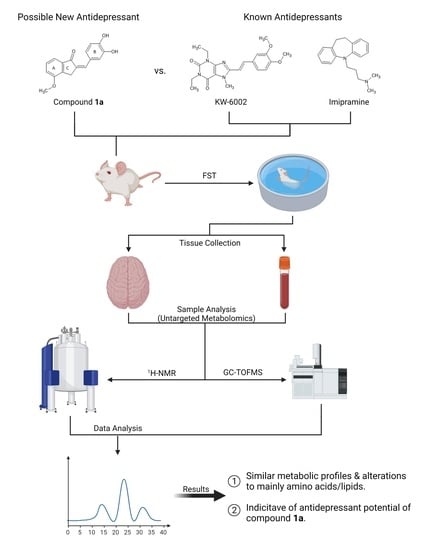
Untargeted Metabolomics Reveals the Potential Antidepressant Activity of a Novel Adenosine Receptor Antagonist
Abstract
:1. Introduction
2. Results
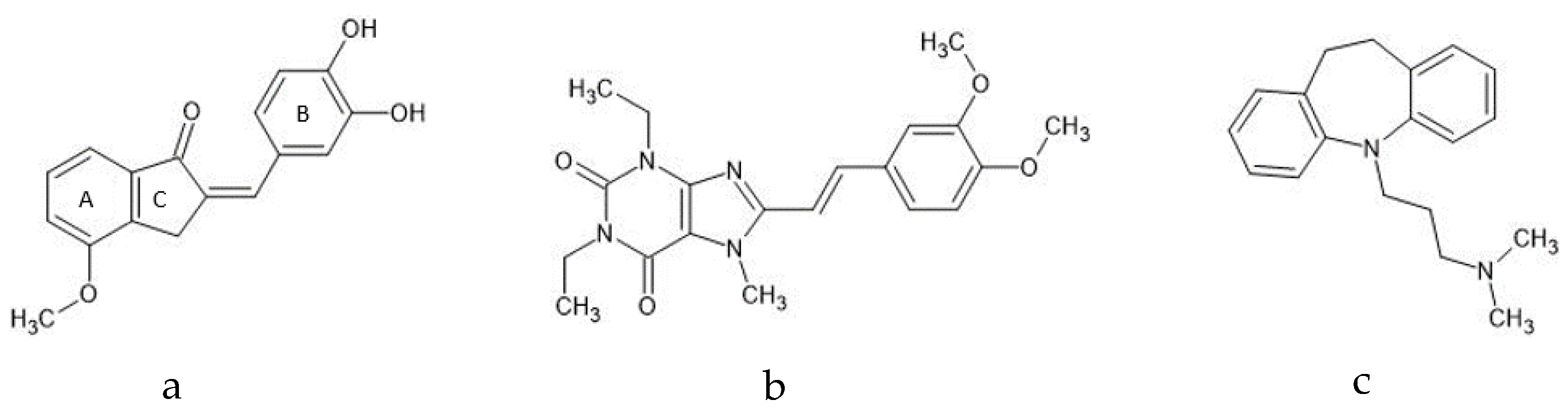
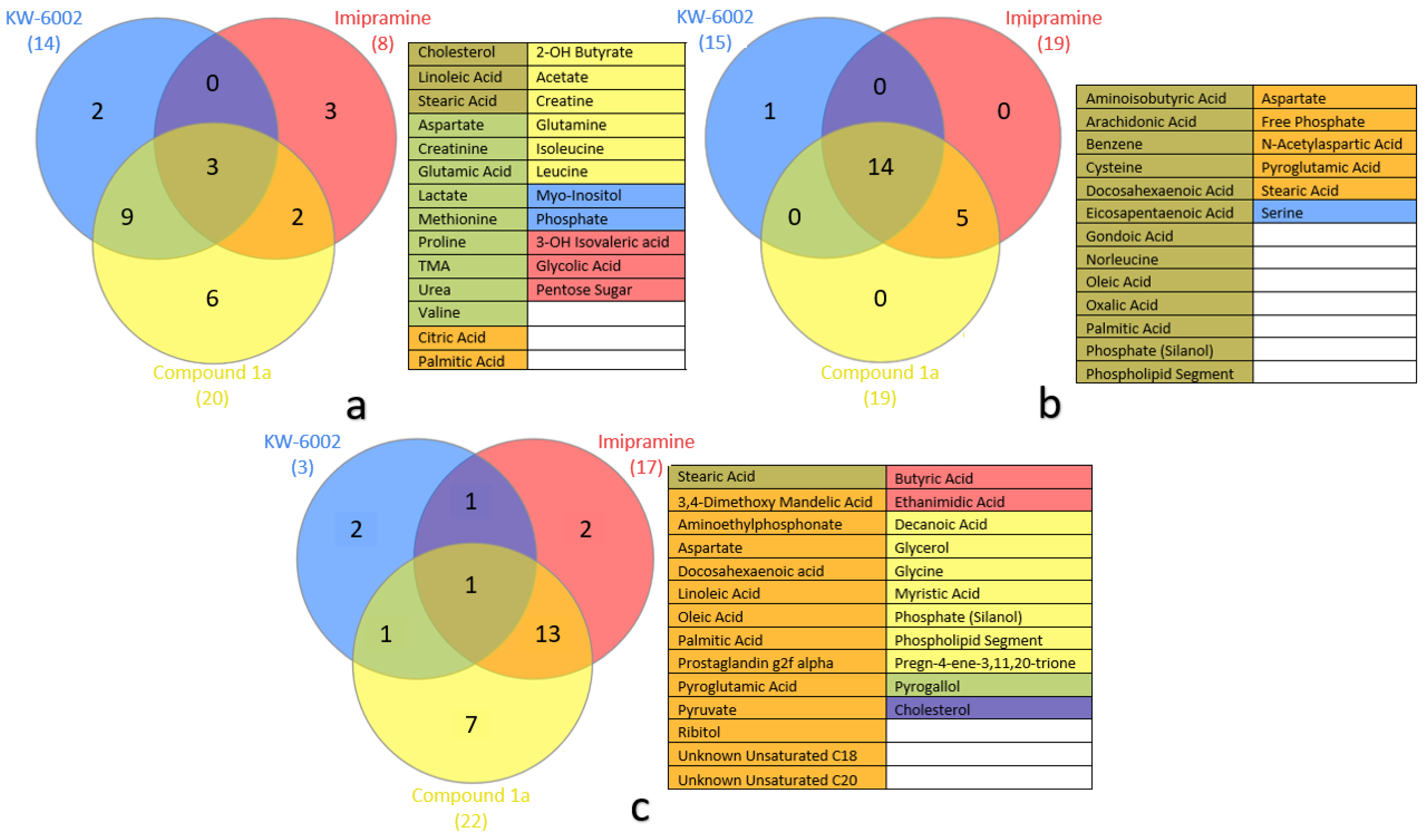
2.1. Compound 1a’s Metabolic Profile
2.2. KW-6002’s Metabolic Profile
2.3. Imipramine Metabolic Profile
2.4. Metabolite Distribution within Serum, Whole Brain (Excluding Striata), and Striata
2.5. General Observations
3. Discussion
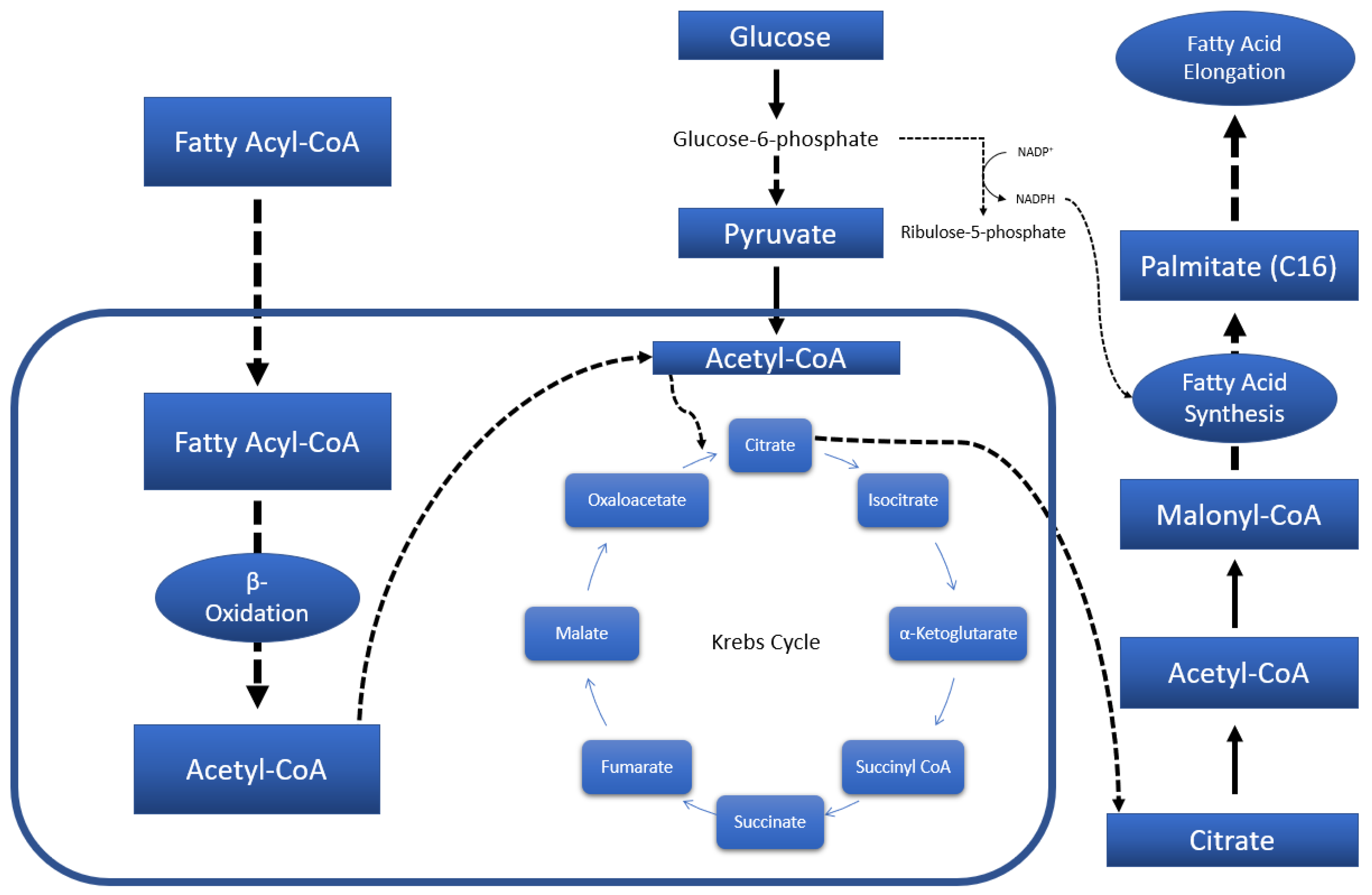
3.1. Systemic Metabolic Profiles—Via Serum
3.2. Neuronal Metabolic Alterations—Via Brain Samples
