales-Mendoza, S.; Alfaro de la Torre, M.C.; Monreal-Escalante, E.; Paz-Maldonado, L.M. Arsenic removal using Chlamydomonas reinhardtii modified with the gene acr3 and enhancement of its performance by decreasing phosphate in the growing media. Int. J. Phytoremed. 2019, 21, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zeng, R.; Chen, D.; Liu, J. A pivotal role of vacuolar H+-ATPase in regulation of lipid production in Phaeodactylum tricornutum. Sci. Rep. 2016, 6, 31319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.I.; Hwang, S.-W.; Kim, J.; Park, B.; Jin, E.; Choi, I.-G.; Sim, S.J. Augmented CO2 tolerance by expressing a single H+-pump enables microalgal valorization of industrial flue gas. Nat. Commun. 2021, 12, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Simon, D.F.; Descombes, P.; Zerges, W.; Wilkinson, K.J. Global expression profiling of Chlamydomonas reinhardtii exposed to trace levels of free cadmium. Environ. Toxicol. Chem. Int. J. 2008, 27, 1668–1675. [Google Scholar] [CrossRef] [PubMed]
- Khatiwada, B.; Hasan, M.T.; Sun, A.; Kamath, K.S.; Mirzaei, M.; Sunna, A.; Nevalainen, H. Proteomic response of Euglena gracilis to heavy metal exposure—Identification of key proteins involved in heavy metal tolerance and accumulation. Algal Res. 2020, 45, 101764. [Google Scholar] [CrossRef]
- Kotrba, P.; Mackova, M.; Macek, T. Transgenic approaches to improve phytoremediation of heavy metal polluted soils. In Biomanagement of Metal-Contaminated Soils; Springer: New York, NY, USA, 2011; pp. 409–438. [Google Scholar]
- Lee, J.; Bae, H.; Jeong, J.; Lee, J.-Y.; Yang, Y.-Y.; Hwang, I.; Martinoia, E.; Lee, Y. Functional expression of a bacterial heavy metal transporter in Arabidopsis enhances resistance to and decreases uptake of heavy metals. Plant Physiol. 2003, 133, 589–596. [Google Scholar] [CrossRef] [Green Version]
- Korenkov, V.; Hirschi, K.; Crutchfield, J.D.; Wagner, G.J. Enhancing tonoplast Cd/H antiport activity increases Cd, Zn, and Mn tolerance, and impacts root/shoot Cd partitioning in Nicotiana tabacum L. Planta 2007, 226, 1379–1387. [Google Scholar] [CrossRef]
- Hirschi, K.D.; Korenkov, V.D.; Wilganowski, N.L.; Wagner, G.J. Expression of Arabidopsis CAX2 in tobacco. Altered metal accumulation and increased manganese tolerance. Plant Physiol. 2000, 124, 125–134. [Google Scholar] [CrossRef] [Green Version]
- Van der Zaal, B.J.; Neuteboom, L.W.; Pinas, J.E.; Chardonnens, A.N.; Schat, H.; Verkleij, J.A.; Hooykaas, P.J. Overexpression of a novel Arabidopsis gene related to putative zinc-transporter genes from animals can lead to enhanced zinc resistance and accumulation. Plant Physiol. 1999, 119, 1047–1056. [Google Scholar] [CrossRef] [Green Version]
- Zheng, C.; Aslam, M.; Liu, X.; Du, H.; Xie, X.; Jia, H.; Huang, N.; Tang, K.; Yang, Y.; Li, P. Impact of Pb on Chlamydomonas reinhardtii at physiological and transcriptional levels. Front. Microbiol. 2020, 11, 1443. [Google Scholar] [CrossRef]
- Puente-Sánchez, F.; Olsson, S.; Aguilera, A. Comparative transcriptomic analysis of the response of Dunaliella acidophila (Chlorophyta) to short-term cadmium and chronic natural metal-rich water exposures. Microb. Ecol. 2016, 72, 595–607. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-S.; Lu, Y.-P.; Zhen, R.-G.; Szczypka, M.; Thiele, D.J.; Rea, P.A. A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis (glutathionato) cadmium. Proc. Natl. Acad. Sci. USA 1997, 94, 42–47. [Google Scholar] [CrossRef] [Green Version]
- Song, W.-Y.; Sohn, E.J.; Martinoia, E.; Lee, Y.J.; Yang, Y.-Y.; Jasinski, M.; Forestier, C.; Hwang, I.; Lee, Y. Engineering tolerance and accumulation of lead and cadmium in transgenic plants. Nat. Biotechnol. 2003, 21, 914–919. [Google Scholar] [CrossRef]
- Yazaki, K.; Yamanaka, N.; Masuno, T.; Konagai, S.; Kaneko, S.; Ueda, K.; Sato, F. Heterologous expression of a mammalian ABC transporter in plant and its application to phytoremediation. Plant Mol. Biol. 2006, 61, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, D.F.; Ruscitti, T.; McCue, K.F.; Ow, D.W. Transport of metal-binding peptides by HMT1, a fission yeast ABC-type vacuolar membrane protein. J. Biol. Chem. 1995, 270, 4721–4728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortiz, D.; Kreppel, L.; Speiser, D.; Scheel, G.; McDonald, G.; Ow, D. Heavy metal tolerance in the fission yeast requires an ATP-binding cassette-type vacuolar membrane transporter. EMBO J. 1992, 11, 3491–3499. [Google Scholar] [CrossRef]
- Cobbett, C.; Goldsbrough, P. Phytochelatins and metallothioneins: Roles in heavy metal detoxification and homeostasis. Annu. Rev. Plant Biol. 2002, 53, 159–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capdevila, M.; Atrian, S. Metallothionein protein evolution: A miniassay. J. Biol. Inorg. Chem. 2011, 16, 977–989. [Google Scholar] [CrossRef] [PubMed]
- Leszczyszyn, O.I.; White, C.R.J.; Blindauer, C.A. The isolated Cys 2 His 2 site in EC metallothionein mediates metal-specific protein folding. Mol. Biosyst. 2010, 6, 1592–1603. [Google Scholar] [CrossRef] [PubMed]
- Ziller, A.; Fraissinet-Tachet, L. Metallothionein diversity and distribution in the tree of life: A multifunctional protein. Metallomics 2018, 10, 1549–1559. [Google Scholar] [CrossRef]
- Gaur, J.; Rai, L. Heavy metal tolerance in algae. In Algal Adaptation to Environmental Stresses; Springer: New York, NY, USA, 2001; pp. 363–388. [Google Scholar]
- Ahner, B.A.; Wei, L.; Oleson, J.R.; Ogura, N. Glutathione and other low molecular weight thiols in marine phytoplankton under metal stress. Mar. Ecol. Prog. Ser. 2002, 232, 93–103. [Google Scholar] [CrossRef] [Green Version]
- Meister, A.; Anderson, M.E. Glutathione. Annu. Rev. Biochem. 1983, 52, 711–760. [Google Scholar] [CrossRef]
- Tsuji, N.; Hirayanagi, N.; Iwabe, O.; Namba, T.; Tagawa, M.; Miyamoto, S.; Miyasaka, H.; Takagi, M.; Hirata, K.; Miyamoto, K. Regulation of phytochelatin synthesis by zinc and cadmium in marine green alga, Dunaliella tertiolecta. Phytochem. 2003, 62, 453–459. [Google Scholar] [CrossRef]
- Hirata, K.; Tsujimoto, Y.; Namba, T.; Ohta, T.; Hirayanagi, N.; Miyasaka, H.; Zenk, M.H.; Miyamoto, K. Strong induction of phytochelatin synthesis by zinc in marine green alga, Dunaliella tertiolecta. J. Biosci. Bioeng. 2001, 92, 24–29. [Google Scholar] [CrossRef]
