Abstract
Quinoline is an N-heterocyclic compound commonly found in wastewater, especially that derived from coal processing, chemical, and pharmaceutical industries. In the present study, the microscopic fungus Curvularia lunata IM 4417, which is known to degrade various xenobiotics, was used. The aim of the research was to study the elimination of quinoline and its influence on fungal phospholipids, which are considered to be excellent indicators of environmental monitoring. Quinoline biodegradation products and phospholipid contents were analyzed using gas chromatography–mass spectrometry and liquid chromatography–tandem mass spectrometry. C. lunata IM 4417 degraded quinoline, which led to the formation of conjugates of glucose with hydroxylated derivatives of the compound. Toxicity tests (Artoxkit M and Microtox assay) indicated that the elimination of lower concentrations of quinoline was efficient and led to a reduction in sample toxicity. The presence of quinoline also significantly affected the profile of fatty acids and phospholipids. The addition of quinoline to a culture of C. lunata IM 4417 caused an increase in the content of phosphatidylcholine (PC) and a decrease in the amount of phosphatidylethanolamine (PE), two major structural lipids. Additionally, decreases in the contents of phosphatidylinositol (PI) and phosphatidylserine (PS), which are responsible for tolerance to toxic substances, cell viability, and signal transduction, were noted. Thus, it can be concluded that the presence of quinoline modifies the membrane composition, and this change may be an important indicator of the presence of N-heterocyclic compounds or other toxins in the environment.
1. Introduction
Quinoline is an N-heterocyclic compound composed of a condensed benzene and pyridine ring and commonly occurs in coal tar, oil shale, crude oil, and petroleum (Figure 1). This compound is used as a raw material or solvent in the pharmaceutical, chemical, dye, and food industries [1,2,3].
Figure 1.
Chemical structure of quinoline.
Due to that fact, quinoline is commonly found in wastewater generated from the coal pyrolysis process, shale oil production, and drug manufacturing. Literature data indicate that quinoline concentration in semi-cooking wastewater is as high as 80 mg/L. High quinoline content has also been reported in soil and groundwater contaminated by creosote [4,5]. The relatively good solubility of quinoline in water, its low biodegradability, and widespread utilization have contributed to the contamination of diverse ecosystems. Moreover, the literature data indicate that quinoline is bioaccumulated by various organisms and can negatively affect different trophic levels. Its ecotoxicity has been demonstrated in tests using organisms such as Aliivibrio fischeri, Pseudomonas putida, Daphnia magna, Desmodesmus subspicatus, and Danio rerio [6,7,8]. Moreover, quinoline and its derivatives have been described to possess genotoxic and mutagenic activity [7]. Because quinoline and its derivatives are present in soil and water ecosystems, it is important to determine its impact on microorganisms which are typical for such habitats and estimate their ability to degrade quinoline, especially in low, environmental concentrations (up to 100 mg/L). Monitoring of intermediates formed during biodegradation and assessing their toxicity are equally important.
In the literature, we can find relevant data addressing the bacterial degradation of quinoline, but little is known about the mechanisms of fungal response to the presence of quinoline and its biodegradation by fungi. Microorganisms from the genus Curvularia are microscopic fungi most often isolated from the soil, although they can also be isolated from plant and animal tissues [9,10]. Strains from this genus are able to undergo steroid transformation, produce compounds with antibacterial and antifungal activity, and degrade polyaromatic hydrocarbons, dyes, and polyethylene [11,12,13,14,15].
In the present work, the influence of quinoline on fatty acid and phospholipid profiles of Curvularia lunata IM 4417 was assessed. Additionally, the ability of C. lunata IM 4417 to degrade and detoxify quinoline was determined using toxicity tests based on organisms from different trophic levels. It is worth mentioning that the C. lunata IM 4417 strain has been previously described as a fungus that is able to 11β-hydroxylate cortexolone and degrade anthracene, phenanthrene, and organotin compounds [16,17,18].
2. Results
2.1. Biodegradation and Detoxification of Quinoline by C. lunata IM 4417
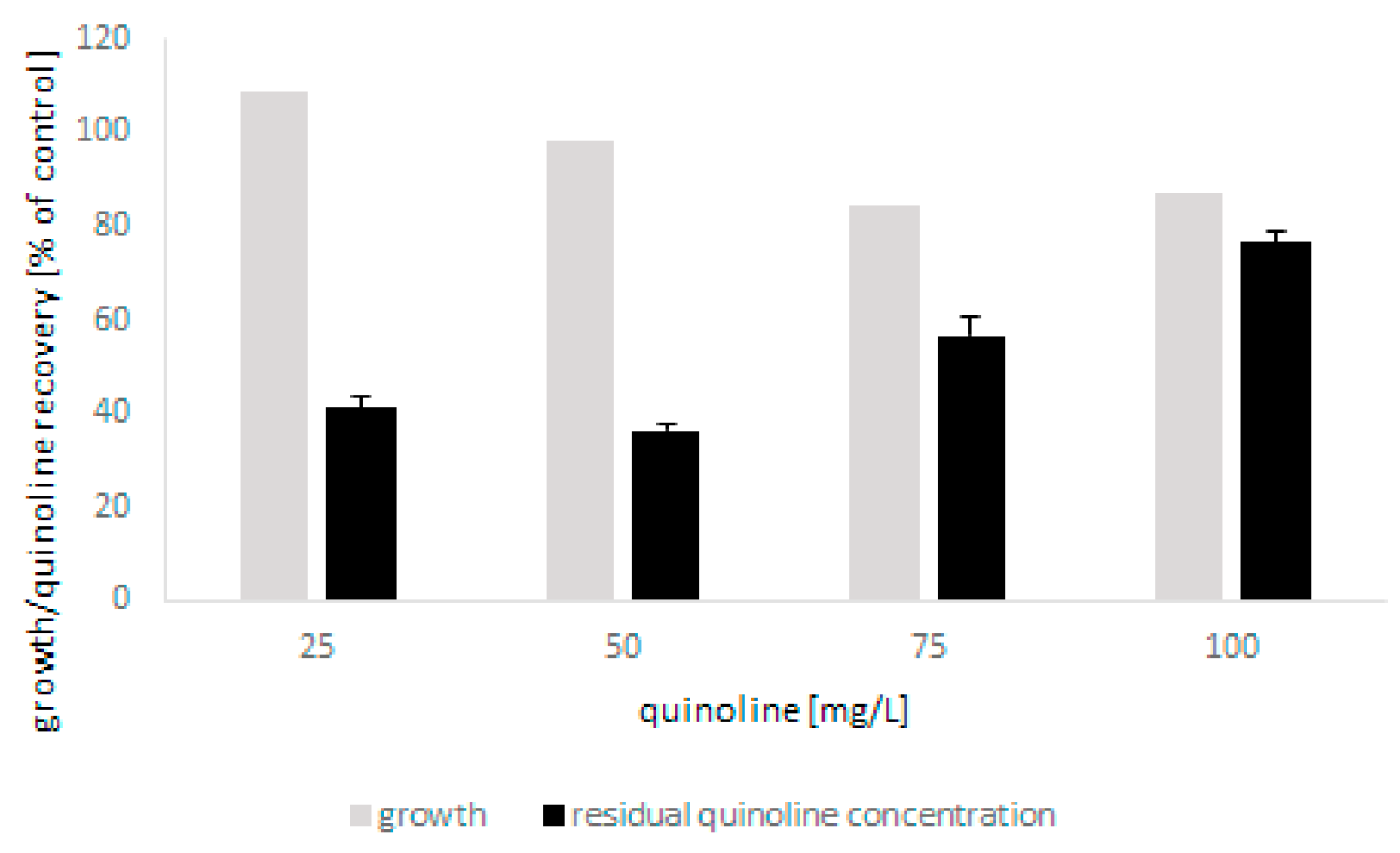
The tests showed that the microscopic fungus was able to grow in the presence of quinoline up to 100 mg/L without a significant limitation of biomass production. The biodegradation of N-heterocyclic compound was found to be closely correlated with the initial concentration of quinoline and was 60% and 20% for the lowest and highest concentrations, respectively (Figure 2). The conducted analyses also showed that bioconversion of quinoline led to the formation of small amounts of 2-hydroxyquinoline. Moreover, detailed chromatographic analysis revealed that during biodegradation, conjugates of glucose and quinoline and its hydroxylated derivatives were formed (Table 1). In the present work, three different glucose and hydroxyquinoline conjugates were determined, which may indicate that metabolites hydroxylated in various positions were formed during bioconversion of the substrate.
