Biochemical and Biophysical Characterisation of the Hepatitis E Virus Guanine-7-Methyltransferase
Abstract
1. Introduction
2. Results
2.1. Expression and Purification of MTase
2.2. MTase Activity of 37 kDa Protein
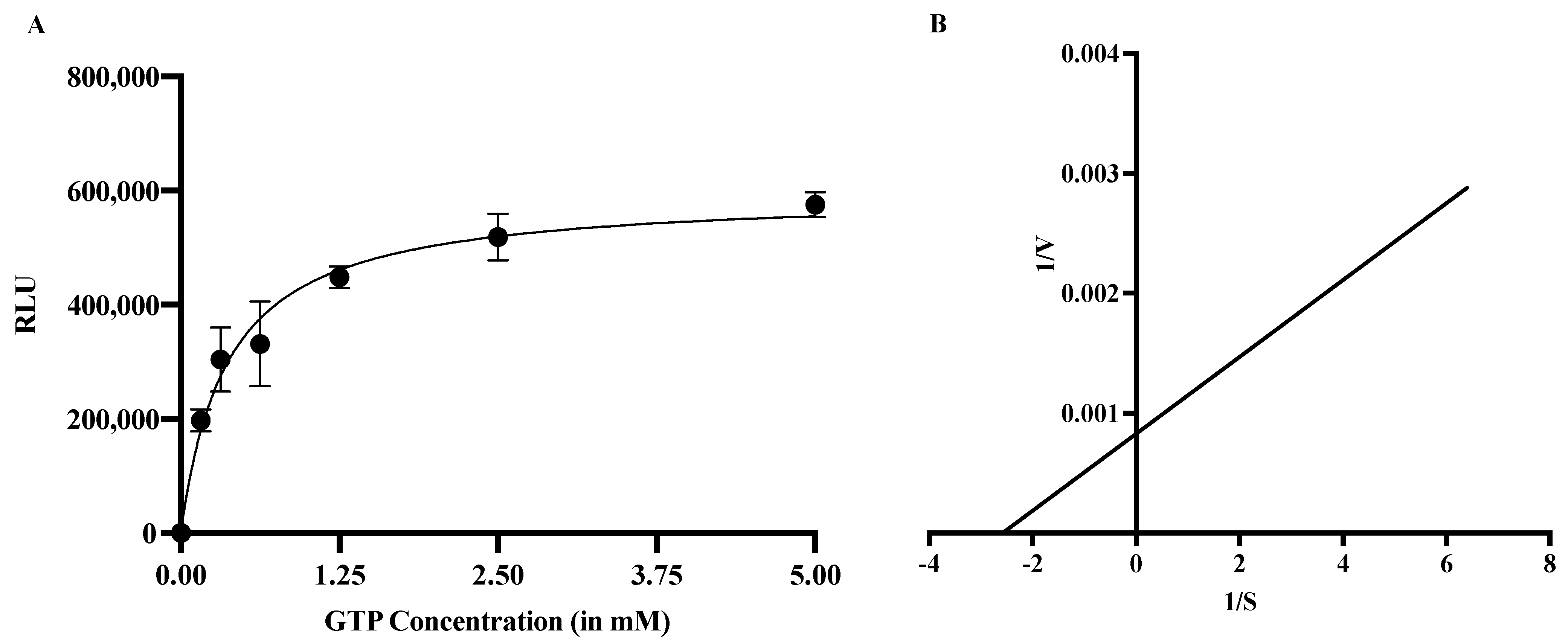
2.2.1. Enzyme Kinetics
2.2.2. MTase Activity in the Presence of Guanosine Triphosphate (GTP) as a Substrate
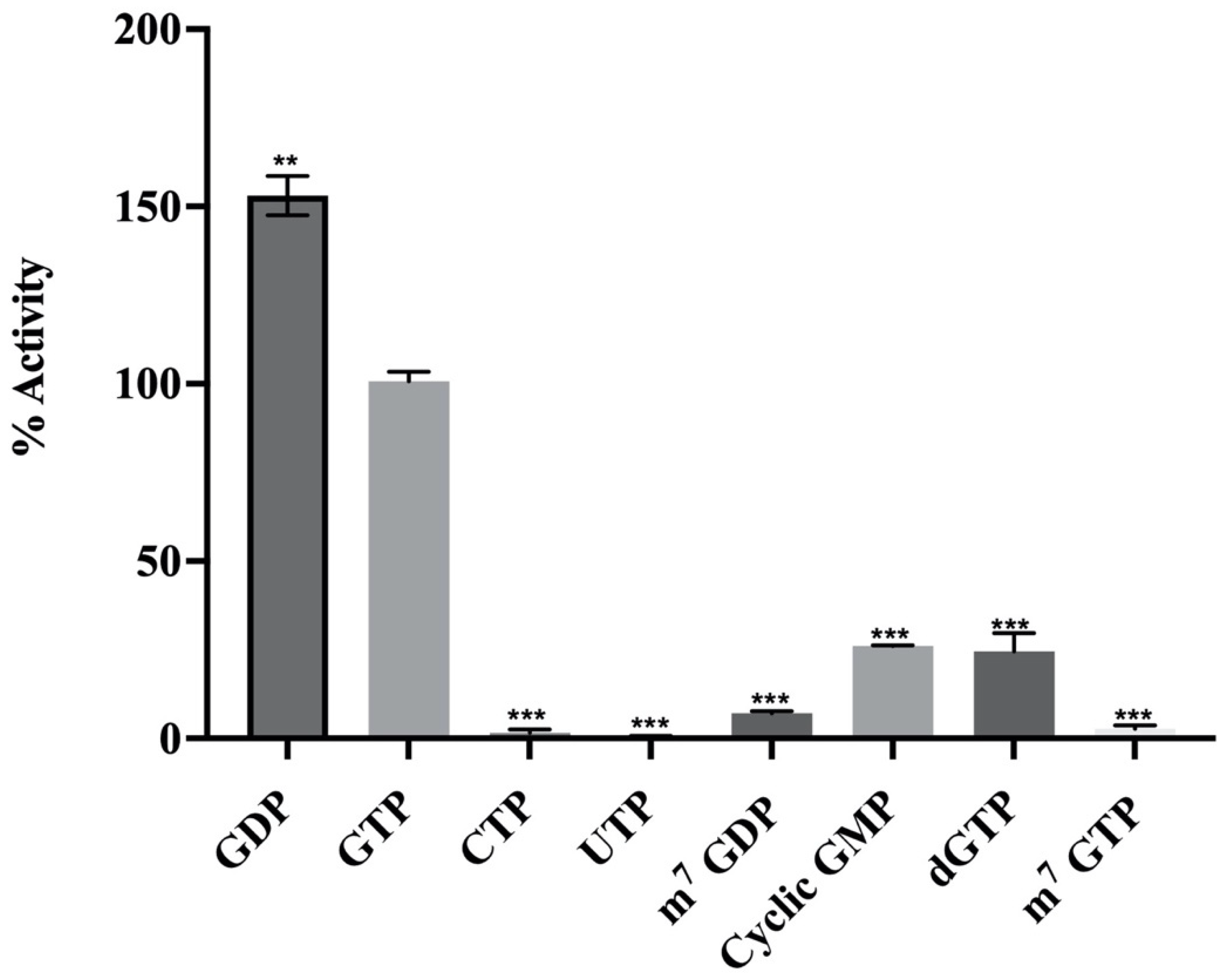
2.2.3. Substrate Specificity of MTase
2.2.4. Effect of Magnesium (Mg2+) on MTase Activity
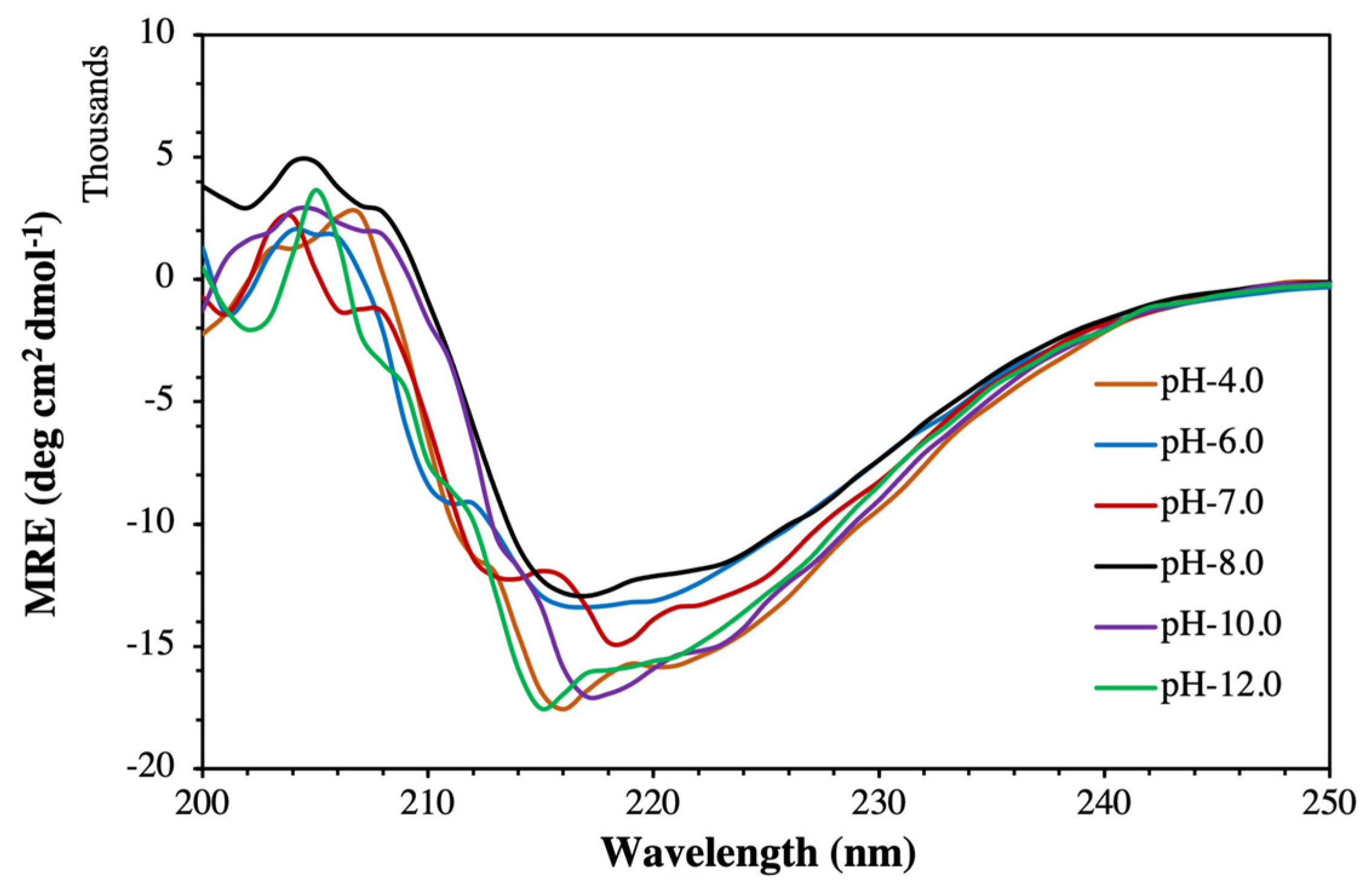
2.3. Circular Dichroism (CD) Analysis
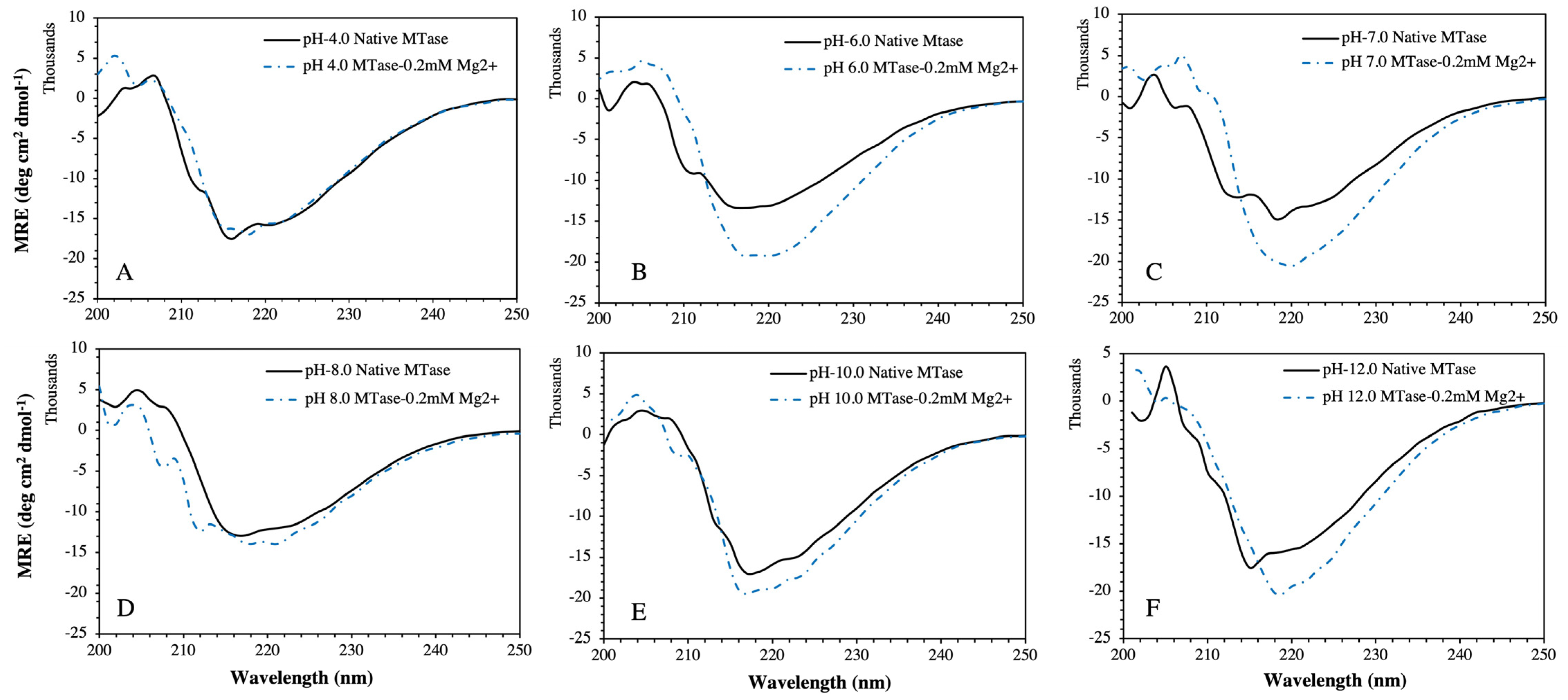
2.4. Structural and Thermal Stability of MTase in the Presence of Mg2+
2.5. Determination of Binding Affinity and Mechanism of Mg2+ Ion with MTase by Fluorescence Quenching Method
2.6. Isothermal Titration Calorimetry (ITC) Analysis
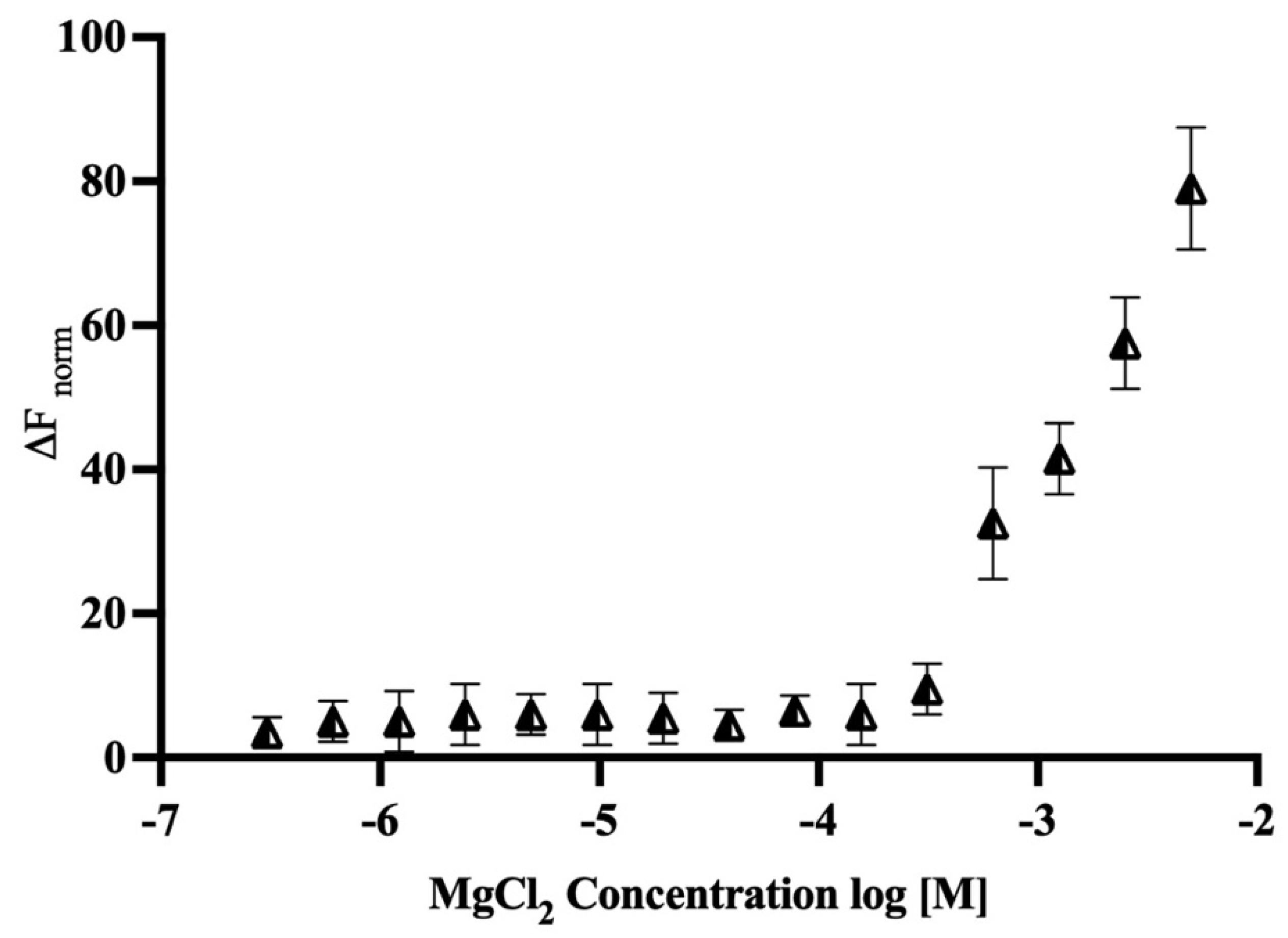
2.7. Binding Study of Magnesium with MTase
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Expression and Purification of MTase
5.2. Functional Aspects of MTase
5.3. Secondary Structure Analysis Using CD Spectroscopy
5.4. Thermal Stability of MTase with Mg2+
5.5. Fluorescence Quenching Measurements
5.6. Binding Affinity Using Isothermal Titration Calorimetry (ITC)
5.7. MicroScale Thermophoresis Measurements
5.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
| HEV-MTase | Hepatitis E Virus Methyltransferase |
| ORFs | Open reading frames |
| FI | Fluorescence Intensity |
| Mg2+ | Magnesium ion |
| ITC | Isothermal Titration Calorimetry |
| CD | Circular Dichroism |
| MST | MicroScale Thermophoresis |
| Tm | Melting point |
References