- Suárez, C.; Torres, E.; Pérez-Rama, M.; Herrero, C.; Abalde, J. Cadmium toxicity on the freshwater microalga Chlamydomonas moewusii Gerloff: Biosynthesis of thiol compounds. Environ. Toxicol. Chem. 2010, 29, 2009–2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Zheng, C.; Fu, H.; Zhai, S.; Hu, F.; Naveed, S.; Zhang, C.; Ge, Y. Contrasting detoxification mechanisms of Chlamydomonas reinhardtii under Cd and Pb stress. Chemosphere 2021, 274, 129771. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, I.; Fujiwara, S.; Saegusa, H.; Inouhe, M.; Matsumoto, H.; Tsuzuki, M. Relief of arsenate toxicity by Cd-stimulated phytochelatin synthesis in the green alga Chlamydomonas reinhardtii. Mar. Biotechnol. 2006, 8, 94–101. [Google Scholar] [CrossRef]
- Abboud, P.; Wilkinson, K.J. Role of metal mixtures (Ca, Cu and Pb) on Cd bioaccumulation and phytochelatin production by Chlamydomonas reinhardtii. Environ. Pollut. 2013, 179, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Pawlik-Skowrońska, B. Correlations between toxic Pb effects and production of Pb-induced thiol peptides in the microalga Stichococcus bacillaris. Environ. Pollut. 2002, 119, 119–127. [Google Scholar] [CrossRef]
- Gómez-Jacinto, V.; García-Barrera, T.; Gómez-Ariza, J.L.; Garbayo-Nores, I.; Vílchez-Lobato, C. Elucidation of the defence mechanism in microalgae Chlorella sorokiniana under mercury exposure. Identification of Hg–phytochelatins. Chem. Biol. Interact. 2015, 238, 82–90. [Google Scholar] [CrossRef]
- Nagalakshmi, N.; Prasad, M. Responses of glutathione cycle enzymes and glutathione metabolism to copper stress in Scenedesmus bijugatus. Plant Sci. 2001, 160, 291–299. [Google Scholar] [CrossRef]
- Gekeler, W.; Grill, E.; Winnacker, E.-L.; Zenk, M.H. Algae sequester heavy metals via synthesis of phytochelatin complexes. Arch. Microbiol. 1988, 150, 197–202. [Google Scholar] [CrossRef]
- Liang Zhu, Y.; Pilon-Smits, E.A.; Jouanin, L.; Terry, N. Overexpression of glutathione synthetase in Indian mustard enhances cadmium accumulation and tolerance. Plant Physiol. 1999, 119, 73–80. [Google Scholar] [CrossRef] [Green Version]
- Piña-Olavide, R.; Paz-Maldonado, L.M.; de La Torre, M.C.A.; García-Soto, M.J.; Ramírez-Rodríguez, A.E.; Rosales-Mendoza, S.; Bañuelos-Hernández, B.; de la-Cruz, R.F.G. Increased removal of cadmium by Chlamydomonas reinhardtii modified with a synthetic gene for γ-glutamylcysteine synthetase. Int. J. Phytoremediat. 2020, 22, 1269–1277. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cheng, Z.Z.; Chen, X.; Zheng, Q.; Yang, Z.M. CrGNAT gene regulates excess copper accumulation and tolerance in Chlamydomonas reinhardtii. Plant Sci. 2015, 240, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Martínez, M.; Bernal, P.; Almela, C.; Vélez, D.; García-Agustín, P.; Serrano, R.; Navarro-Aviñó, J. An engineered plant that accumulates higher levels of heavy metals than Thlaspi caerulescens, with yields of 100 times more biomass in mine soils. Chemosphere 2006, 64, 478–485. [Google Scholar] [CrossRef]
- Chaurasia, N.; Mishra, Y.; Rai, L.C. Cloning expression and analysis of phytochelatin synthase (pcs) gene from Anabaena sp. PCC 7120 offering multiple stress tolerance in Escherichia coli. Biochem. Biophys. Res. Commun. 2008, 376, 225–230. [Google Scholar] [CrossRef]
- Picault, N.; Cazalé, A.; Beyly, A.; Cuiné, S.; Carrier, P.; Luu, D.; Forestier, C.; Peltier, G. Chloroplast targeting of phytochelatin synthase in Arabidopsis: Effects on heavy metal tolerance and accumulation. Biochimie 2006, 88, 1743–1750. [Google Scholar] [CrossRef]
- Pal, R.; Rai, J. Phytochelatins: Peptides involved in heavy metal detoxification. Appl. Biochem. Biotechnol. 2010, 160, 945–963. [Google Scholar] [CrossRef]
- Leszczyszyn, O.I.; Imam, H.T.; Blindauer, C.A. Diversity and distribution of plant metallothioneins: A review of structure, properties and functions. Metallomics 2013, 5, 1146–1169. [Google Scholar] [CrossRef]
- Gutierrez, J.C.; Amaro, F.; Martín-González, A. From heavy metal-binders to biosensors: Ciliate metallothioneins discussed. BioEssays 2009, 31, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.C.; Amaro, F.; Diaz, S.; de Francisco, P.; Cubas, L.; Martín-González, A. Ciliate metallothioneins: Unique microbial eukaryotic heavy-metal-binder molecules. J. Biol. Inorg. Chem. 2011, 16, 1025–1034. [Google Scholar] [CrossRef]
- Kawakami, S.K.; Gledhill, M.; Achterberg, E.P. Production of phytochelatins and glutathione by marine phytoplankton in response to metal stress 1. J. Phycol. 2006, 42, 975–989. [Google Scholar] [CrossRef]
- Cai, X.-H.; Brown, C.; Adhiya, J.; Traina, S.J.; Sayre, R.T. Growth and heavy metal binding properties of transgenic Chlamydomonas expressing a foreign metallothionein gene. Int. J. Phytoremediat. 1999, 1, 53–65. [Google Scholar] [CrossRef]
- He, Z.; Siripornadulsil, S.; Sayre, R.T.; Traina, S.J.; Weavers, L.K. Removal of mercury from sediment by ultrasound combined with biomass (transgenic Chlamydomonas reinhardtii). Chemosphere 2011, 83, 1249–1254. [Google Scholar] [CrossRef]
- Domínguez-Solís, J.R.; López-Martín, M.C.; Ager, F.J.; Ynsa, M.D.; Romero, L.C.; Gotor, C. Increased cysteine availability is essential for cadmium tolerance and accumulation in Arabidopsis thaliana. Plant Biotechnol. J. 2004, 2, 469–476. [Google Scholar] [CrossRef] [Green Version]
- Kawashima, C.G.; Noji, M.; Nakamura, M.; Ogra, Y.; Suzuki, K.T.; Saito, K. Heavy metal tolerance of transgenic tobacco plants over-expressing cysteine synthase. Biotechnol. Lett. 2004, 26, 153–157. [Google Scholar] [CrossRef]
- Freeman, J.L.; Salt, D.E. The metal tolerance profile of Thlaspi goesingense is mimicked in Arabidopsis thaliana heterologously expressing serine acetyl-transferase. BMC Plant Biol. 2007, 7, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Wangeline, A.L.; Burkhead, J.L.; Hale, K.L.; Lindblom, S.D.; Terry, N.; Pilon, M.; Pilon-Smits, E.A. Overexpression of ATP sulfurylase in Indian mustard: Effects on tolerance and accumulation of twelve metals. J. Environ. Qual. 2004, 33, 54–60. [Google Scholar] [CrossRef] [Green Version]
- Torres, E.; Cid, A.; Fidalgo, P.; Herrero, C.; Abalde, J. Long-chain class III metallothioneins as a mechanism of cadmium tolerance in the marine diatom Phaeodactylum tricornutum Bohlin. Aquat. Toxicol. 1997, 39, 231–246. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Rama, M.; López, C.H.; Alonso, J.A.; Vaamonde, E.T. Class III metallothioneins in response to cadmium toxicity in the marine microalga Tetraselmis suecica (Kylin) Butch. Environ. Toxicol. Chem. Int. J. 2001, 20, 2061–2066. [Google Scholar] [CrossRef]