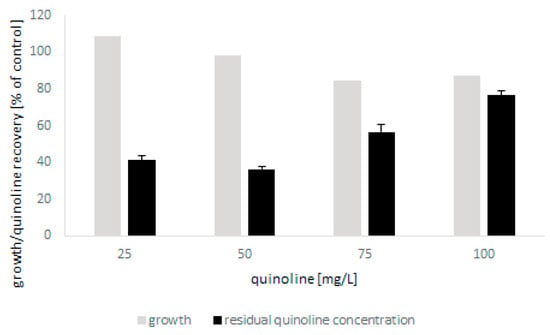
Figure 2.
The growth of C. lunata IM 4417 in the presence of quinoline and its elimination during 48 h of incubation.

Table 1.
LC-MS/MS results of quinoline biotransformation by C. lunata IM 4417.
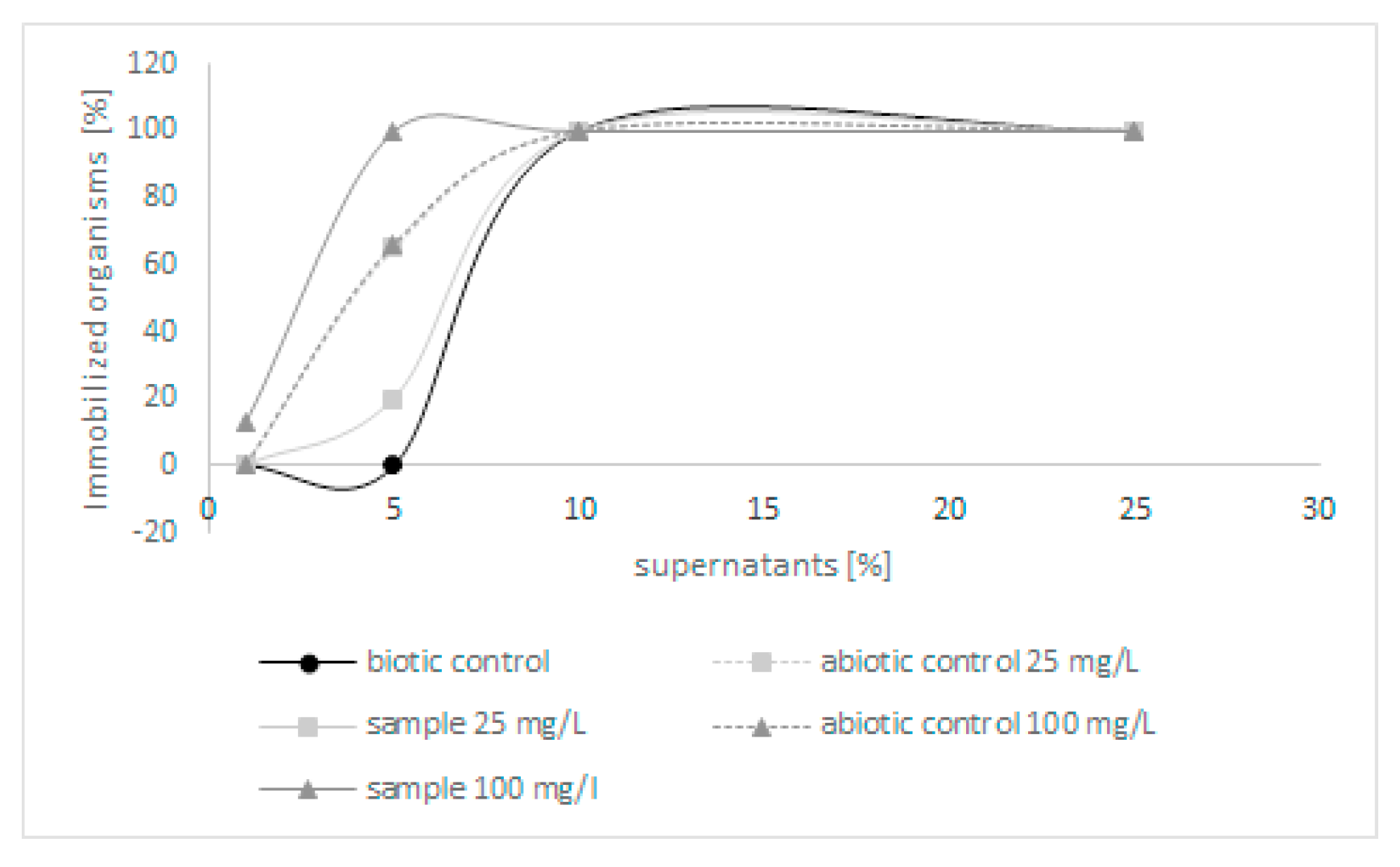
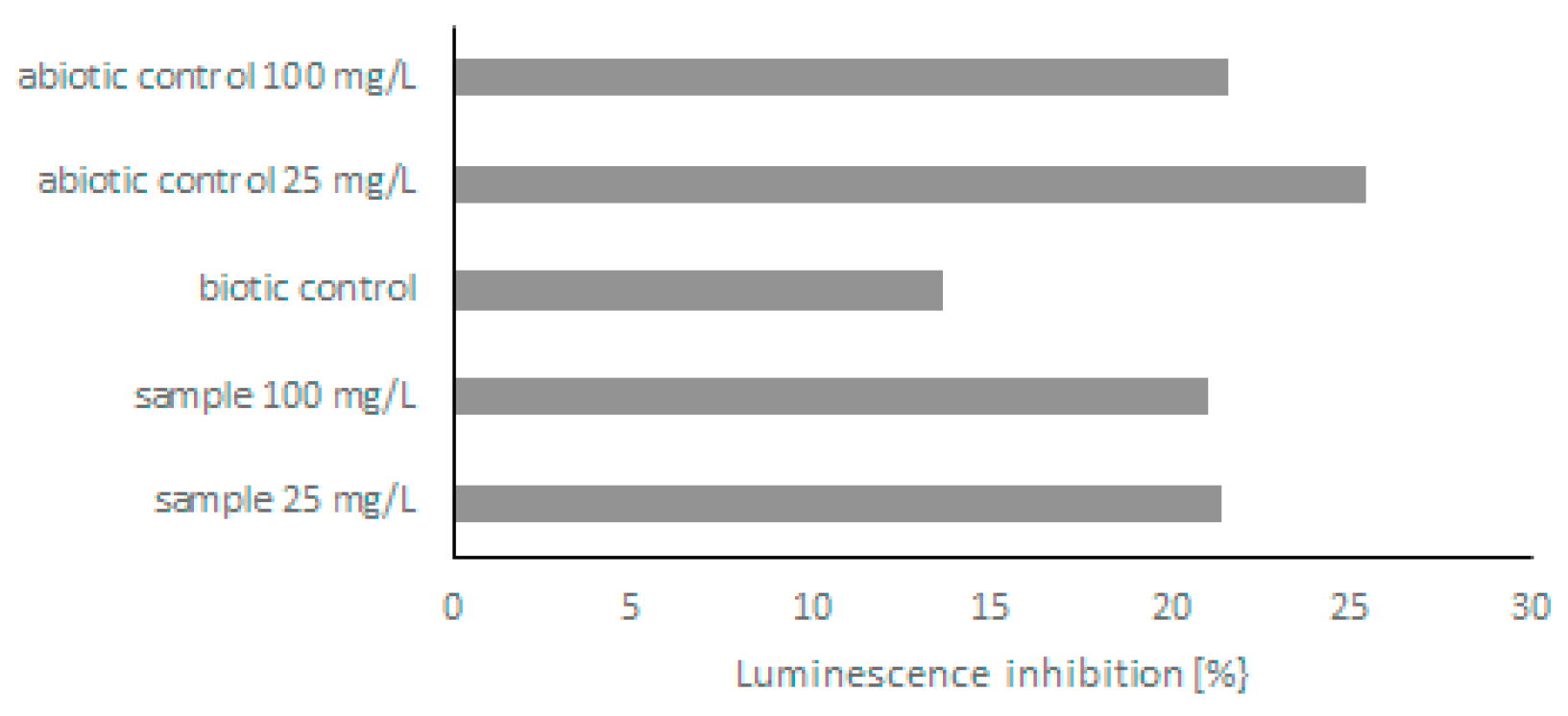
Because few derivatives were formed during the biodegradation of quinoline, in the next step, the toxicity of post-culture liquids was assessed. The ability of C. lunata IM 4417 to detoxify quinoline was determined using two different Toxkits: Artoxkit M and Microtox assay. Based on the results obtained from the Artoxkit assay, it can be stated that the fluids obtained after the incubation of C. lunata IM 4417 with 25 mg/L quinoline showed much lower toxicity than the corresponding abiotic controls. Such a relationship was not found for the probes containing the highest tested concentrations of N-heterocyclic compounds (Figure 3). Additionally, the Microtox test showed that, in the samples containing 25 mg/L quinoline and C. lunata IM 4417, the inhibition of luminescence was lower than that in analogous abiotic controls. In the case of samples containing 100 mg/L of N-heterocyclic compound, the toxicity levels of samples with fungus and abiotic control were the same (Figure 4).
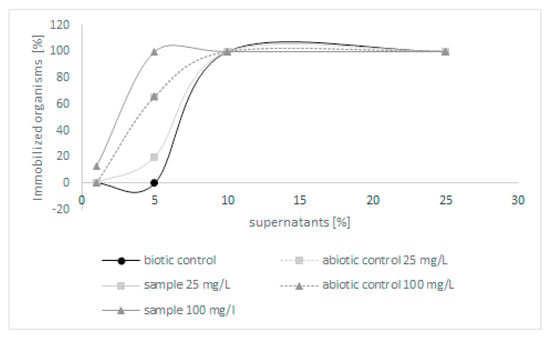
Figure 3.
The toxicity of abiotic controls and post-culture liquids obtained after 48 h of incubation of C. lunata IM 4417 with quinoline.
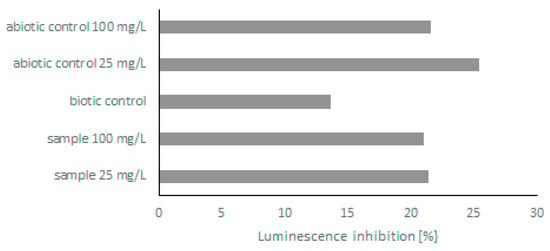
Figure 4.
Bioluminescence reduction by abiotic control and post-culture liquids obtained after 48 h of incubation of C. lunata IM 4417 with quinoline.