- Chandra, V.; Taneja, S.; Kalia, M.; Jameel, S. Molecular biology and pathogenesis of hepatitis E virus. J. Biosci. 2008, 33, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Parvez, M.K. Molecular characterization of hepatitis E virus ORF1 gene supports a papain-like cysteine protease (PCP)-domain activity. Virus Res. 2013, 178, 553–556. [Google Scholar] [CrossRef] [PubMed]
- Kamar, N.; Izopet, J.; Tripon, S.; Bismuth, M.; Hillaire, S.; Dumortier, J.; Radenne, S.; Coilly, A.; Garrigue, V.; D’Alteroche, L.; et al. Ribavirin for chronic hepatitis E virus infection in transplant recipients. N. Engl. J. Med. 2014, 370, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Parvez, M.K.; Khan, A.A. Molecular modeling and analysis of hepatitis E virus (HEV) papain-like cysteine protease. Virus Res. 2014, 179, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Navaneethan, U.; Al Mohajer, M.; Shata, M.T. Hepatitis E and pregnancy: Understanding the pathogenesis. Liver Int. 2008, 28, 1190–1199. [Google Scholar] [CrossRef]
- Emerson, S.U.; Zhang, M.; Meng, X.J.; Nguyen, H.; St. Claire, M.; Govindarajan, S.; Huang, Y.K.; Purcell, R.H. Recombinant hepatitis E virus genomes infectious for primates: Importance of capping and discovery of a cis-reactive element. Proc. Natl. Acad. Sci. USA 2001, 98, 15270–15275. [Google Scholar] [CrossRef]
- Emerson, S.U.; Nguyen, H.; Graff, J.; Stephany, D.A.; Brockington, A.; Purcell, R.H. In Vitro Replication of Hepatitis E Virus (HEV) Genomes and of an HEV Replicon Expressing Green Fluorescent Protein. J. Virol. 2004, 78, 4838–4846. [Google Scholar] [CrossRef]