- Tsednee, M.; Castruita, M.; Salomé, P.A.; Sharma, A.; Lewis, B.E.; Schmollinger, S.R.; Strenkert, D.; Holbrook, K.; Otegui, M.S.; Khatua, K. Manganese co-localizes with calcium and phosphorus in Chlamydomonas acidocalcisomes and is mobilized in manganese-deficient conditions. J. Biol. Chem. 2019, 294, 17626–17641. [Google Scholar] [CrossRef] [PubMed]
- Goodenough, U.; Heiss, A.A.; Roth, R.; Rusch, J.; Lee, J.-H. Acidocalcisomes: Ultrastructure, biogenesis, and distribution in microbial eukaryotes. Protist 2019, 170, 287–313. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, F.A.; Marchesini, N.; Seufferheld, M.; Docampo, R. The polyphosphate bodies of Chlamydomonas reinhardtii possess a proton-pumping pyrophosphatase and are similar to acidocalcisomes. J. Biol. Chem. 2001, 276, 46196–46203. [Google Scholar] [CrossRef] [Green Version]
- Sanz-Luque, E.; Bhaya, D.; Grossman, A.R. Polyphosphate: A multifunctional metabolite in cyanobacteria and algae. Front. Plant Sci. 2020, 11, 938. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-X.; Dei, R.C. Metal stoichiometry in predicting Cd and Cu toxicity to a freshwater green alga Chlamydomonas reinhardtii. Environ. Pollut. 2006, 142, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Rao, N.N.; Gómez-García, M.R.; Kornberg, A. Inorganic polyphosphate: Essential for growth and survival. Annu. Rev. Biochem. 2009, 78, 605–647. [Google Scholar] [CrossRef]
- Gao, F.; Wu, H.; Zeng, M.; Huang, M.; Feng, G. Overproduction, purification, and characterization of nanosized polyphosphate bodies from Synechococcus sp. PCC 7002. Microb. Cell Factories 2018, 17, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.; Loubéry, S.; Broger, L.; Zhang, Y.; Lorenzo-Orts, L.; Utz-Pugin, A.; Fernie, A.R.; Young-Tae, C.; Hothorn, M. A genetically validated approach for detecting inorganic polyphosphates in plants. Plant J. 2020, 102, 507–516. [Google Scholar] [CrossRef]
- Gerasimaitė, R.; Sharma, S.; Desfougeres, Y.; Schmidt, A.; Mayer, A. Coupled synthesis and translocation restrains polyphosphate to acidocalcisome-like vacuoles and prevents its toxicity. J. Cell Sci. 2014, 127, 5093–5104. [Google Scholar] [CrossRef] [Green Version]
- Aksoy, M.; Pootakham, W.; Grossman, A.R. Critical function of a Chlamydomonas reinhardtii putative polyphosphate polymerase subunit during nutrient deprivation. Plant Cell 2014, 26, 4214–4229. [Google Scholar] [CrossRef] [Green Version]
- Chia, M.A.; Lombardi, A.T.; da Graça Gama Melão, M.; Parrish, C.C. Combined nitrogen limitation and cadmium stress stimulate total carbohydrates, lipids, protein and amino acid accumulation in Chlorella vulgaris (Trebouxiophyceae). Aquat. Toxicol. 2015, 160, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-L.; Lan, S.-S.; Gong, M. Hydrogen peroxide-induced proline and metabolic pathway of its accumulation in maize seedlings. J. Plant Physiol. 2009, 166, 1694–1699. [Google Scholar] [CrossRef]
- Szabados, L.; Savouré, A. Proline: A multifunctional amino acid. Trends Plant Sci. 2010, 15, 89–97. [Google Scholar] [CrossRef]
- Farago, M.; ME, F.; WA, M. Plants which accumulate metals. IV. A possible copper-proline complex from the roots of Armeria maritima. Inorg. Chim. Acta 1979, 32, L93–L94. [Google Scholar] [CrossRef]
- Siripornadulsil, S.; Traina, S.; Verma, D.P.S.; Sayre, R.T. Molecular mechanisms of proline-mediated tolerance to toxic heavy metals in transgenic microalgae. Plant Cell 2002, 14, 2837–2847. [Google Scholar] [CrossRef] [PubMed]
- Kolodyazhnaya, Y.S.; Titov, S.; Kochetov, A.; Trifonova, E.; Romanova, A.; Komarova, M.; Koval, V.; Shumny, V. Tobacco transformants expressing antisense sequence of proline dehydrogenase gene possess tolerance to heavy metals. Russ. J. Genet. 2007, 43, 825–828. [Google Scholar] [CrossRef]
- Zheng, Q.; Cheng, Z.Z.; Yang, Z.M. HISN3 mediates adaptive response of Chlamydomonas reinhardtii to excess nickel. Plant Cell Physiol. 2013, 54, 1951–1962. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.F.; Ryan, P.R.; Delhaize, E. Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci. 2001, 6, 273–278. [Google Scholar] [CrossRef]
- Boominathan, R.; Doran, P.M. Organic acid complexation, heavy metal distribution and the effect of ATPase inhibition in hairy roots of hyperaccumulator plant species. J. Biotechnol. 2003, 101, 131–146. [Google Scholar] [CrossRef]
- Berg, C.V.d.; Wong, P.; Chau, Y. Measurement of complexing materials excreted from algae and their ability to ameliorate copper toxicity. J. Fish. Board Can. 1979, 36, 901–905. [Google Scholar] [CrossRef]
- McKnight, D.M.; Morel, F.M. Release of weak and strong copper-complexing agents by algae 1. Limnol. Oceanogr. 1979, 24, 823–837. [Google Scholar] [CrossRef]
- Yen, H.-W.; Chen, P.-W.; Hsu, C.-Y.; Lee, L. The use of autotrophic Chlorella vulgaris in chromium (VI) reduction under different reduction conditions. J. Taiwan Inst. Chem. Eng. 2017, 74, 1–6. [Google Scholar] [CrossRef]
- Lee, L.; Hsu, C.-Y.; Yen, H.-W. The effects of hydraulic retention time (HRT) on chromium (VI) reduction using autotrophic cultivation of Chlorella vulgaris. Bioprocess Biosyst. Eng. 2017, 40, 1725–1731. [Google Scholar] [CrossRef] [PubMed]
- Dhankher, O.P.; Li, Y.; Rosen, B.P.; Shi, J.; Salt, D.; Senecoff, J.F.; Sashti, N.A.; Meagher, R.B. Engineering tolerance and hyperaccumulation of arsenic in plants by combining arsenate reductase and γ-glutamylcysteine synthetase expression. Nat. Biotechnol. 2002, 20, 1140–1145. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Wang, L.; Duan, G.; Sun, G. Characterization of arsenate transformation and identification of arsenate reductase in a green alga Chlamydomonas reinhardtii. J. Environ. Sci. 2011, 23, 1186–1193. [Google Scholar] [CrossRef]
- Arora, N.; Gulati, K.; Tripathi, S.; Pruthi, V.; Poluri, K.M. Algae as a budding tool for mitigation of arsenic from aquatic systems. In Mechanisms of Arsenic Toxicity and Tolerance in Plants; Springer: Berlin/Heidelberg, Germany, 2018; pp. 269–297. [Google Scholar]
- Wang, Y.; Wang, S.; Xu, P.; Liu, C.; Liu, M.; Wang, Y.; Wang, C.; Zhang, C.; Ge, Y. Review of arsenic speciation, toxicity and metabolism in microalgae. Rev. Environ. Sci. Bio/Technol. 2015, 14, 427–451. [Google Scholar] [CrossRef]