2.2. Modification of Fatty Acid Profile and Phospholipids of C. lunata IM 4417 during Incubation with Quinoline
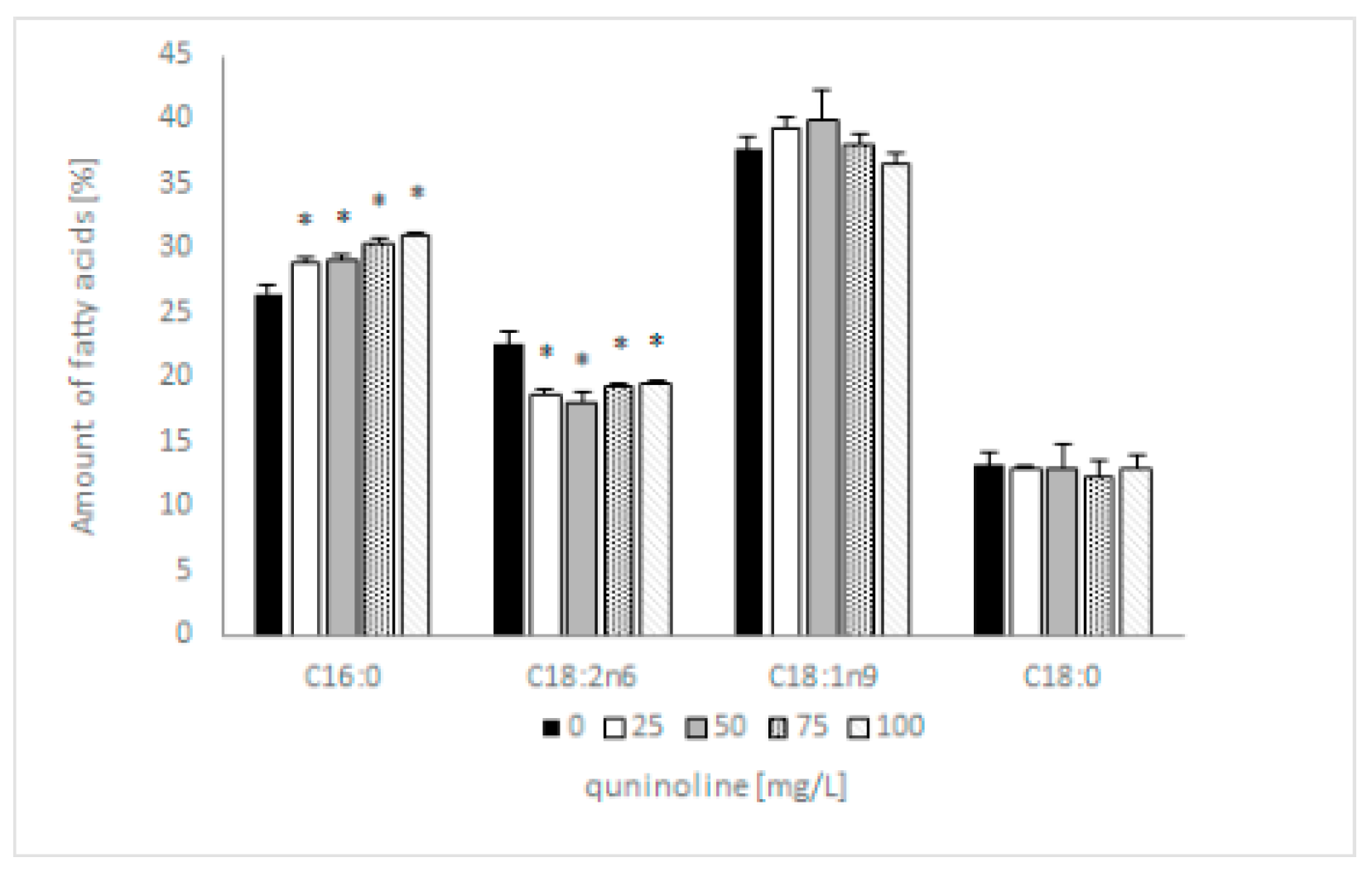
Previous research showed that 4 out of 12 identified fatty acids constitute 95% of total lipids and can be considered dominant. These were two saturated (C16:0; C18:0) and two unsaturated fatty acids (C18:1n9; C18:2n6). The addition of quinoline does not affect the type of synthesized fatty acids but influences the content of individual acids. In samples containing 100 mg/L quinoline, the content of C16:0 increased by 5% and C18:2 decreased by 3% in comparison with the control (Figure 5). The amount of C18:1 in the tested samples depended on the quinoline concentration, whereas the content of C18:0 remained constant regardless of the concentration of quinoline. The discussed changes influenced the value of the saturation index, which increased from 0.66 to 0.78 for the biotic control and culture with 100 mg/L quinoline. To expand our knowledge regarding the effects of quinoline in C. lunata IM 4417, phospholipid analysis of the mycelium was performed in the next set of studies.
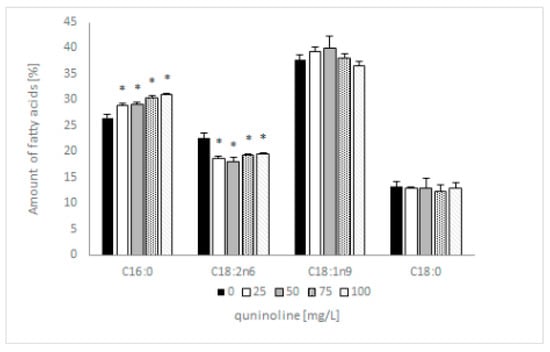
Figure 5.
Influence of quinoline on the fatty acid profile of C. lunata IM 4417 determined in the stationary phase of growth. Asterisk (p < 0.05) indicates values that differ significantly from the control.