- Magden, J.; Takeda, N.; Li, T.; Auvinen, P.; Ahola, T.; Miyamura, T.; Merits, A.; Kaariainen, L. Virus-Specific mRNA Capping Enzyme Encoded by Hepatitis E Virus. J. Virol. 2001, 75, 6249–6255. [Google Scholar] [CrossRef]
- Koonin, E.V.; Gorbalenya, A.E.; Purdy, M.A.; Rozanov, M.N.; Reyes, G.R.; Bradley, D.W. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: Delineation of an additional group of positive-strand RNA plant and animal viruses. Proc. Natl. Acad. Sci. USA 1992, 89, 8259–8263. [Google Scholar] [CrossRef]
- Zhao, Y.; Soh, T.S.; Zheng, J.; Chan, K.W.K.; Phoo, W.W.; Lee, C.C.; Tay, M.Y.F.; Swaminathan, K.; Cornvik, T.C.; Lim, S.P.; et al. A Crystal Structure of the Dengue Virus NS5 Protein Reveals a Novel Inter-domain Interface Essential for Protein Flexibility and Virus Replication. PLOS Pathog. 2015, 11, e1004682. [Google Scholar] [CrossRef]
- Krafcikova, P.; Silhan, J.; Nencka, R.; Boura, E. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat. Commun. 2020, 11, 3717. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Fink, K.; Züst, R.; Lim, S.P.; Qin, C.-F.; Shi, P.-Y. Flavivirus RNA methylation. J. Gen. Virol. 2014, 95, 763–778. [Google Scholar] [CrossRef]
- Venkatesan, S.; Moss, B. Eukaryotic mRNA capping enzyme-guanylate covalent intermediate. Proc. Natl. Acad. Sci. USA 1982, 79, 340–344. [Google Scholar] [CrossRef]
- Coutard, B.; Barral, K.; Lichière, J.; Selisko, B.; Martin, B.; Aouadi, W.; Lombardia, M.O.; Debart, F.; Vasseur, J.-J.; Guillemot, J.C.; et al. Zika Virus Methyltransferase: Structure and Functions for Drug Design Perspectives. J. Virol. 2017, 91, 1190–1199. [Google Scholar] [CrossRef]