3.2.1. Striatal Metabolic Alterations
3.2.2. Whole-Brain (Excluding Striata) Metabolic Profiles
4. Materials and Methods
4.1. Ethics
4.2. Animal Model and Sampling
4.3. GC-TOFMS Sample Preparation and Analysis
4.3.1. Serum Deproteinization
4.3.2. Metabolome Extraction from Brain Tissue Samples
4.3.3. Derivatization: Oximation and Silylation
4.3.4. GC-TOFMS Analysis
4.3.5. GC-MS Data Extraction and Processing
4.4. 1H-NMR Sample Preparation and Analysis
4.4.1. Serum, Whole-Brain (Excluding Striata), and Striata Sample Preparation
4.4.2. 1H-NMR Analysis
4.4.3. 1H-NMR Data Extraction and Processing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- WHO. Depression and Other Common Mental Disorders Global Health Estimates; World Health Organization: Geneva, Switzerland, 2017.
- Stein, D.J.; Szatmari, P.; Gaebel, W.; Berk, M.; Vieta, E.; Maj, M.; de Vries, Y.A.; Roest, A.M.; de Jonge, P.; Maercker, A.; et al. Mental, behavioral and neurodevelopmental disorders in the ICD-11: An international perspective on key changes and controversies. BMC Med. 2020, 18, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayo-Clinic. Depression (Major Depressive Disorder). Available online: https://www.mayoclinic.org/diseases-conditions/depression/symptoms-causes/syc-20356007 (accessed on 5 April 2021).