- Levy, J.L.; Stauber, J.L.; Adams, M.S.; Maher, W.A.; Kirby, J.K.; Jolley, D.F. Toxicity, biotransformation, and mode of action of arsenic in two freshwater microalgae (Chlorella sp. and Monoraphidium arcuatum). Environ. Toxicol. Chem. Int. J. 2005, 24, 2630–2639. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, H.; Sohrin, Y.; Seki, K.; Sato, M.; Norisuye, K.; Naito, K.; Matsui, M. Biosynthesis and release of methylarsenic compounds during the growth of freshwater algae. Chemosphere 2001, 43, 265–272. [Google Scholar] [CrossRef] [Green Version]
- Karadjova, I.B.; Slaveykova, V.I.; Tsalev, D.L. The biouptake and toxicity of arsenic species on the green microalga Chlorella salina in seawater. Aquat. Toxicol. 2008, 87, 264–271. [Google Scholar] [CrossRef]
- Kelly, D.J.; Budd, K.; Lefebvre, D.D. Biotransformation of mercury in pH-stat cultures of eukaryotic freshwater algae. Arch. Microbiol. 2007, 187, 45–53. [Google Scholar] [CrossRef]
- Huang, C.-C.; Chen, M.-W.; Hsieh, J.-L.; Lin, W.-H.; Chen, P.-C.; Chien, L.-F. Expression of mercuric reductase from Bacillus megaterium MB1 in eukaryotic microalga Chlorella sp. DT: An approach for mercury phytoremediation. Appl. Microbiol. Biotechnol. 2006, 72, 197–205. [Google Scholar] [CrossRef] [Green Version]
- Bizily, S.P.; Kim, T.; Kandasamy, M.K.; Meagher, R.B. Subcellular targeting of methylmercury lyase enhances its specific activity for organic mercury detoxification in plants. Plant Physiol. 2003, 131, 463–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bizily, S.P.; Rugh, C.L.; Meagher, R.B. Phytodetoxification of hazardous organomercurials by genetically engineered plants. Nat. Biotechnol. 2000, 18, 213–217. [Google Scholar] [CrossRef]
- Yang, H.; Nairn, J.; Ozias-Akins, P. Transformation of peanut using a modified bacterial mercuric ion reductase gene driven by an actin promoter from Arabidopsis thaliana. J. Plant Physiol. 2003, 160, 945–952. [Google Scholar] [CrossRef]
- Heaton, A.C.; Rugh, C.L.; Kim, T.; Wang, N.J.; Meagher, R.B. Toward detoxifying mercury-polluted aquatic sediments with rice genetically engineered for mercury resistance. Environ. Toxicol. Chem. Int. J. 2003, 22, 2940–2947. [Google Scholar] [CrossRef] [PubMed]
- Czakó-Vér, K.; Batiè, M.; Raspor, P.; Sipiczki, M.; Pesti, M. Hexavalent chromium uptake by sensitive and tolerant mutants of Schizosaccharomyces pombe. FEMS Microbiol. Lett. 1999, 178, 109–115. [Google Scholar] [CrossRef]
- Rugh, C.L.; Wilde, H.D.; Stack, N.M.; Thompson, D.M.; Summers, A.O.; Meagher, R.B. Mercuric ion reduction and resistance in transgenic Arabidopsis thaliana plants expressing a modified bacterial merA gene. Proc. Natl. Acad. Sci. USA 1996, 93, 3182–3187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rugh, C.L.; Senecoff, J.F.; Meagher, R.B.; Merkle, S.A. Development of transgenic yellow poplar for mercury phytoremediation. Nat. Biotechnol. 1998, 16, 925–928. [Google Scholar] [CrossRef]
- Che, D.; Meagher, R.B.; Heaton, A.C.; Lima, A.; Rugh, C.L.; Merkle, S.A. Expression of mercuric ion reductase in Eastern cottonwood (Populus deltoides) confers mercuric ion reduction and resistance. Plant Biotechnol. J. 2003, 1, 311–319. [Google Scholar] [CrossRef]
- Lyyra, S.; Meagher, R.B.; Kim, T.; Heaton, A.; Montello, P.; Balish, R.S.; Merkle, S.A. Coupling two mercury resistance genes in Eastern cottonwood enhances the processing of organomercury. Plant Biotechnol. J. 2007, 5, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.; Zaidi, A.; Goel, R.; Musarrat, J. Biomanagement of Metal-Contaminated Soils, 1st ed.; Springer: New York, NY, USA, 2011; pp. 409–438. [Google Scholar]
- Hussein, H.S.; Ruiz, O.N.; Terry, N.; Daniell, H. Phytoremediation of mercury and organomercurials in chloroplast transgenic plants: Enhanced root uptake, translocation to shoots, and volatilization. Environ. Sci. Technol. 2007, 41, 8439–8446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz, O.N.; Hussein, H.S.; Terry, N.; Daniell, H. Phytoremediation of organomercurial compounds via chloroplast genetic engineering. Plant Physiol. 2003, 132, 1344–1352. [Google Scholar] [CrossRef] [Green Version]
- Ruiz, O.N.; Daniell, H. Genetic engineering to enhance mercury phytoremediation. Curr. Opin. Biotechnol. 2009, 20, 213–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silver, S.; Phung, L.T. A bacterial view of the periodic table: Genes and proteins for toxic inorganic ions. J. Ind. Microbiol. Biotechnol. 2005, 32, 587–605. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.D.; Li, M.; Lee, S.-B.; Schnell, D.; Daniell, H. Arabidopsis Tic40 expression in tobacco chloroplasts results in massive proliferation of the inner envelope membrane and upregulation of associated proteins. Plant Cell 2008, 20, 3405–3417. [Google Scholar] [CrossRef] [Green Version]
- Neuhierl, B.; Thanbichler, M.; Lottspeich, F.; Bock, A. A family of S-methylmethionine-dependent thiol/selenol methyltransferases: Role in selenium tolerance and evolutionary relation. J. Biol. Chem. 1999, 274, 5407–5414. [Google Scholar] [CrossRef] [Green Version]
- Bañuelos, G.; Leduc, D.L.; Pilon-Smits, E.A.; Terry, N. Transgenic Indian mustard overexpressing selenocysteine lyase or selenocysteine methyltransferase exhibit enhanced potential for selenium phytoremediation under field conditions. Environ. Sci. Technol. 2007, 41, 599–605. [Google Scholar] [CrossRef]
- LeDuc, D.L.; Tarun, A.S.; Montes-Bayon, M.; Meija, J.; Malit, M.F.; Wu, C.P.; Abdel-Samie, M.; Chiang, C.-Y.; Tagmount, A.; de Souza, M. Overexpression of selenocysteine methyltransferase in Arabidopsis and Indian mustard increases selenium tolerance and accumulation. Plant Physiol. 2004, 135, 377–383. [Google Scholar] [CrossRef] [Green Version]
- Rai, A.; Singh, R.; Shirke, P.A.; Tripathi, R.D.; Trivedi, P.K.; Chakrabarty, D. Expression of rice CYP450-like gene (Os08g01480) in Arabidopsis modulates regulatory network leading to heavy metal and other abiotic stress tolerance. PLoS ONE 2015, 10, e0138574. [Google Scholar] [CrossRef]
- Deng, L.; Wang, H.; Deng, N. Photoreduction of chromium (VI) in the presence of algae, Chlorella vulgaris. J. Hazard. Mater. 2006, 138, 288–292. [Google Scholar] [CrossRef]
- Chaudhary, R.; Nawaz, K.; Khan, A.K.; Hano, C.; Abbasi, B.H.; Anjum, S. An overview of the algae-mediated biosynthesis of nanoparticles and their biomedical applications. Biomolecules 2020, 10, 1498. [Google Scholar] [CrossRef] [PubMed]
- Priyadarshini, E.; Priyadarshini, S.S.; Pradhan, N. Heavy metal resistance in algae and its application for metal nanoparticle synthesis. Appl. Microbiol. Biotechnol. 2019, 103, 3297–3316. [Google Scholar] [CrossRef] [PubMed]