- Sutto-Ortiz, P.; Tcherniuk, S.; Ysebaert, N.; Abeywickrema, P.; Noël, M.; Decombe, A.; Debart, F.; Vasseur, J.-J.; Canard, B.; Roymans, D.; et al. The methyltransferase domain of the Respiratory Syncytial Virus L protein catalyzes cap N7 and 2′-O-methylation. PLoS Pathog. 2021, 17, e1009562. [Google Scholar] [CrossRef] [PubMed]
- Martin, B.; Coutard, B.; Guez, T.; Paesen, G.C.; Canard, B.; Debart, F.; Vasseur, J.-J.; Grimes, J.M.; Decroly, E. The methyltransferase domain of the Sudan ebolavirus L protein specifically targets internal adenosines of RNA substrates, in addition to the cap structure. Nucleic Acids Res. 2018, 46, 7902–7912. [Google Scholar] [CrossRef] [PubMed]
- Bouvet, M.; Debarnot, C.; Imbert, I.; Selisko, B.; Snijder, E.J.; Canard, B.; Decroly, E. In Vitro Reconstitution of SARS-Coronavirus mRNA Cap Methylation. PLoS Pathog. 2010, 6, e1000863. [Google Scholar] [CrossRef]
- Issur, M.; Geiss, B.J.; Bougie, I.; Picard-Jean, F.; Despins, S.; Mayette, J.; Hobdey, S.E.; Bisaillon, M. The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA 2009, 15, 2340–2350. [Google Scholar] [CrossRef]
- Bougie, I.; Charpentier, S.; Bisaillon, M. Characterization of the Metal Ion Binding Properties of the Hepatitis C Virus RNA Polymerase. J. Biol. Chem. 2003, 278, 3868–3875. [Google Scholar] [CrossRef]
- Decroly, E.; Debarnot, C.; Ferron, F.; Bouvet, M.; Coutard, B.; Imbert, I.; Gluais, L.; Papageorgiou, N.; Sharff, A.; Bricogne, G.; et al. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011, 7, e1002059. [Google Scholar] [CrossRef]