- Santomauro, D.F.; Mantilla Herrera, A.M.; Shadid, J.; Zheng, P.; Ashbaugh, C.; Pigott, D.M.; Abbafati, C.; Adolph, C.; Amlag, J.O.; Aravkin, A.Y.; et al. Global prevalence and burden of depressive and anxiety disorders in 204 countries and territories in 2020 due to the COVID-19 pandemic. Lancet 2021, 398, 1700–1712. [Google Scholar] [CrossRef]
- Gautam, S.; Jain, A.; Gautam, M.; Vahia, V.N.; Grover, S. Clinical Practice Guidelines for the management of Depression. Indian J. Psychiatry 2017, 59, S34–S50. [Google Scholar] [CrossRef] [PubMed]
- Brenner, P.; Brandt, L.; Li, G.; DiBernardo, A.; Bodén, R.; Reutfors, J. Substance use disorders and risk for treatment resistant depression: A population-based, nested case-control study. Addiction 2020, 115, 768–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemke, M.R. Depressive symptoms in Parkinson’s disease. Eur. J. Neurol. 2008, 15 (Suppl. S1), 21–25. [Google Scholar] [CrossRef] [PubMed]
- Reijnders, J.S.A.M.; Ehrt, U.; Weber, W.E.J.; Aarsland, D.; Leentjens, A.F.G. A systematic review of prevalence studies of depression in Parkinson’s disease. Mov. Disord. 2008, 23, 183–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.-F.; Eltzschig, H.K.; Fredholm, B.B. Adenosine receptors as drug targets—What are the challenges? Nat. Rev. Drug Discov. 2013, 12, 265–286. [Google Scholar] [CrossRef] [Green Version]
- Cieślak, M.; Komoszyński, M.; Wojtczak, A. Adenosine A 2A receptors in Parkinson’s disease treatment. Purinergic Signal. 2008, 4, 305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espinosa, A.; Alegret, M.; Valero, S.; Vinyes-Junqué, G.; Hernández, I.; Mauleón, A.; Rosende-Roca, M.; Ruiz, A.; López, O.; Tárraga, L.; et al. A Longitudinal Follow-Up of 550 Mild Cognitive Impairment Patients: Evidence for Large Conversion to Dementia Rates and Detection of Major Risk Factors Involved. J. Alzheimer’s Dis. 2013, 34, 769–780. [Google Scholar] [CrossRef] [Green Version]
- Antonioli, L.; Blandizzi, C.; Csóka, B.; Pacher, P.; Haskó, G. Adenosine signalling in diabetes mellitus—pathophysiology and therapeutic considerations. Nat. Rev. Endocrinol. 2015, 11, 228–241. [Google Scholar] [CrossRef] [PubMed]
- Gessi, S.; Merighi, S.; Sacchetto, V.; Simioni, C.; Borea, P.A. Adenosine receptors and cancer. Biochim. Biophys. Acta (BBA)-Biomembr. 2011, 1808, 1400–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yacoubi, M.E.; Ledent, C.; Parmentier, M.; Bertorelli, R.; Ongini, E.; Costentin, J.; Vaugeois, J.M. Adenosine A2A receptor antagonists are potential antidepressants: Evidence based on pharmacology and A2A receptor knockout mice. Br. J. Pharmacol. 2001, 134, 68–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, K.; Kobayashi, M.; Mori, A.; Jenner, P.; Kanda, T. Antidepressant-like activity of the adenosine A2A receptor antagonist, istradefylline (KW-6002), in the forced swim test and the tail suspension test in rodents. Pharmacol. Biochem. Behav. 2013, 114–115, 23–30. [Google Scholar] [CrossRef]
- Yamada, K.; Kobayashi, M.; Kanda, T. Chapter Fifteen-Involvement of Adenosine A2A Receptors in Depression and Anxiety. In International Review of Neurobiology; Mori, A., Ed.; Academic Press: Cambridge, MA, USA, 2014; Volume 119, pp. 373–393. [Google Scholar]
- van der Walt, M.M.; Terre’Blanche, G. Selected C8 two-chain linkers enhance the adenos ine A1/A2A receptor affinity and selectivity of caffeine. Eur. J. Med. Chem. 2017, 125, 652–656. [Google Scholar] [CrossRef] [PubMed]
- López-Cruz, L.; Salamone, J.D.; Correa, M. Caffeine and Selective Adenosine Receptor Antagonists as New Therapeutic Tools for the Motivational Symptoms of Depression. Front. Pharmacol. 2018, 9, 526. [Google Scholar] [CrossRef]