- Hamida, R.S.; Ali, M.A.; Redhwan, A.; Bin-Meferij, M.M. Cyanobacteria—A promising platform in green nanotechnology: A review on nanoparticles fabrication and their prospective applications. Int. J. Nanomed. 2020, 15, 6033. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhu, Y.; Hu, Z.; Lei, A.; Wang, J. Towards elucidation of the toxic mechanism of copper on the model green alga Chlamydomonas reinhardtii. Ecotoxicology 2016, 25, 1417–1425. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.-L.; Guo, S.-N.; Wen, F.; Zhang, X.-L.; Wang, C.-C.; Si, L.-F.; Zheng, J.-L.; Liu, J. Transcriptional and physiological responses of Dunaliella salina to cadmium reveals time-dependent turnover of ribosome, photosystem, and ROS-scavenging pathways. Aquat. Toxicol. 2019, 207, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Elleuch, J.; Hmani, R.; Drira, M.; Michaud, P.; Fendri, I.; Abdelkafi, S. Potential of three local marine microalgae from Tunisian coasts for cadmium, lead and chromium removals. Sci. Total Environ. 2021, 799, 149464. [Google Scholar] [CrossRef] [PubMed]
- Athanasiadis, K. Effects of Heavy Metal Stress on Chlamydomonas reinhardtii: A Comprehensive Study. Master’s Thesis, University of Antwerp, Antwerp, Belgium, September 2017. [Google Scholar]
- Ireland, H.E.; Harding, S.J.; Bonwick, G.A.; Jones, M.; Smith, C.J.; Williams, J.H. Evaluation of heat shock protein 70 as a biomarker of environmental stress in Fucus serratus and Lemna minor. Biomarkers 2004, 9, 139–155. [Google Scholar] [CrossRef]
- Neumann, D.; Lichtenberger, O.; Günther, D.; Tschiersch, K.; Nover, L. Heat-shock proteins induce heavy-metal tolerance in higher plants. Planta 1994, 194, 360–367. [Google Scholar] [CrossRef]
- Lewis, S.; Donkin, M.; Depledge, M. Hsp70 expression in Enteromorpha intestinalis (Chlorophyta) exposed to environmental stressors. Aquat. Toxicol. 2001, 51, 277–291. [Google Scholar] [CrossRef]
- Vierling, E. The roles of heat shock proteins in plants. Annu. Rev. Plant Biol. 1991, 42, 579–620. [Google Scholar] [CrossRef]
- Sathasivam, R.; Ki, J.-S. Heat shock protein genes in the green alga Tetraselmis suecica and their role against redox and non-redox active metals. Eur. J. Protistol. 2019, 69, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Kimura, T.; Itoh, N.; Andrews, G.K. Mechanisms of heavy metal sensing by metal response element-binding transcription factor-1. J. Health Sci. 2009, 55, 484–494. [Google Scholar] [CrossRef] [Green Version]
- Lichtlen, P.; Schaffner, W. Putting its fingers on stressful situations: The heavy metal-regulatory transcription factor MTF-1. Bioessays 2001, 23, 1010–1017. [Google Scholar] [CrossRef]
- Langmade, S.J.; Ravindra, R.; Daniels, P.J.; Andrews, G.K. The transcription factor MTF-1 mediates metal regulation of the mouse ZnT1 gene. J. Biol. Chem. 2000, 275, 34803–34809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Günther, V.; Lindert, U.; Schaffner, W. The taste of heavy metals: Gene regulation by MTF-1. Biochim. Biophys. Acta Mol. Cell Res. 2012, 1823, 1416–1425. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Georgiev, O.; Hagmann, M.; Günes, C.A.; Cramer, M.; Faller, P.; Vasák, M.; Schaffner, W. Activity of metal-responsive transcription factor 1 by toxic heavy metals and H2O2 in vitro is modulated by metallothionein. Mol. Cell. Biol. 2003, 23, 8471–8485. [Google Scholar] [CrossRef] [Green Version]
- Song, C.; Yan, Y.; Rosado, A.; Zhang, Z.; Castellarin, S.D. ABA alleviates uptake and accumulation of zinc in grapevine (Vitis vinifera L.) by inducing expression of ZIP and detoxification-related genes. Front. Plant Sci. 2019, 10, 872. [Google Scholar] [CrossRef]
- Ai, T.N.; Naing, A.H.; Yun, B.-W.; Lim, S.H.; Kim, C.K. Overexpression of RsMYB1 enhances anthocyanin accumulation and heavy metal stress tolerance in transgenic petunia. Front. Plant Sci. 2018, 9, 1388. [Google Scholar] [CrossRef] [Green Version]
- Sheng, Y.; Yan, X.; Huang, Y.; Han, Y.; Zhang, C.; Ren, Y.; Fan, T.; Xiao, F.; Liu, Y.; Cao, S. The WRKY transcription factor, WRKY13, activates PDR8 expression to positively regulate cadmium tolerance in Arabidopsis. Plant Cell Environ. 2019, 42, 891–903. [Google Scholar] [CrossRef]
- Merchant, S.S.; Schmollinger, S.; Strenkert, D.; Moseley, J.L.; Blaby-Haas, C.E. From economy to luxury: Copper homeostasis in Chlamydomonas and other algae. Biochim. Biophys. Acta M Mol. Cell Res. 2020, 1867, 118822. [Google Scholar] [CrossRef]
- Sapara, K.K.; Khedia, J.; Agarwal, P.; Gangapur, D.R.; Agarwal, P.K. SbMYB15 transcription factor mitigates cadmium and nickel stress in transgenic tobacco by limiting uptake and modulating antioxidative defence system. Funct. Plant Biol. 2019, 46, 702–714. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wang, R.; Ju, Q.; Li, W.; Tran, L.-S.P.; Xu, J. The R2R3-MYB transcription factor MYB49 regulates cadmium accumulation. Plant Physiol. 2019, 180, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Yadav, B.S.; Mani, A. Analysis of bHLH coding genes of Cicer arietinum during heavy metal stress using biological network. Physiol. Mol. Biol. Plants 2019, 25, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Liu, X.; He, X.; Xu, L.; Huang, Y.; Shao, H.; Zhang, D.; Tang, B.; Ma, H. The soybean basic helix-loop-helix transcription factor ORG3-like enhances cadmium tolerance via increased iron and reduced cadmium uptake and transport from roots to shoots. Front. Plant Sci. 2017, 8, 1098. [Google Scholar] [CrossRef] [Green Version]
- Romanenko, K.; Kosakovskaya, I.; Romanenko, P. Phytohormones of microalgae: Biological role and involvement in the regulation of physiological processes. Int. J. Algae 2016, 18, 179–201. [Google Scholar] [CrossRef]
- Piotrowska-Niczyporuk, A.; Bajguz, A.; Kotowska, U.; Zambrzycka-Szelewa, E.; Sienkiewicz, A. Auxins and cytokinins regulate phytohormone homeostasis and thiol-mediated detoxification in the green alga Acutodesmus obliquus exposed to lead stress. Sci. Rep. 2020, 10, 10193. [Google Scholar] [CrossRef]
- Nguyen, T.Q.; Sesin, V.; Kisiala, A.; Emery, R.N. The role of phytohormones in enhancing metal remediation capacity of algae. Bull. Environ. Contam. Toxicol. 2020, 105, 671–678. [Google Scholar] [CrossRef]