In the present work, the following phospholipids (PLs) were analyzed: phosphatidic acid (PA), phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), phosphatidylserine (PS), and phosphatidylglycerol (PG). Among the analyzed PLs, PC and PE played dominant roles, and their sum was approximately 80% in the control samples (Table 2). On the basis of the obtained results, it can be concluded that the addition of quinoline essentially affects the two most important groups of phospholipids, i.e., PE and PC. In samples containing 25 mg/L quinoline, a four-fold decrease in the content of PA and a two-fold increase in the level of PC were observed. At the same time, a more than six-fold reduction in PI and PS content was noted. Additionally, it was found that the content of PE also decreased in the mentioned samples. Similar changes were observed in all probes containing quinoline, which resulted in a modification of the PC/PE ratio. The mentioned index reached values of 0.52, 1.51, and 2.48 for the control, the sample containing 25 mg/L quinoline, and the probe with 100 mg/L quinoline, respectively.

Table 2.
The percentage of total phospholipids and their characteristic index values. Asterisk (p < 0.05) indicates values that differ significantly from the control.
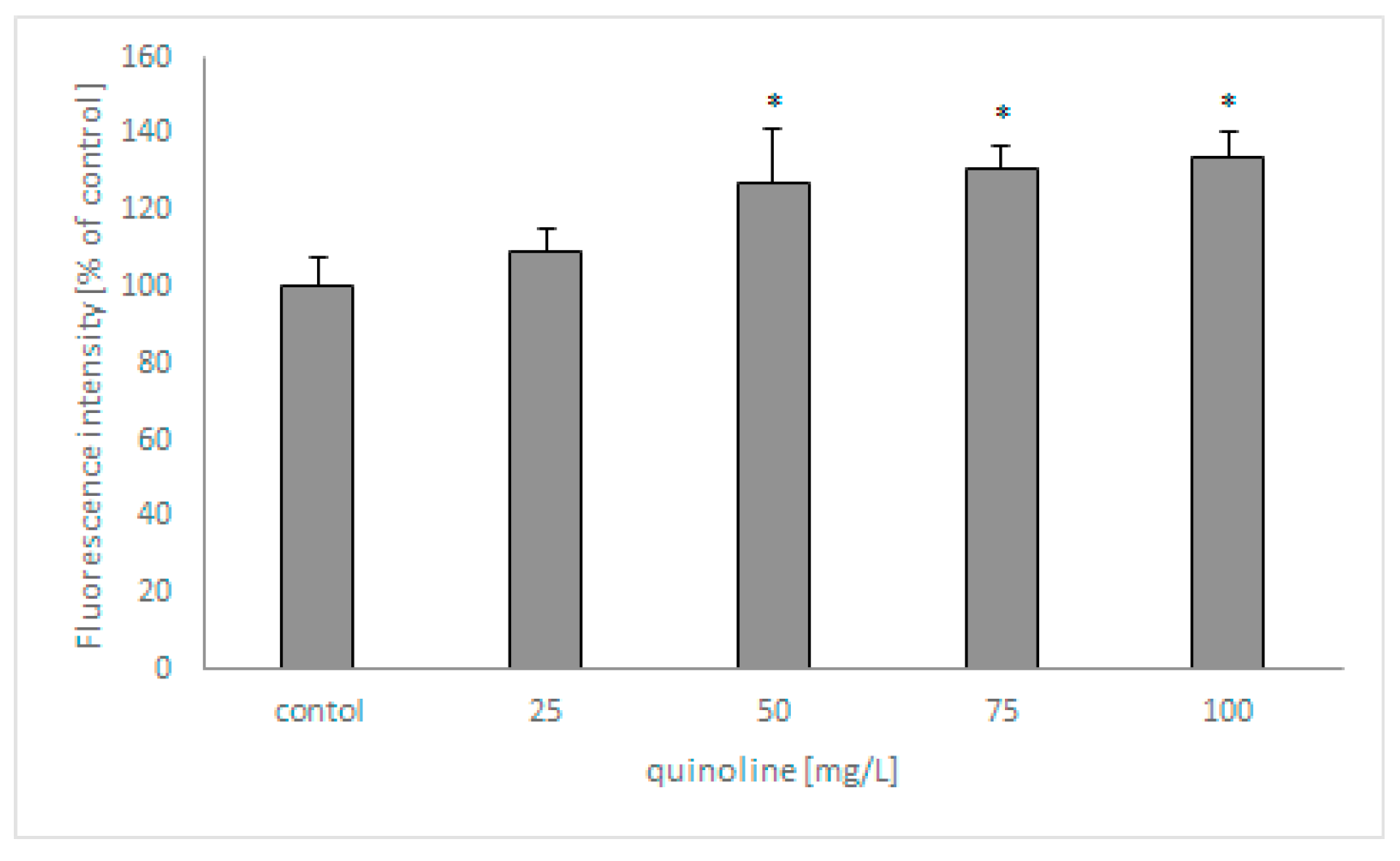
In the next set of studies, the influence of quinoline on fungal membrane permeabilization was determined using SYTOX Green (Figure 6). The analysis revealed that the fluorescence of the samples increased with increasing concentrations of quinoline and reached 135% in samples containing the studied compound at 100 mg/L.