- Szopa, A.; Poleszak, E.; Bogatko, K.; Wyska, E.; Wośko, S.; Doboszewska, U.; Świąder, K.; Wlaź, A.; Dudka, J.; Wróbel, A. DPCPX, a selective adenosine A1 receptor antagonist, enhances the antidepressant-like effects of imipramine, escitalopram, and reboxetine in mice behavioral tests. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2018, 391, 1361–1371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Legoabe, L.J.; Van der Walt, M.M.; Terre’Blanche, G. Evaluation of 2-benzylidene-1-tetralone derivatives as antagonists of A1 and A2A adenosine receptors. Chem. Biol. Drug Des. 2018, 91, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Janse van Rensburg, H.D.; Terre’Blanche, G.; van der Walt, M.M.; Legoabe, L.J. 5-Substituted 2-benzylidene-1-tetralone analogues as A1 and/or A2A antagonists for the potential treatment of neurological conditions. Bioorganic Chem. 2017, 74, 251–259. [Google Scholar] [CrossRef]
- van Rensburg, H.J.; Legoabe, L.; Terre’Blanche, G.; Van der Walt, M. 2–Benzylidene–1–Indanone Analogues as Dual Adenosine A1/A2a Receptor Antagonists for the Potential Treatment of Neurological Conditions. Drug Res. 2019, 69, 382–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janse van Rensburg, H.D.; Legoabe, L.J.; Terre’Blanche, G.; Van der Walt, M.M. Methoxy substituted 2-benzylidene-1-indanone derivatives as A1 and/or A2A AR antagonists for the potential treatment of neurological conditions. Medchemcomm 2019, 10, 300–309. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying Small Molecules via High Resolution Mass Spectrometry: Communicating Confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Kobayashi, M.; Shiozaki, S.; Ohta, T.; Mori, A.; Jenner, P.; Kanda, T. Antidepressant activity of the adenosine A2A receptor antagonist, istradefylline (KW-6002) on learned helplessness in rats. Psychopharmacology 2014, 231, 2839. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, K.A.; Gao, Z.-G. Adenosine receptors as therapeutic targets. Nat. Rev. Drug Discov. 2006, 5, 247–264. [Google Scholar] [CrossRef] [Green Version]
- Shao, W.-H.; Chen, J.-J.; Fan, S.-H.; Lei, Y.; Xu, H.-B.; Zhou, J.; Cheng, P.-F.; Yang, Y.-T.; Rao, C.-L.; Wu, B. Combined metabolomics and proteomics analysis of major depression in an animal model: Perturbed energy metabolism in the chronic mild stressed rat cerebellum. Omics A J. Integr. Biol. 2015, 19, 383–392. [Google Scholar] [CrossRef] [Green Version]
- Shi, B.; Tian, J.; Xiang, H.; Guo, X.; Zhang, L.; Du, G.; Qin, X. A 1H-NMR plasma metabonomic study of acute and chronic stress models of depression in rats. Behav. Brain Res. 2013, 241, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Jung, Y.H.; Jang, C.G.; Lee, J.; Chun, K.H.; Kwon, S.W. Metabolomic identification of biochemical changes induced by fluoxetine and imipramine in a chronic mild stress mouse model of depression. Sci. Rep. 2015, 5, 8890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Zheng, X.; Du, G.; Li, Z.; Qin, X. Brain metabonomics study of the antidepressant-like effect of Xiaoyaosan on the CUMS-depression rats by 1H NMR analysis. J. Ethnopharmacol. 2019, 235, 141–154. [Google Scholar] [CrossRef]
- Van der Walt, M.M.; Terre’Blanche, G. 1,3,7-Triethyl-substituted xanthines—possess nanomolar affinity for the adenosine A1 receptor. Bioorganic Med. Chem. 2015, 23, 6641–6649. [Google Scholar] [CrossRef] [PubMed]
- Gray, L.R.; Tompkins, S.C.; Taylor, E.B. Regulation of pyruvate metabolism and human disease. Cell Mol. Life Sci. 2014, 71, 2577–2604. [Google Scholar] [CrossRef] [Green Version]
- Tracey, T.J.; Steyn, F.J.; Wolvetang, E.J.; Ngo, S.T. Neuronal Lipid Metabolism: Multiple Pathways Driving Functional Outcomes in Health and Disease. Front. Mol. Neurosci. 2018, 11, 10. [Google Scholar] [CrossRef] [Green Version]
- Abbott, S.K.; Else, P.L.; Atkins, T.A.; Hulbert, A.J. Fatty acid composition of membrane bilayers: Importance of diet polyunsaturated fat balance. Biochim. Biophys. Acta (BBA)-Biomembr. 2012, 1818, 1309–1317. [Google Scholar] [CrossRef] [Green Version]
- Myristic Acid (HMDB0000806). Available online: https://hmdb.ca/metabolites/HMDB0000806 (accessed on 26 January 2021).
- Cholesterol (HMDB0000067). Available online: https://hmdb.ca/metabolites/HMDB0000067 (accessed on 26 January 2021).