- Piotrowska-Niczyporuk, A.; Bajguz, A.; Zambrzycka, E.; Godlewska-Żyłkiewicz, B. Phytohormones as regulators of heavy metal biosorption and toxicity in green alga Chlorella vulgaris (Chlorophyceae). Plant Physiol. Biochem. 2012, 52, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Piotrowska-Niczyporuk, A.; Bajguz, A.; Talarek, M.; Bralska, M.; Zambrzycka, E. The effect of lead on the growth, content of primary metabolites, and antioxidant response of green alga Acutodesmus obliquus (Chlorophyceae). Environ. Sci. Pollut. Res. 2015, 22, 19112–19123. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, H.-P.; Han, B.; Yu, X. Coupling of abiotic stresses and phytohormones for the production of lipids and high-value by-products by microalgae: A review. Bioresour. Technol. 2019, 274, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Talarek-Karwel, M.; Bajguz, A.; Piotrowska-Niczyporuk, A. Hormonal response of Acutodesmus obliquus exposed to combined treatment with 24-epibrassinolide and lead. J. Appl. Phycol. 2020, 32, 2903–2914. [Google Scholar] [CrossRef]
- Bajguz, A.; Piotrowska-Niczyporuk, A. Interactive effect of brassinosteroids and cytokinins on growth, chlorophyll, monosaccharide and protein content in the green alga Chlorella vulgaris (Trebouxiophyceae). Plant Physiol. Biochem. 2014, 80, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Vo, T.-T.; Lee, C.; Han, S.-I.; Kim, J.Y.; Kim, S.; Choi, Y.-E. Effect of the ethylene precursor, 1-aminocyclopropane-1-carboxylic acid on different growth stages of Haematococcus pluvialis. Bioresour. Technol. 2016, 220, 85–93. [Google Scholar] [CrossRef]
- Raman, V.; Ravi, S. Effect of salicylic acid and methyl jasmonate on antioxidant systems of Haematococcus pluvialis. Acta Physiol. Plant. 2011, 33, 1043–1049. [Google Scholar] [CrossRef]
- Kim, S.-H.; Lim, S.R.; Hong, S.-J.; Cho, B.-K.; Lee, H.; Lee, C.-G.; Choi, H.-K. Effect of ethephon as an ethylene-releasing compound on the metabolic profile of Chlorella vulgaris. J. Agric. Food Chem. 2016, 64, 4807–4816. [Google Scholar] [CrossRef]
- Thomas, J.C.; Perron, M.; LaRosa, P.C.; Smigocki, A.C. Cytokinin and the regulation of a tobacco metallothionein-like gene during copper stress. Physiol. Plant. 2005, 123, 262–271. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, C.; Cao, H.; Chen, Z.; Yang, J.; Cao, S.; Wei, Z. The role of cytokinin in selenium stress response in Arabidopsis. Plant Sci. 2019, 281, 122–132. [Google Scholar] [CrossRef]
- Mohan, T.C.; Castrillo, G.; Navarro, C.; Zarco-Fernández, S.; Ramireddy, E.; Mateo, C.; Zamarreño, A.M.; Paz-Ares, J.; Muñoz, R.; García-Mina, J.M. Cytokinin determines thiol-mediated arsenic tolerance and accumulation. Plant Physiol. 2016, 171, 1418–1426. [Google Scholar]
- Tian, B.J.; Wang, Y.; Zhu, Y.R.; Lü, X.Y.; Huang, K.; Shao, N.; Beck, C.F. Synthesis of the photorespiratory key enzyme serine: Glyoxylate aminotransferase in C. reinhardtii is modulated by the light regime and cytokinin. Physiol. Plant. 2006, 127, 571–582. [Google Scholar] [CrossRef]
- Piotrowska, A.; Czerpak, R. Cellular response of light/dark-grown green alga Chlorella vulgaris Beijerinck (Chlorophyceae) to exogenous adenine- and phenylurea-type cytokinins. Acta Physiol. Plant. 2009, 31, 573–585. [Google Scholar] [CrossRef]
- Souza, J.M.; Yokoya, N.S. Effects of cytokinins on physiological and biochemical responses of the agar-producing red alga Gracilaria caudata (Gracilariales, Rhodophyta). J. Appl. Phycol. 2016, 28, 3491–3499. [Google Scholar] [CrossRef]
- Udayan, A.; Kathiresan, S.; Arumugam, M. Kinetin and gibberellic acid (GA3) act synergistically to produce high value polyunsaturated fatty acids in Nannochloropsis oceanica CASA CC201. Algal Res. 2018, 32, 182–192. [Google Scholar] [CrossRef]
- Du, H.; Ahmed, F.; Lin, B.; Li, Z.; Huang, Y.; Sun, G.; Ding, H.; Wang, C.; Meng, C.; Gao, Z. The effects of plant growth regulators on cell growth, protein, carotenoid, PUFAs and lipid production of Chlorella pyrenoidosa ZF strain. Energies 2017, 10, 1696. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.-J.; Sun, J.; Sun, Y.-Q.; Zheng, J.-Y.; Wang, Z. Metabolomics analysis of phytohormone gibberellin improving lipid and DHA accumulation in Aurantiochytrium sp. Biochem. Eng. J. 2016, 112, 258–268. [Google Scholar] [CrossRef]
- Sulochana, S.B.; Arumugam, M. Influence of abscisic acid on growth, biomass and lipid yield of Scenedesmus quadricauda under nitrogen starved condition. Bioresour. Technol. 2016, 213, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Noble, A.; Kisiala, A.; Galer, A.; Clysdale, D.; Emery, R.N. Euglena gracilis (Euglenophyceae) produces abscisic acid and cytokinins and responds to their exogenous application singly and in combination with other growth regulators. Eur. J. Phycol. 2014, 49, 244–254. [Google Scholar] [CrossRef]
- Contreras-Pool, P.Y.; Peraza-Echeverria, S.; Ku-González, Á.F.; Herrera-Valencia, V.A. The phytohormone abscisic acid increases triacylglycerol content in the green microalga Chlorella saccharophila (Chlorophyta). Algae 2016, 31, 267–276. [Google Scholar] [CrossRef]
- Bajguz, A.; Piotrowska-Niczyporuk, A. Synergistic effect of auxins and brassinosteroids on the growth and regulation of metabolite content in the green alga Chlorella vulgaris (Trebouxiophyceae). Plant Physiol. Biochem. 2013, 71, 290–297. [Google Scholar] [CrossRef]
- Lu, Y.; Xu, J. Phytohormones in microalgae: A new opportunity for microalgal biotechnology? Trends Plant Sci. 2015, 20, 273–282. [Google Scholar] [CrossRef]
- Ding, Y.; Ding, L.; Xia, Y.; Wang, F.; Zhu, C. Emerging roles of microRNAs in plant heavy metal tolerance and homeostasis. J. Agric. Food Chem. 2020, 68, 1958–1965. [Google Scholar] [CrossRef]
- Noman, A.; Aqeel, M. miRNA-based heavy metal homeostasis and plant growth. Environ. Sci. Pollut. Res. 2017, 24, 10068–10082. [Google Scholar] [CrossRef] [PubMed]
- Lou, S.; Zhu, X.; Zeng, Z.; Wang, H.; Jia, B.; Li, H.; Hu, Z. Identification of microRNAs response to high light and salinity that is involved in beta-carotene accumulation in microalga Dunaliella salina. Algal Res. 2020, 48, 101925. [Google Scholar] [CrossRef]
- Azaman, S.N.A.; Satharasinghe, D.A.; Tan, S.W.; Nagao, N.; Yusoff, F.M.; Yeap, S.K. Identification and analysis of microRNAs in Chlorella sorokiniana using high-throughput sequencing. Genes 2020, 11, 1131. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Miao, X.; Chen, G.; Cui, Y.; Sun, F.; Fan, J.; Gao, Z.; Meng, C. Identification of microRNAs involved in astaxanthin accumulation responding to high light and high sodium acetate (NaAC) stresses in Haematococcus pluvialis. Algal Res. 2021, 54, 102179. [Google Scholar] [CrossRef]