- Viswanathan, T.; Misra, A.; Chan, S.-H.; Qi, S.; Dai, N.; Arya, S.; Martinez-Sobrido, L.; Gupta, Y.K. A metal ion orients SARS-CoV-2 mRNA to ensure accurate 2′-O methylation of its first nucleotide. Nat. Commun. 2021, 12, 3287. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Soh, T.S.; Lim, S.P.; Chung, K.Y.; Swaminathan, K.; Vasudevan, S.G.; Shi, P.-Y.; Lescar, J.; Luo, D. Molecular basis for specific viral RNA recognition and 2′-O-ribose methylation by the dengue virus nonstructural protein 5 (NS5). Proc. Natl. Acad. Sci. USA 2015, 112, 14834–14839. [Google Scholar] [CrossRef] [PubMed]
- Kelly, S.M.; Jess, T.J.; Price, N.C. How to study proteins by circular dichroism. Biochim. Biophys. Acta-Proteins Proteom. 2005, 1751, 119–139. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Yang, J.T.; Martinez, H.M. Determination of the Secondary Structures of Proteins by Circular Dichroism and Optical Rotatory Dispersion. Biochemistry 1972, 11, 4120–4131. [Google Scholar] [CrossRef] [PubMed]
- Marszalek, M.; Konarska, A.; Szajdzinska-Pietek, E.; Wolszczak, M. Interaction of cationic protoberberine alkaloids with human serum albumin. No spectroscopic evidence on binding to Sudlow’s site 1. J. Phys. Chem. B 2013, 117, 15987–15993. [Google Scholar] [CrossRef]
- Zolfagharzadeh, M.; Pirouzi, M.; Asoodeh, A.; Saberi, M.R.; Chamani, J. A comparison investigation of DNP-binding effects to HSA and HTF by spectroscopic and molecular modeling techniques. J. Biomol. Struct. Dyn. 2014, 32, 1936–1952. [Google Scholar] [CrossRef]
- Ishtikhar, M.; Chandel, T.I.; Ahmad, A.; Ali, M.S.; Allohadan, H.A.; Atta, A.M.; Khan, R.H. Rosin surfactant QRMAE can be utilized as an amorphous aggregate inducer: A case study of mammalian serum albumin. PLoS ONE 2015, 10, e0139027. [Google Scholar] [CrossRef]