- Stone, V.; Kudo, K.; August, P.; Marcelino, T.; Matté, C. Polyols accumulated in ribose-5-phosphate isomerase deficiency increase mitochondrial superoxide production and improve antioxidant defenses in rats’ prefrontal cortex. Int. J. Dev. Neurosci. 2014, 37, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Palmer, B.F.; Alpern, R.J. CHAPTER 12—Metabolic Acidosis. In Comprehensive Clinical Nephrology, 4th ed.; Floege, J., Johnson, R.J., Feehally, J., Eds.; Mosby: Philadelphia, PA, USA, 2010; pp. 155–166. [Google Scholar]
- Mandelic Acid (HMDB0000703). Available online: https://hmdb.ca/metabolites/HMDB0000703 (accessed on 26 January 2021).
- Tallima, H.; El Ridi, R. Arachidonic acid: Physiological roles and potential health benefits—A review. J. Adv. Res. 2017, 11, 33–41. [Google Scholar] [CrossRef]
- Dalangin, R.; Kim, A.; Campbell, R.E. The Role of Amino Acids in Neurotransmission and Fluorescent Tools for Their Detection. Int. J. Mol. Sci. 2020, 21, 6197. [Google Scholar] [CrossRef]
- Patri, M. Synaptic Transmission and Amino Acid Neurotransmitters; IntechOpen: London, UK, 2019. [Google Scholar]
- Hetherington, H.; Petroff, O.; Jackson, G.D.; Kuzniecky, R.I.; Briellmann, R.S.; Wellard, R.M. CHAPTER 13—Magnetic Resonance Spectroscopy. In Magnetic Resonance in Epilepsy, 2nd ed.; Kuzniecky, R.I., Jackson, G.D., Eds.; Academic Press: Burlington, NJ, USA, 2005; pp. 333–383. [Google Scholar]
- Scherma, M.; Masia, P.; Satta, V.; Fratta, W.; Fadda, P.; Tanda, G. Brain activity of anandamide: A rewarding bliss? Acta Pharmacol. Sin. 2019, 40, 309–323. [Google Scholar] [CrossRef]
- Yankelevitch-Yahav, R.; Franko, M.; Huly, A.; Doron, R. The forced swim test as a model of depressive-like behavior. J. Vis. Exp. 2015, 97, 52587. [Google Scholar] [CrossRef] [PubMed]
- Lindeque, J.Z.; Hidalgo, J.; Louw, R.; van der Westhuizen, F.H. Systemic and organ specific metabolic variation in metallothionein knockout mice challenged with swimming exercise. Metab. Off. J. Metab. Soc. 2013, 9, 418. [Google Scholar] [CrossRef]
- Venter, L.; van Rensburg, P.J.; Loots, D.T.; Lindeque, J.Z.; Vosloo, A. Untargeted Metabolite Profiling of Abalone Using Gas Chromatography Mass Spectrometry. Food Anal. Methods 2016, 9, 1254–1261. [Google Scholar] [CrossRef]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef] [PubMed]
- Bester, R.; Stander, Z.; Mason, S.; Keane, K.M.; Howatson, G.; Clifford, T.; Stevenson, E.J.; Loots, D.T. Characterizing Marathon-Induced Metabolic Changes Using 1H-NMR Metabolomics. Metabolites 2021, 11, 656. [Google Scholar] [CrossRef] [PubMed]
- Mason, S.; Terburgh, K.; Louw, R. Miniaturized 1 H-NMR method for analyzing limited-quantity samples applied to a mouse model of Leigh disease. Metabolomics 2018, 14, 74. [Google Scholar] [CrossRef] [PubMed]


| Associated Metabolism | Metabolite + | p-Value | Alteration | ||
|---|---|---|---|---|---|
| Whole Brain α | Striata δ | Serum | |||
| Amino Acid | Methionine | 0.0018 | ↓ | ||