- Srivastava, S.; Srivastava, A.K.; Suprasanna, P.; D’souza, S. Identification and profiling of arsenic stress-induced microRNAs in Brassica juncea. J. Exp. Bot. 2013, 64, 303–315. [Google Scholar] [CrossRef] [Green Version]
- Remans, T.; Opdenakker, K.; Guisez, Y.; Carleer, R.; Schat, H.; Vangronsveld, J.; Cuypers, A. Exposure of Arabidopsis thaliana to excess Zn reveals a Zn-specific oxidative stress signature. Environ. Exp. Bot. 2012, 84, 61–71. [Google Scholar] [CrossRef]
- Huang, S.Q.; Xiang, A.L.; Che, L.L.; Chen, S.; Li, H.; Song, J.B.; Yang, Z.M. A set of miRNAs from Brassica napus in response to sulphate deficiency and cadmium stress. Plant Biotechnol. J. 2010, 8, 887–899. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, H. Molecular identification and analysis of arsenite stress-responsive miRNAs in rice. J. Agric. Food Chem. 2012, 60, 6524–6536. [Google Scholar] [CrossRef]
- Zhou, Z.S.; Song, J.B.; Yang, Z.M. Genome-wide identification of Brassica napus microRNAs and their targets in response to cadmium. J. Exp. Bot. 2012, 63, 4597–4613. [Google Scholar] [CrossRef] [Green Version]
- Pilon, M. The copper microRNAs. New Phytol. 2017, 213, 1030–1035. [Google Scholar] [CrossRef]
- Ding, Y.-F.; Zhu, C. The role of microRNAs in copper and cadmium homeostasis. Biochem. Biophys. Res. Commun. 2009, 386, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Soto, A.B.; Sánchez, F.; Hernández, G. MicroRNAs as regulators in plant metal toxicity response. Front. Plant Sci. 2012, 3, 105. [Google Scholar] [CrossRef] [Green Version]
- Paul, S.; Datta, S.K.; Datta, K. miRNA regulation of nutrient homeostasis in plants. Front. Plant Sci. 2015, 6, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jagadeeswaran, G.; Li, Y.F.; Sunkar, R. Redox signaling mediates the expression of a sulfate-deprivation-inducible micro RNA 395 in Arabidopsis. Plant J. 2014, 77, 85–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Y.; Chen, Z.; Zhu, C. Microarray-based analysis of cadmium-responsive microRNAs in rice (Oryza sativa). J. Exp. Bot. 2011, 62, 3563–3573. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.W.; Song, J.B.; Shu, X.X.; Zhang, Y.; Yang, Z.M. miR395 is involved in detoxification of cadmium in Brassica napus. J. Hazard. Mater. 2013, 250, 204–211. [Google Scholar] [CrossRef]
- Zeng, Q.-Y.; Yang, C.-Y.; Ma, Q.-B.; Li, X.-P.; Dong, W.-W.; Nian, H. Identification of wild soybean miRNAs and their target genes responsive to aluminum stress. BMC Plant Biol. 2012, 12, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Wang, T.; Zhao, M.; Tian, Q.; Zhang, W.-H. Identification of aluminum-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. Planta 2012, 235, 375–386. [Google Scholar] [CrossRef]
- Halder, S. Bioremediation of heavy metals through fresh water microalgae: A review. Sch. Acad. J. Biosci. 2014, 2, 825–830. [Google Scholar]
- Wang, Y.; Selvamani, V.; Yoo, I.-K.; Kim, T.W.; Hong, S.H. A novel strategy for the microbial removal of heavy metals: Cell-surface display of peptides. Biotechnol. Bioprocess Eng. 2021, 26, 1–9. [Google Scholar] [CrossRef]
- Yang, T.; Chen, M.-L.; Wang, J.-H. Genetic and chemical modification of cells for selective separation and analysis of heavy metals of biological or environmental significance. Trends Anal. Chem. 2015, 66, 90–102. [Google Scholar] [CrossRef]
- Kuroda, K.; Ueda, M. Molecular design of the microbial cell surface toward the recovery of metal ions. Curr. Opin. Biotechnol. 2011, 22, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, K.; Shibasaki, S.; Ueda, M.; Tanaka, A. Cell surface-engineered yeast displaying a histidine oligopeptide (hexa-His) has enhanced adsorption of and tolerance to heavy metal ions. Appl. Microbiol. Biotechnol. 2001, 57, 697–701. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, K.; Ueda, M.; Shibasaki, S.; Tanaka, A. Cell surface-engineered yeast with ability to bind, and self-aggregate in response to copper ion. Appl. Microbiol. Biotechnol. 2002, 59, 259–264. [Google Scholar]
- Kuroda, K.; Ueda, M. Bioadsorption of cadmium ion by cell surface-engineered yeasts displaying metallothionein and hexa-His. Appl. Microbiol. Biotechnol. 2003, 63, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, K.; Ueda, M. Effective display of metallothionein tandem repeats on the bioadsorption of cadmium ion. Appl. Microbiol. Biotechnol. 2006, 70, 458–463. [Google Scholar] [CrossRef] [PubMed]
- Kotrba, P.; Ruml, T. Surface display of metal fixation motifs of bacterial P1-type ATPases specifically promotes biosorption of Pb2+ by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2010, 76, 2615–2622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bae, W.; Wu, C.H.; Kostal, J.; Mulchandani, A.; Chen, W. Enhanced mercury biosorption by bacterial cells with surface-displayed MerR. Appl. Environ. Microbiol. 2003, 69, 3176–3180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sousa, C.; Kotrba, P.; Ruml, T.; Cebolla, A.; de Lorenzo, V. Metalloadsorption by Escherichia coli cells displaying yeast and mammalian metallothioneins anchored to the outer membrane protein LamB. J. Bacteriol. 1998, 180, 2280–2284. [Google Scholar] [CrossRef] [Green Version]
- Krishnaswamy, R.; Wilson, D.B. Construction and characterization of an Escherichia coli strain genetically engineered for Ni(II) bioaccumulation. Appl. Environ. Microbiol. 2000, 66, 5383–5386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, N.; Birungi, Z.S.; Chirwa, E. Selective biosorption of precious metals by cell-surface engineered microalgae. Chem. Eng. Trans. 2017, 61, 25–30. [Google Scholar]
- Kuroda, K.; Ebisutani, K.; Iida, K.; Nishitani, T.; Ueda, M. Enhanced adsorption and recovery of uranyl ions by NikR mutant-displaying yeast. Biomolecules 2014, 4, 390–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishitani, T.; Shimada, M.; Kuroda, K.; Ueda, M. Molecular design of yeast cell surface for adsorption and recovery of molybdenum, one of rare metals. Appl. Microbiol. Biotechnol. 2010, 86, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, K.; Nishitani, T.; Ueda, M. Specific adsorption of tungstate by cell surface display of the newly designed ModE mutant. Appl. Microbiol. Biotechnol. 2012, 96, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Fakhrullin, R.F.; Lvov, Y.M.; Choi, I.S. Future of cell surface engineering. In Cell Surface Engineering, 1st ed.; Royal Society of Chemistry: London, UK, 2014; pp. 240–245. [Google Scholar]