- Ishtikhar, M.; Ali, M.S.; Atta, A.M.; Al-Lohedan, H.A.; Nigam, L.; Subbarao, N.; Hasan Khan, R. Interaction of biocompatible natural rosin-based surfactants with human serum albumin: A biophysical study. J. Lumin. 2015, 167, 399–407. [Google Scholar] [CrossRef]
- Ishtikhar, M.; Rabbani, G.; Khan, R.H. Interaction of 5-fluoro-5′-deoxyuridine with human serum albumin under physiological and non-physiological condition: A biophysical investigation. Colloids Surf. B Biointerfaces 2014, 123, 469–477. [Google Scholar] [CrossRef]
- Scheidel, L.M.; Durbin, R.K.; Stollar, V. SVLM21, a Sindbis virus mutant resistant to methionine deprivation, encodes an altered methyltransferase. Virology 1989, 173, 408–414. [Google Scholar] [CrossRef]
- Ahola, T.; den Boon, J.A.; Ahlquist, P. Helicase and Capping Enzyme Active Site Mutations in Brome Mosaic Virus Protein 1a Cause Defects in Template Recruitment, Negative-Strand RNA Synthesis, and Viral RNA Capping. J. Virol. 2000, 74, 8803–8811. [Google Scholar] [CrossRef] [PubMed]
- Kabrane-Lazizi, Y.; Meng, X.-J.; Purcell, R.H.; Emerson, S.U. Evidence that the Genomic RNA of Hepatitis E Virus Is Capped. J. Virol. 1999, 73, 8848–8850. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Hooda, P.; Khanna, M.; Patel, U.; Sehgal, D. Development of BacMam Induced Hepatitis E Virus Replication Model in Hepatoma Cells to Study the Polyprotein Processing. Front. Microbiol. 2020, 11, 1347. [Google Scholar] [CrossRef] [PubMed]
- Saraswat, S.; Chaudhary, M.; Sehgal, D. Hepatitis E Virus Cysteine Protease Has Papain Like Properties Validated by in silico Modeling and Cell-Free Inhibition Assays. Front. Cell. Infect. Microbiol. 2020, 9, 478. [Google Scholar] [CrossRef] [PubMed]