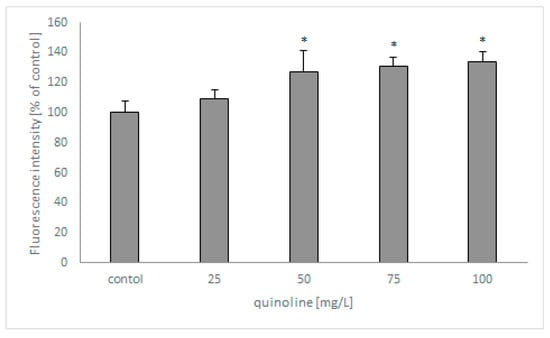
Figure 6.
The effect of quinoline on the membrane permeabilization of C. lunata IM 4417. Asterisk (p < 0.05) indicates values that differ significantly from the control.
3. Discussion
Quinoline, a typical N-heterocyclic compound, is one of the main pollutants formed during the processing of creosote and coal tar. Its good solubility, high toxicity, and low biodegradability has made it a serious threat to humans and nature. Despite the widespread presence of quinoline in the environment, the knowledge of its effects on fungi and their metabolism is still limited. Due to the fact that fungi are considered as useful models of mammalian metabolism, the Curvularia lunata strain was selected for the study. Abundant data in the literature indicate that fungi from the genus Curvularia have the ability to degrade various xenobiotics, such as phenanthrene, fluorene, or organotin compounds [17,18]. The present study focuses not only on the ability of Curvularia lunata IM 4417 to degrade and detoxify quinoline but also on adaptive changes occurring in the presence of the N-heterocyclic compound. Importantly, in the present work, quinoline concentrations typical for wastewater and ground water contaminated by creosote were used.
Many types of bacteria are characterized by high tolerance to quinoline and have the ability to degrade it, including Pseudomonas, Rhodococcus, Streptomyces, Comamonas, Burkholderia, and Bacillus strains [19,20,21,22]. The bacterial degradation of quinoline carried out under aerobic conditions is based on numerous hydroxylation reactions leading to the formation of dihydroxy derivatives and the breakdown of the rings that build the quinoline molecule. Moreover, literature data confirm that the first metabolite that is formed, regardless of the type of pathway, is 2-hydroxyquinoline.
Among microscopic filamentous fungi, only a few have been reported to grow in the presence of quinoline and have been described as capable of its elimination. Moreover, information on the biodegradation of quinoline by fungi is scarce. Similarly, little is known about the metabolites that are formed during this process. Literature data indicate that the ability to grow in the presence of quinoline is demonstrated by the white rot fungus Pleurotus ostreatus and Cunninghamella elegans 21Gp strain [23,24]. Additionally, analysis of extracts from P. ostreatus culture showed the presence of two unidentified metabolites of quinoline. In the case of C. elegans 21Gp, studies revealed that the tested microorganism converted quinoline into two hydroxylated derivatives: 2-hydroxyquinoline and 3-hydroxyquinoline. Sutherland et al. [25] also showed that a strain of C. elegans was capable of performing N-oxidation of quinoline.
The conducted analyses revealed that C. lunata IM 4417 shows a high tolerance in relation to quinoline and has the ability to eliminate the studied compound. The highest removal of quinoline was noted in samples containing up to 50 mg/L of quinoline, and in these probes, no inhibition of biomass production was observed. In addition, during the biodegradation process, trace amounts of hydroxylated derivatives of quinoline and three conjugates of hydroxyquinoline with glucose were formed. The mentioned metabolites confirm that during the degradation of quinoline by C. lunata IM 4417, hydroxylation of the studied compound first occurs, which may not be confined to the C-2 position. In the analyzed samples, N-glucose-quinoline was also identified. The presented data are in accordance with the conclusions presented in the paper of Felczak et al. [24] and confirm the fact that the hydroxylation of the tested compound may take place at several positions in the ring. Moreover, to our knowledge, this is the first paper describing the ability of microscopic filamentous fungi to produce hydroxyquinoline-glucose conjugates.
N-heterocyclic compounds, including quinoline, due to the presence of a nitrogen atom in their structure, are characterized by greater water solubility, mobility, and bioavailability than their homocyclic analogues. These features favor the accumulation of the mentioned substance in groundwater and soil, which in turn leads to increased exposure to quinoline organisms representing various trophic levels [6,26]. Because different derivatives are formed during the degradation of quinoline, including a few conjugates with glucose, in the next stage of studies, the toxicity of post-culture liquids was assessed. The research was carried out on the marine crustacean Artemia franciscana and bacteria Aliivibrio fischeri using the Artoxkit M and Microtox assays, respectively. Both tests are commonly used to determine the toxicity of various chemicals, waters, and wastewater. Additionally, the use of the mentioned toxicity models enabled us to obtain information about the influence of quinoline on different trophic levels.
Analysis of post-culture fluids containing 25 mg/L of quinoline using Artoxkit M showed that they were less toxic than the corresponding abiotic control. In these samples, good mycelium growth and high quinoline elimination was accomplished with the production of glucose conjugates with quinoline derivatives. This indicated that degradation of quinoline leads to detoxification of the N-heterocyclic compound. Additionally, the Microtox assay indicated that samples containing the lowest concentration of N-heterocyclic compound after incubation with C. lunata IM 4417 exhibited lower toxicity. To our knowledge, this is the first report showing the ability of C. lunata IM 4417 to degrade and detoxify quinolone.