| Genetic and Metabolic Engineering Tools | Type | Specifics | Advantages and Disadvantages |
|---|---|---|---|
| CRISPR/Cas systems | Gene editing | A single guide RNA (sgRNA) drives the Cas protein to a matching DNA sequence on the host cell genome—which is degraded by Cas protein to create knockout and deletion mutants; and replaced by a new gene to generate insertion and knockdown mutants, with the aid of non-homologous, end-joining machinery | Allows manipulation of any DNA sequences; multiple mutations are possible; comparably more whole genome data are required; possibility of off-target events; large size of Cas makes cell delivery challenging |
| ZFNs | Gene editing | An array of site-specific DNA-binding domains that recognizes two sequences flanking a specific site, attached to the endonuclease domain of bacterial FokI restriction enzyme; upon binding, FokI domains dimerize and cleave DNA at the site, which will be repaired by the DNA repair machinery of the cell | Comparably high probability of off-target events; complicated programming is required; limited possible genomic target sites; only single mutations are possible |
| TALENs | Gene editing | Tandem arrays with 10 to 30 repeats that bind and recognize extended DNA sequences, attached to the endonuclease domain of bacterial FokI restriction enzyme; upon binding, FokI domains dimerize and cleave DNA at the site, which will be repaired by the DNA repair machinery of the cell | Comparably high probability of off-target events; complicated programming is required; limited possible genomic target sites. large TALENs hard to express or transfect into the cell; only single mutations are possible |
| Cre/loxP recombination systems | Gene editing | Two loxP sequences flanking the target gene interact with Cre recombinase for insertion of a new gene fragment or deletion of the one targeted | Simple and efficient; allows multiple genome integration; occasional off-target events are possible; toxicity of Cre for Cre-expressing cells |
| Modular cloning systems | Synthetic biology | Design and construction of expression vectors by providing a library of genetic building blocks (e.g., promoters, UTRs, terminators, tags, reporters, antibiotic resistance genes, introns) | Simple and efficient; high flexibility; expression of multiple transgenes possible; developed for limited range of species |
| RNAi technology | Gene silencing | Small RNA molecules bind to target mRNAs to form double-stranded RNAs, which are degraded by RNA-induced silencing complex (RISC)—and cause sequence-specific suppression of gene expression, through translational or transcriptional repression | Simple and efficient; occasional off-target effects; produces hypomorphic phenotypes, which do not always mirror the complete loss-of-function that often occurs with genetic mutation; nuclear transcripts—e.g., long non-coding RNAs or lncRNAs, more difficult to effectively target |
| Multiomics technologies | Omics | Analysis of whole-cell biochemical information of cell through genomics, transcriptomics, proteomics, metabolomics, interactomics, phenomics, meta-omics, etc. | Provide snapshot of response of cell or whole ecosystem to environmental changes; improves data comparability; time-consuming; high cost; sophisticated, expensive equipment required |
| Online databases | Bioinformatics | Integrated online platforms e.g., ChlamyCys, Greenhouse, Diatom EST database, Alga-PrAs, Cyan-Omics, and KEGG | Functional interpretation of genes and elucidation of their underlying biological themes via integrated annotation and expression data; freely available |
| Flux balance analysis | Systems | All relevant metabolic information of an organism (e.g., genes, enzymes, reactions) are collected, and analyzed with the aid of a mathematical model within the perspective of the entire network, and applied to make predictions and genome reconstruction | Allows tailor-made design of cell factories aimed at maximum efficiency; time-consuming; still in its infancy |
| Illumina microarray technology | Sequencing | Tiny silica microbeads are housed in carefully etched microwells, and coated with multiple copies of an oligonucleotide probe targeting a specific DNA or RNA sequence; upon excitation by laser, binding of each probe to a complementary sequence in sample results in signal that conveys information to the detector | Fast and robust; no prior knowledge of gene sequences required; potent means for gene sequence and expression analysis; high cost |
| Approach | Targets |
|---|---|
| Metal transportation | NRAMP, ZRT, IRT, ZIP, FTR, CTR, CDF, HMA, FPN, Ccc1/VIT1, PTA, AQP, MTP, PMA, V-ATPase, V-PPase, MRP, ATM/HMT, PDR, YSL, CAX, MFS |
| Metal chelation | MTs, PCs, GSH, PPK, VTC, PPX, Pro (P5CS), His (HISN3), Cys (HAL2), Ser (SDC1), glycine-betaine, and organic acids |
| Metal biotransformation | ChrR, arsC, CrACR2s, MerA/B/P/C/T/F, SMT, CYPs |
| Oxidative stress response regulation | Trx, HSPs, carotenoids, SOD, POD, CAT, GPX, GST |
| Metal stress response regulation | MTF-1, C2H2, AP2, MYB, bHLH, YABBY, bZIP, SBP, HB, WRKY13, CRR1, ABI5, GATA, CKs (IPT and CKX1), GA, ABA, BRs, JA, ET, SA, miRNAs (miR398, miR319, miR390, miR393, miR171, miR395, miR397, miR408, and miR857) |
| Cell-surface bioengineering | MTs, PCs, 6x-His, CXXEE, MerR, GST, NikRm, ModE |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ranjbar, S.; Malcata, F.X. Is Genetic Engineering a Route to Enhance Microalgae-Mediated Bioremediation of Heavy Metal-Containing Effluents? Molecules 2022, 27, 1473. https://doi.org/10.3390/molecules27051473
Ranjbar S, Malcata FX. Is Genetic Engineering a Route to Enhance Microalgae-Mediated Bioremediation of Heavy Metal-Containing Effluents? Molecules. 2022; 27(5):1473. https://doi.org/10.3390/molecules27051473
Chicago/Turabian StyleRanjbar, Saeed, and Francisco Xavier Malcata. 2022. "Is Genetic Engineering a Route to Enhance Microalgae-Mediated Bioremediation of Heavy Metal-Containing Effluents?" Molecules 27, no. 5: 1473. https://doi.org/10.3390/molecules27051473
APA StyleRanjbar, S., & Malcata, F. X. (2022). Is Genetic Engineering a Route to Enhance Microalgae-Mediated Bioremediation of Heavy Metal-Containing Effluents? Molecules, 27(5), 1473. https://doi.org/10.3390/molecules27051473