- Swift, R.V.; Ong, C.D.; Amaro, R.E. Magnesium-Induced Nucleophile Activation in the Guanylyltransferase mRNA Capping Enzyme. Biochemistry 2012, 51, 10236–10243. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lakowicz, J.R. Quenching of Fluorescence BT-Principles of Fluorescence Spectroscopy; Lakowicz, J.R., Ed.; Springer: Boston, MA, USA, 1983; pp. 257–301. ISBN 978-1-4615-7658-7. [Google Scholar]
- Anand, U.; Jash, C.; Mukherjee, S. Spectroscopic probing of the microenvironment in a protein-surfactant assembly. J. Phys. Chem. B 2010, 114, 15839–15845. [Google Scholar] [CrossRef]
- Feng, X.; Bai, C.; Lin, Z.; Wang, N.; Wang, C. The interaction between acridine orange and bovine serum albumin. Fenxi Huaxue 1998, 26, 154–157. [Google Scholar] [CrossRef]
- Lissi, E.; Calderón, C.; Campos, A. Evaluation of the number of binding sites in proteins from their intrinsic fluorescence: Limitations and pitfalls. Photochem. Photobiol. 2013, 89, 1413–1416. [Google Scholar] [CrossRef]



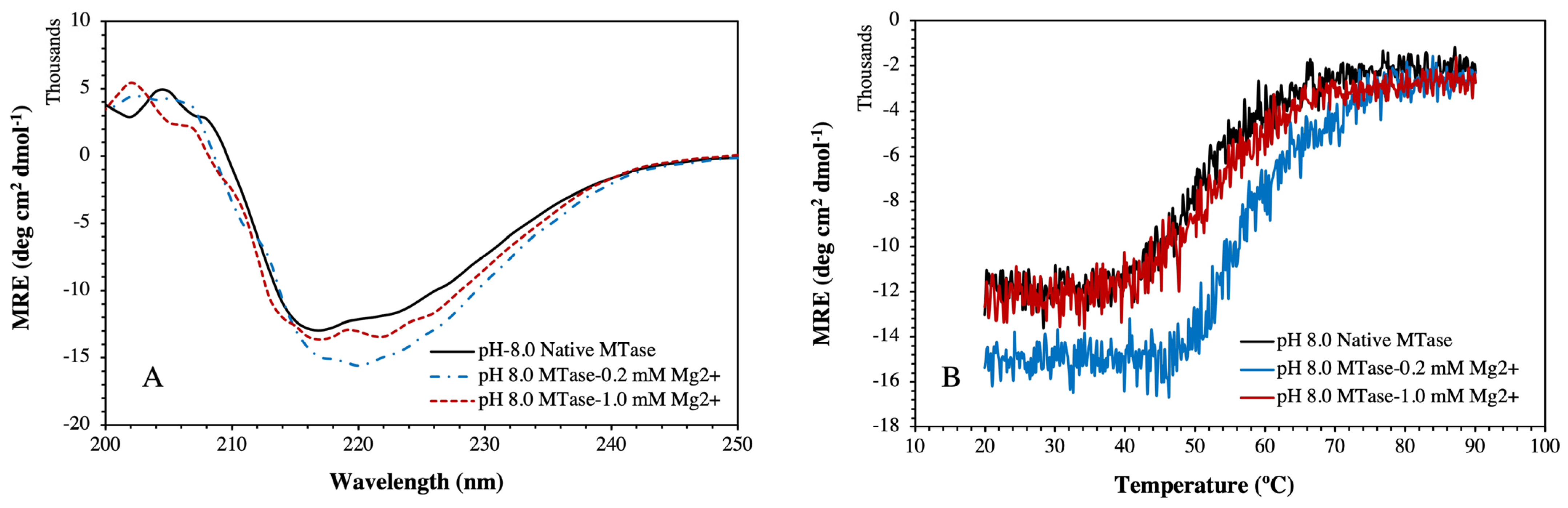
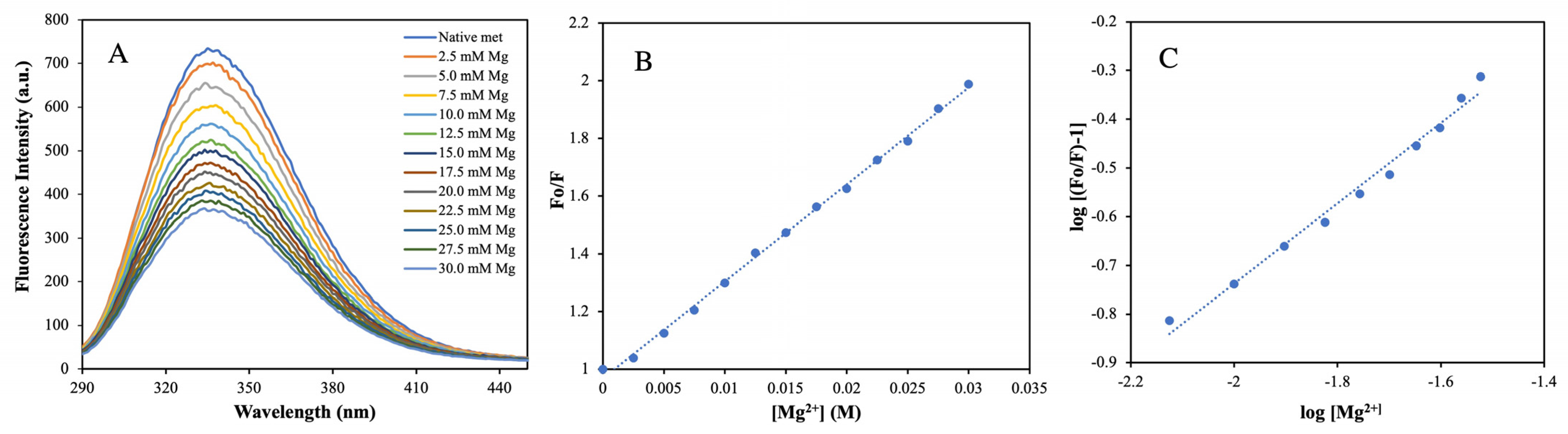

| Sample System | % α-Helices | % β-Sheets | % Random Coils and Other Sec. Structures | Tm (°C) |
|---|---|---|---|---|
| Native MTase | 27.08 | 23.03 | 48.09 | 58.83 |
| MTase + 0.2 mM Mg2+ | 22.64 | 36.96 | 40.40 | 61.57 |
| MTase + 1.0 mM Mg2+ | 25.93 | 24.87 | 49.20 | 59.73 |
| Systems | Ksv (M−1) | Kq × 10−9 (M−1S−1) | N | Kb (M−1) | ΔG (kcal mol−1) |
|---|---|---|---|---|---|
| HEV MTase–Mg2+ | 33.61± 0.02 | 5.81 ± 0.02 | 0.96 | 8.11 ± 0.59 | −1.239 ± 0.010 |