| Amino Acid | Isoleucine | 0.002 | ↓ | ||
| Amino Acid | Leucine | 0.003 | ↓ | ||
| Amino Acid | Proline | β * | ↓ | ||
| Amino Acid | Glutamine | 0.009 | ↓ | ||
| Amino Acid | Valine | 0.007 | ↓ | ||
| Amino Acid | Cysteine | 0.0032 | ↑ | ||
| Amino Acid | Norleucine | 0.0019 | ↑ | ||
| Amino Acid | Glycine | 0.0022 | ↓ | ||
| Amino Acid | Aspartate | β *; 0.004; β * | ↓ | ↓ | ↓ |
| Amino Acid | 2-OH Butyrate | 0.0026 | ↓ | ||
| Amino Acid | Creatine | 0.0004 | ↓ | ||
| Amino Acid | Creatinine | 0.0029 | ↓ | ||
| Amino Acid | N-Acetylaspartic Acid (2) | β * | ↑ | ||
| Amino Acid | Aminoisobutyric Acid (2) | β * | ↑ | ||
| Amino Acid | Glutamic Acid | 0.018 | ↓ | ||
| Lipid | Stearic Acid | 0.019; 0.006; β * | ↑ | ↓ | ↓ |
| Lipid | Palmitic Acid | β *; 0.013; β * | ↑ | ↓ | ↓ |
| Lipid | Linoleic Acid | 0.0014; 0.0081 | ↓ | ↓ | |
| Lipid | Cholesterol | 0.013 | ↓ | ||
| Lipid | Gondoic Acid (11-Eicosenoic Acid) | 0.0066 | ↑ | ||
| Lipid | Eicosapentaenoic Acid (cis-5,8,11,14,17) | β * | ↑ | ||
| Lipid | Arachidonic Acid | β * | ↑ | ||
| Lipid | Docosahexaenoic acid (cis-4,7,10,13,16,19) | β *; 0.012 | ↑ | ↓ | |
| Lipid | Oleic Acid | β *; β * | ↑ | ↓ | |
| Lipid | Nonadecanoic acid | β * | ↓ | ||
| Lipid | Unknown Unsaturated C18 (2) | β * | ↓ | ||
| Lipid | 3,4-Dimethoxy Mandelic Acid | β * | ↓ | ||
| Lipid | Unknown Unsaturated C20 (3) | 0.0053 | ↓ | ||
| Lipid | Decanoic Acid (2) | 0.0046 | ↓ | ||
| Lipid | Glycerol | 0.0033 | ↓ | ||
| Lipid | Myristic Acid | 0.0016 | ↓ | ||
| Lipid | Phospholipid Segment | β * | ↑ | ↑ | |
| Lipid | Free Phosphate | 0.0012 | ↑ | ||
| Lipid | Phosphate (Silanol) | β *; β * | ↑ | ↓ | |
| Glutathione | Pyroglutamic Acid | β *; 0.0016 | ↓ | ↓ | |
| Inflammation | Prostaglandin g2f alpha (3) | β * | ↓ | ||
| Pentose Phosphate | Ribitol (2) | 0.0038 | ↓ | ||
| Steroid Hormone | Pregn-4-ene-3,11,20-trione (3) | 0.0043 | ↑ | ||
| Central Carbon | Lactate | β * | ↓ | ||
| Central Carbon | Acetate | β * | ↓ | ||
| Central Carbon | Pyruvate | β * | ↓ | ||
| Central Carbon | Citric Acid | 0.0036 | ↓ | ||
| Urea | Urea | 0.009 | ↓ | ||
| Microbiota | TMA | 0.0047 | ↓ | ||
| Microbiota | Benzene (3) | β * | ↑ | ||
| Other | Oxalic Acid (3) | β * | ↑ | ||
| Other | Aminoethylphosphonate | 0.0037 | ↓ | ||
| Other | Pyrogallol | β * | ↓ | ||
| Associated Metabolism | Metabolite + | p-Value | Alteration | ||
|---|---|---|---|---|---|
| Whole Brain α | Striata δ | Serum | |||
| Amino Acid | Methionine | 0.0018 | ↓ | ||
| Amino Acid | Glutamic Acid | 0.018 | ↓ | ||
| Amino Acid | Proline | β * | ↓ | ||
| Amino Acid | Valine | 0.007 | ↓ | ||
| Amino Acid | Cysteine | 0.0032 | ↑ | ||
| Amino Acid | Norleucine | 0.0019 | ↑ | ||
| Amino Acid | Serine | β * | ↑ | ||
| Amino Acid | Aspartate | β * | ↓ | ||
| Amino Acid | Creatinine | 0.0029 | ↓ | ||
| Amino Acid | Aminoisobutyric acid (2) | β * | ↑ | ||
| Lipid | Stearic Acid | β * | ↓ | ↓ | |
| Lipid | Linoleic Acid | 0.0081 | ↓ | ||