It is well known that quinoline and its derivatives are ecotoxic compounds. Neuwoehnet et al. [6] found that the experimental EC50 values in Daphnia and Aliivibrio fischeri tests for quinoline reached 31.8 and 6.7 mg/L, respectively. The same authors also showed that the EC50 for 2-hydroxyquinoline was 63.4 mg/L for the Daphnia test and 0.9 mg/L for the Aliivibrio fischeri assay. Oberoi and Philip [27] studied the toxicity of quinoline and its hydroxylated derivatives and indicated that these compounds caused acute toxicity in E. coli and P. fluorescence tests. The EC50 values in the P. fluorescence assay were 49.4 and 75.0 mg/L for quinoline and 2–hydroxyquinoline, respectively. The literature data also indicate that biotransformation of quinoline does not always have to be associated with toxicity reduction, and hydroxylated derivatives may show even higher toxicity than parent compounds [6].
Because many toxic substances present in the environment interfere with components of cell structures, the fatty acid composition and the phospholipid profile are extremely important ecological indicators. Among membrane lipids, glycerophospholipids are the largest group and constitute approximately 70% of total membrane lipids. They are extremely sensitive to any changes in the environment and are responsible for the development of adaptations to various stress factors. PLs are the main structural components of the cell membrane, and they determine its stability, fluidity, and proper functioning. Some of them may also be involved in signal transduction or secretion [28].
The properties of the membrane are determined by the type of fatty acids, PL profile, acyl chain composition, and saturation index, and they are responsible for its proper functioning. The presented results indicate that the addition of quinoline to C. lunata IM 4417 causes a reduction in the C:18:2 content, with a simultaneous increase in the amount of C:16:0. Because linoleic acid is considered one of most important biomarker fatty acids in fungi [29], the significant decrease in C:18:2 content seems to be very important in an ecological context. Changes in fatty acids influenced the saturation index, which increased by 20%.
Literature data indicate that the presence of other xenobiotics can cause similar changes. Bernat and Długoński [30] indicated that the addition of tributyltin to C. elegans 21 Gp culture increases the content of saturated fatty acids. Additionally, studies investigating the influence of octyltin on the fatty acid profile of C. lunata showed that the presence of organotin compounds causes a decrease in the unsaturation index [18]. On the other hand, salt stress can cause both an increase in the content of unsaturated fatty acids or a decrease, depending on the strain tested, as demonstrated by Turk et al. [31], who studied yeast-like fungi.
Analysis of PL composition indicated that the main lipids in C. lunata IM 4417 biomass are PCs and PEs. This result is in accordance with published data, which show that the mentioned lipids account for over 50% of all PLs in fungi [32,33]. Both PC and PE are major structural lipids and determine membrane fluidity, wherein PCs are typical bilayer lipids and PEs are nonbilayer molecules. On the basis of the presented results, it can be concluded that the addition of quinoline causes an increase in PC content and a decrease in PE amount. These PLs are crucial for vegetative fungal growth [33]. Thus, this change may be essential to maintaining the proper functioning of the cell in the presence of quinoline and can lead to increased membrane fluidity. This hypothesis is confirmed by the results obtained in the SYTOX Green test, which has also shown that in the presence of quinoline, the membrane permeability increases. Increased membrane permeabilization in fungal cells was also shown in the presence of the antimicrobial peptide Pv D1. Moreover, a change in membrane permeability was closely correlated with fungal growth inhibition [34].
In samples containing quinoline, decreases in the contents of PIs and PSs were also noted. PIs are considered signal molecules and are a precursor in the synthesis of sphingolipids. Published data indicated that the decrease in the content of these lipids in Saccharomyces cerevisiae reduced cell viability. Additionally, PS are believed to play a crucial role in growth and tolerance to toxic compounds in some yeast strains [35].
In conclusion, understanding the influence of various xenobiotics on the PL profile seems to be crucial for understanding the adaptive mechanisms of fungi and may be an important indicator of the state of the environment in which these microorganisms live.
4. Materials and Methods
4.1. Microorganism
The filamentous fungus Curvularia lunata IM 4417, from the Culture Collection of the Department of Industrial Microbiology and Biotechnology, University of Lodz, Lodz, Poland, was used in the study.
4.2. Preparation of Inoculum
The spores from 14-day-old cultures on ZT slants were washed with 5 mL of Sabouraud medium (Difco) and transferred to 10 mL of fresh Sabouraud medium. The obtained inoculum was incubated at 28 °C on a rotary shaker (140 rpm) for 48 h.
4.3. Dry Weight Assessment
Biomass production was determined by filtering the samples through Whatman filter no. 1 and drying the mycelium at 105 °C to obtain a constant weight.
4.4. Lipid Profile Determination
Mycelium obtained after filtration was mixed with 10 mL of methanol and homogenized on a Fast-Prep-24 h Instrument (MP Biomedicals, Eschwege, Germany). In the next step, the supernatant was collected, and after supplementation with 2 mL of 0.9% NaCl, it was extracted with chloroform. The organic phase was collected, evaporated, dissolved in a methanol/chloroform mixture (4:1, v/v), and analyzed on an Agilent 1200 HPLC system with a 3200 QTRAP (Sciex Framingham, MA, USA) mass spectrometer [28].
4.5. Elimination of Quinoline
The preparation of samples included transfer of 2 mL of inoculum to 18 mL of fresh Sabouraud medium containing appropriate amounts of quinoline (concentrations ranged from 25 to 100 mg/L). Additionally, biotic controls (without xenobiotics) and abiotic controls (without inoculum) were prepared. All samples were incubated on a rotary shaker (140 rpm) at 28 °C for 48 h.
4.6. Quinoline Determination
Quinoline and its derivative contents were analyzed using gas chromatography–mass spectrometry (GC/MS) and liquid chromatography–tandem mass spectrometry (LC-MS/MS). A detailed description of the sample preparation and chromatographic methods is included in Felczak et al. [24]. Additionally, qualitative analysis involving the loss of the neutral glucose molecule (162 Da) was carried out using an Eksigent microLC 200 System (Sciex Framingham, MA, USA) connected with an AB Sciex 4500 QTRAP mass spectrometer (Sciex Framingham, MA, USA). A C18 column (50 mm × 0.5 mm, particle size: 2.7 μm, (Sciex Framingham, MA, USA)) maintained at 45 °C was used for the chromatographic separation of quinoline metabolites. The eluents with a constant flow of 15 μL/min were water (A) and acetonitrile (B). Both eluents were supplemented with 0.1% formic acid. The injection volume was 5 µL. The mass spectra of quinoline metabolites were collected at collision energy settings of 30 ± 15 and 50 ± 10 V. The gradient of eluents and the microESI ion source parameters are presented in Table 3. The negative mode was retained for quinoline metabolite analysis. Analyst™ version 1.6.2 software (Sciex Framingham, MA, USA) was used in the qualitative analysis of quinolone metabolites.