| Systems | K1 (M−1) | ∆H1 (kcal/mol) | T∆S1 (kcal/mol) | ∆G1 (kcal/mol) | K2 (M−1) | ∆H2 (kcal/mol) | T∆S2 (kcal/mol) | ∆G2 (kcal/mol) |
|---|---|---|---|---|---|---|---|---|
| HEV MTase–Mg2+ | 6.7 × 104 ± 1.15 × 104 | −0.08 ± 0.06 | 6.49 ± 0.10 | −6.57 ± 0.16 | 300 ± 55 | 4.00 ± 0.36 | 7.39 ± 0.44 | −3.39 ± 0.09 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hooda, P.; Ishtikhar, M.; Saraswat, S.; Bhatia, P.; Mishra, D.; Trivedi, A.; Kulandaisamy, R.; Aggarwal, S.; Munde, M.; Ali, N.; et al. Biochemical and Biophysical Characterisation of the Hepatitis E Virus Guanine-7-Methyltransferase. Molecules 2022, 27, 1505. https://doi.org/10.3390/molecules27051505
Hooda P, Ishtikhar M, Saraswat S, Bhatia P, Mishra D, Trivedi A, Kulandaisamy R, Aggarwal S, Munde M, Ali N, et al. Biochemical and Biophysical Characterisation of the Hepatitis E Virus Guanine-7-Methyltransferase. Molecules. 2022; 27(5):1505. https://doi.org/10.3390/molecules27051505
Chicago/Turabian StyleHooda, Preeti, Mohd Ishtikhar, Shweta Saraswat, Pooja Bhatia, Deepali Mishra, Aditya Trivedi, Rajkumar Kulandaisamy, Soumya Aggarwal, Manoj Munde, Nemat Ali, and et al. 2022. "Biochemical and Biophysical Characterisation of the Hepatitis E Virus Guanine-7-Methyltransferase" Molecules 27, no. 5: 1505. https://doi.org/10.3390/molecules27051505
APA StyleHooda, P., Ishtikhar, M., Saraswat, S., Bhatia, P., Mishra, D., Trivedi, A., Kulandaisamy, R., Aggarwal, S., Munde, M., Ali, N., AlAsmari, A. F., Rauf, M. A., Inampudi, K. K., & Sehgal, D. (2022). Biochemical and Biophysical Characterisation of the Hepatitis E Virus Guanine-7-Methyltransferase. Molecules, 27(5), 1505. https://doi.org/10.3390/molecules27051505