Table 3.
Eluent gradient and the parameters of the ion source used during LC-MS/MS analysis.
4.7. Artoxkit M
The Artoxkit M assay, which is based on Artemia franciscana crustaceans, was used to determine the ability of C. lunata IM 4417 to detoxify quinoline. The fluids from 48 h cultures of fungus containing 25 mg/L and 100 mg/L quinoline were filtered and analyzed. In addition, the toxicities of the parent abiotic and biotic controls were determined. The test was performed according to the standard ASTM E1440-91.
4.8. Microtox Assay
Assessment of C. lunata IM 4417 post-culture liquids toxicity was performed using the Aliivibrio fischeri DSM 7151 test in accordance with ISO 11348-1: 2007. The mentioned test allowed us to determine the effect of a selected xenobiotic on bioluminescence emissions, which in turn is closely related to cellular respiration and metabolism. The study was carried out with the use of post-culture fluids, obtained after biomass separation from samples containing 25 mg/L and 100 mg/L of quinoline, appropriate abiotic controls, and biotic probes. Post-culture fluids were diluted so that their final concentration was 1%.
4.9. Estimation of Fungal Membrane Permeability
The influence of quinoline on fungal membrane permeability was determined using a SYTOX Green assay. SYTOX Green nucleic acid stain is a substance that penetrates through damaged cell membranes. The spores of C. lunata IM 4417 were washed from Sabouraud slants, resuspended to a final concentration (5 × 105 spores/mL) in Sabouraud medium, and supplemented with quinoline. After incubation (48 h at 28 °C), SYTOX Green was added to the studied samples, and the probes were incubated for 15 min in the dark. The fluorescence of fungal biomass was measured on a FLUOstar OMEGA at an excitation of 485 nm and emission of 535 nm. The obtained results are expressed as the percentage of fluorescence of the biotic control.
4.10. Statistical Analysis
All experiments were performed at least in triplicate and were analyzed individually. One-way analysis of variation was used to estimate the statistical significance of the results.
5. Conclusions
In summary, the conducted analyses showed that C. lunata IM 4417 has the ability to eliminate quinoline, and this process is most efficient at low concentrations. Quinoline bioconversion leads to the formation of conjugates of hydroxylated derivatives of the tested compound and glucose, as well as to a reduction in the toxicity of post-culture liquids. Furthermore, the addition of quinoline fundamentally influenced the fatty acid content of the tested strain. The presence of the compound affects the content of linoleic acid, which is an important ecological indicator and significantly modifies the phospholipid profile of C. lunata IM 4417. The addition of quinoline affects the content of PC and PE, two groups of lipids responsible for the proper functioning of the membrane and vegetative growth. These changes led to an increase in membrane permeability, which was confirmed by the SYTOX Green assay. Moreover, the changes in the content of PI and PS, which play a crucial role in signal transduction, cell viability, and tolerance to toxic compounds, were noted in the presence of quinoline.
Author Contributions
Conceptualization, A.F.; methodology, A.F., K.Z. and P.B.; investigation, A.F., K.Z., M.N.-L. and P.B.; writing—original draft preparation, A.F.; writing—review and editing, A.F. and K.L.; supervision, K.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the grant of University of Lodz for the Young Scientist nr B141000000741.02(dec.5811E-345/M/2014-1).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are not available from the authors.
References
- Fetzner, S. Bacterial degradation of pyridine, indole, quinoline, and their derivatives under different redox conditions. Appl. Microbiol. Biotechnol. 1998, 49, 237–250. [Google Scholar] [CrossRef]
- Shi, J.; Xu, C.; Han, Y.; Han, H. Enhanced anaerobic biodegradation efficiency and mechanism of quinoline, pyridine, and indole in coal gasification wastewater. Chem. Eng. J. 2019, 361, 1019–1029. [Google Scholar] [CrossRef]
- Kang, W.; Cui, Y.; Yang, Y.; Zhao, Z.; Wang, X.; Liu, X. An acid induction strategy to construct an ultralight and durable amino-functionalized graphene oxide aerogel for enhanced quinoline pollutants extraction from coking wastewater. Chem. Eng. J. 2021, 412, 128686. [Google Scholar] [CrossRef]
- Gao, Y.; Kong, X.; Zhou, A.; Yue, X.; Luo, Y.; Defemur, Z. Enhanced degradation of quinoline by coupling microbial electrolysis cell with anaerobic digestion simultaneous. Bioresour. Technol. 2020, 306, 123077. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Y.; Hou, S.; Zhou, J. Induction conditions and kinetic properties of quinoline biodegradation by a newly isolated bacterial strain. Pol. J. Environ. Stud. 2020, 29, 1715–1724. [Google Scholar] [CrossRef]
- Neuwoehner, J.; Reineke, A.K.; Hollender, J.; Eisentraeger, A. Ecotoxicity of quinoline and hydroxylated derivatives and their occurrence in groundwater of a tar-contaminated field site. Ecotoxicol. Environ. Saf. 2009, 72, 819–827. [Google Scholar] [CrossRef]
- Eisentraeger, A.; Brinkmann, C.; Hollert, H.; Sagner, A.; Tiehm, A.; Neuwoehner, J. Heterocyclic compounds: Toxic effects using algae, daphnids, and the Salmonella/microsome test taking methodical quantitative aspects into account. Environ. Toxicol. Chem. 2008, 27, 1590–1596. [Google Scholar] [CrossRef] [PubMed]
- Peddinghaus, S.; Brinkmann, M.; Bluhm, K.; Sagner, A.; Hinger, G.; Braunbeck, T.; Eisenträger, A.; Tiehm, A.; Hollert, H.; Keiter, S.H. Quantitative assessment of the embryotoxic potential of NSO-heterocyclic compounds using zebrafish (Danio rerio). Reprod. Toxicol. 2012, 33, 24–32. [Google Scholar] [CrossRef]
- Paraszkiewicz, K.; Długonski, J. Wykorzystanie grzybow mikroskopowych z rodzaju Curvularia w badaniach podstawowych i biotechnologii. Post. Mikrobiol. 2006, 45, 107–118. [Google Scholar]
- Manamgoda, D.S.; Rossman, A.Y.; Castlebury, L.A.; Chukeatirote, E.; Hyde, K.D. A taxonomic and phylogenetic re-appraisal of the genus Curvularia (Pleosporaceae): Human and plant pathogens. Phytotaxa 2015, 212, 175–198. [Google Scholar] [CrossRef] [Green Version]
- Kollerov, V.V.; Fokina, V.V.; Sukhodolskaya, G.V.; Shutov, A.A.; Donova, M.V. 11β-Hydroxylation of 6α-fluoro-16α-methyl-deoxycorticosterone 21-acetate by filamentous fungi. Appl. Microbiol. Biotechnol. 2015, 51, 169–179. [Google Scholar] [CrossRef]
- Chomcheon, P.; Wiyakrutta, S.; Aree, T.; Sriubolmas, N.; Ngamrojanavanich, N.; Mahidol, C.; Ruchirawat, S.; Kittakoop, P. Curvularides A–E: Antifungal hybrid peptide-polyketides from the endophytic fungus Curvularia geniculata. Chemistry 2010, 16, 11178–11185. [Google Scholar] [CrossRef] [PubMed]
- Han, W.B.; Lu, Y.H.; Zhang, A.H.; Zhang, G.F.; Mei, Y.N.; Jiang, N.; Lei, X.; Song, Y.C.; Ng, S.W.; Tan, R.X. Curvulamine, a new antibacterial alkaloid incorporating two undescribed units from a Curvularia species. Org. Lett. 2014, 16, 5366–5369. [Google Scholar] [CrossRef] [PubMed]
- Senthilkumar, S.; Perumalsamy, M.; Prabhu, H.J.; Thajudin, N. Optimization of process parameters for efficient decolorization of Congo red by novel fungus Curvularia sp. using Box–Behnken design. Desalin. Water Treat. 2015, 54, 1708–1716. [Google Scholar] [CrossRef]
- Raut, S.; Raut, S.; Sharma, M.; Srivastav, C.; Adhikari, B.; Sen, S.K. Enhancing degradation of low density polyethylene films by Curvularia lunata SG1 using particle swarm optimization strategy. Indian J. Microbiol. 2015, 55, 258–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lisowska, K.; Długoński, J. Removal of anthracene and phenanthrene by filamentous fungi capable of cortexolone 11-hydroxylation. J. Basic Microbiol. 1999, 39, 117–125. [Google Scholar] [CrossRef]
- Bernat, P.; Szewczyk, R.; Krupiński, M.; Długoński, J. Butyltins degradation by Cunninghamella elegans and Cochliobolus lunatus co-culture. J. Hazard. Mater. 2013, 246, 277–282. [Google Scholar] [CrossRef]
- Felczak, A.; Bernat, P.; Długoński, J. Biodegradation of octyltin compounds by Cochliobolus lunatus and influence of xenobiotics on fungal fatty acid composition. Process Biochem. 2014, 49, 295–300. [Google Scholar] [CrossRef]
- Jianlong, W.; Xiangchun, Q.; Liping, H.; Yi, Q.; Hegemann, W. Microbial degradation of quinoline by immobilized cells of Burkholderia pickettii. Water Res. 2002, 36, 2288–2296. [Google Scholar] [CrossRef]
- Felczak, A.; Zawadzka, K.; Lisowska, K. Efficient biodegradation of quinolone–Factors determining the process. Int. Biodeterior. Biodegrad. 2014, 96, 127–134. [Google Scholar] [CrossRef]
- Liu, J.; Yu, Y.; Chang, Y.; Li, B.; Bian, D.; Yang, W.; Huo, H.; Huo, M.; Zhu, S. Enhancing quinolone and phenol removal by adding Comamonas testosteroni bdg06 in treatment of accidental dye wastewater. Int. Biodeterior. Biodegrad. 2016, 115, 74–82. [Google Scholar] [CrossRef]
- Wang, P.Y.; Chen, H.; Wang, Y.; Lyu, Y.K. Quinoline biodegradation characteristics of a new quinoline-degrading strain, Pseudomonas citronellolis PY1. J. Chem. Technol. Biotechnol. 2020, 95, 2171–2179. [Google Scholar] [CrossRef]
- Zhang, X.; Yan, K.; Ren, D.; Wang, H. Studies on quinoline biodegradation by a white rot fungus (Pleurotus ostreatus BP) in liquid and solid state substrates. Fresenius Environ. Bull. 2007, 16, 632–638. [Google Scholar]
- Felczak, A.; Bernat, P.; Różalska, S.; Lisowska, K. Quinoline biodegradation by filamentous fungus Cunninghamella elegans and adaptive modifications of the fungal membrane composition. Environ. Sci. Pollut. Res. 2016, 23, 8872–8880. [Google Scholar] [CrossRef] [Green Version]
- Sutherland, J.B.; Freeman, J.P.; Williams, A.J.; Cerniglia, C.E. N-oxidation of quinoline and isoquinoline by Cunninghamella elegans. Exp. Mycol. 1994, 18, 271–274. [Google Scholar] [CrossRef]
- Lin, Q.; Jianlong, W. Biodegradation characteristics of quinoline by Pseudomonas putida. Bioresour. Technol. 2010, 101, 7683–7686. [Google Scholar] [CrossRef] [PubMed]
- Oberoi, A.S.; Philip, L. Variation in toxicity during the biodegradation of various heterocyclic and homocyclic aromatic hydrocarbons in single and multi-substrate systems. Ecotoxicol. Environ. Saf. 2017, 135, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Bernat, P.; Gajewska, E.; Szewczyk, R.; Słaba, M.; Długoński, J. Tributyltin (TBT) induces oxidative stress and modifies lipid profile in the filamentous fungus Cunninghamella elegans. Environ. Sci. Pollut. Res. 2014, 21, 4228–4235. [Google Scholar] [CrossRef] [Green Version]
- Kaur, A.; Chaudhary, A.; Kaur, A.; Choudhary, R.; Kaushik, R. Phospholipid fatty acid–a bioindicator of environment monitoring and assessment in soil ecosystem. Curr. Sci. 2005, 89, 1103–1112. Available online: https://www.currentscience.ac.in/Volumes/89/07/1103.pdf (accessed on 24 February 2005).
- Bernat, P.; Długoński, J. Tributyltin chloride interactions with fatty acids composition and degradation ability of the filamentous fungus Cunninghamella elegans. Int. Biodeterior. Biodegrad. 2007, 6, 133–136. [Google Scholar] [CrossRef]
- Turk, M.; Mejanelle, L.; Šentjurc, M.; Grimalt, J.O.; Gunde-Cimerman, N.; Plemenitaš, A. Salt-induced changes in lipid composition and membrane fluidity of halophilic yeast-like melanized fungi. Extremophiles 2004, 8, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Renne, M.F.; de Kroon, A.I. The role of phospholipid molecular species in determining the physical properties of yeast membranes. FEBS Lett. 2018, 592, 1330–1345. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wang, H.; Zhang, C.; Wu, T.; Ma, Z.; Chen, Y. Phospholipid homeostasis plays an important role in fungal development, fungicide resistance and virulence in Fusarium graminearum. Phytopathol. Res. 2019, 1, 16. [Google Scholar] [CrossRef] [Green Version]
- Mello, E.O.; Ribeiro, S.F.F.; Carvalho, A.O.; Santos, I.S.; Da Cunha, M.; Santa-Catarina, C.; Gomes, V.M. Antifungal activity of PvD1 defensin involves plasma membrane permeabilization, inhibition of medium acidification, and induction of ROS in fungi cells. Curr. Microbiol. 2011, 62, 1209–1217. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Jones, A.D.; Lau, M.W.; Yuan, Y.J.; Dale, B.E.; Balan, V. Comparative lipidomic profiling of xylose-metabolizing S. cerevisiae and its parental strain in different media reveals correlations between membrane lipids and fermentation capacity. Biotechnol. Bioeng. 2011, 108, 12–21. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).