| Lipid | Cholesterol | 0.006; 0.013 | ↓ | ↓ | |
| Lipid | Gondoic Acid (11-Eicosenoic Acid) | 0.0066 | ↑ | ||
| Lipid | Eicosapentaenoic Acid (cis-5,8,11,14,17) (3) | β * | ↑ | ||
| Lipid | Arachidonic Acid | β * | ↑ | ||
| Lipid | Docosahexaenoic acid (cis-4,7,10,13,16,19) | β * | ↑ | ||
| Lipid | Palmitic Acid | β * | ↑ | ||
| Lipid | Oleic Acid | β * | ↑ | ||
| Lipid | Nonadecanoic acid | β * | ↓ | ||
| Lipid | Phosphate | β * | ↑ | ||
| Lipid | Phospholipid Segment | β * | ↑ | ||
| Lipid | Phosphate (Silanol) | β * | ↑ | ||
| Central Carbon | Lactate | β * | ↓ | ||
| Urea | Urea | 0.009 | ↓ | ||
| Microbiota | TMA | 0.0047 | ↓ | ||
| Microbiota | Benzene (3) | β * | ↑ | ||
| Other | Myo-Inositol | β * | ↓ | ||
| Other | Oxalic Acid (3) | β * | ↑ | ||
| Other | Pyrogallol | β * | ↓ | ||
| Associated Metabolism | Metabolite + | p-Value | Alteration | ||
|---|---|---|---|---|---|
| Whole Brain α | Striata δ | Serum | |||
| Amino Acid | Cysteine | 0.0032 | ↑ | ||
| Amino Acid | Norleucine | 0.0019 | ↑ | ||
| Amino Acid | 3-OH Isovaleric acid | 0.0015 | ↑ | ||
| Amino Acid | Aspartate | β *; 0.004 | ↓ | ↓ | |
| Amino Acid | N-Acetylaspartic Acid (2) | β * | ↑ | ||
| Amino Acid | Aminoisobutyric Acid (2) | β * | ↑ | ||
| Carbon | Glycolic Acid | β * | ↑ | ||
| Lipid | Stearic Acid | 0.019; 0.006; β * | ↑ | ↓ | ↓ |
| Lipid | Palmitic Acid | β *; 0.013; β * | ↑ | ↓ | ↓ |
| Lipid | Linoleic Acid | 0.0014; 0.0081 | ↓ | ↓ | |
| Lipid | Cholesterol | 0.006; 0.013 | ↓ | ↓ | |
| Lipid | Gondoic Acid (11-Eicosenoic Acid) | 0.0066 | ↑ | ||
| Lipid | Eicosapentaenoic Acid (cis-5,8,11,14,17) (3) | β * | ↑ | ||
| Lipid | Arachidonic Acid | β * | ↑ | ||
| Lipid | Docosahexaenoic acid (cis-4,7,10,13,16,19) | β *; 0.012 | ↑ | ↓ | |
| Lipid | Oleic Acid | β *; β * | ↑ | ↓ | |
| Lipid | Nonadecanoic acid | β * | ↓ | ||
| Lipid | Unknown Unsaturated C18 (2) | β * | ↓ | ||
| Lipid | 3,4 Dimethoxy Mandelic Acid | β * | ↓ | ||
| Lipid | Unknown Unsaturated C20 (3) | 0.0053 | ↓ | ||
| Lipid | Butyric Acid | 0.0056 | ↓ | ||
| Lipid | Ethanimidic Acid (3) | 0.013 | ↓ | ||
| Lipid | Phospholipid Segment | β * | ↑ | ||
| Lipid | Free Phosphate | 0.0012 | ↑ | ||
| Lipid | Phosphate (Silanol) | β * | ↑ | ||
| Glutathione | Pyroglutamic Acid | β *; 0.0016 | ↓ | ↓ | |
| Inflammation | Prostaglandiene g2f alpha (3) | β * | ↓ | ||
| Pentose Phosphate | Ribitol (2) | 0.0038 | ↓ | ||
| Central Carbon | Pentose Sugar | 0.013 | ↑ | ||
| Central Carbon | Pyruvate | β * | ↓ | ||
| Central Carbon | Citric Acid | 0.0036 | ↓ | ||
| Microbiota | Benzene (3) | β * | ↑ | ||
| Other | Oxalic Acid | β * | ↑ | ||
| Other | Aminoethylphosphonate | 0.0037 | ↓ | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, A.P.; Lindeque, J.Z.; van der Walt, M.M. Untargeted Metabolomics Reveals the Potential Antidepressant Activity of a Novel Adenosine Receptor Antagonist. Molecules 2022, 27, 2094. https://doi.org/10.3390/molecules27072094
Smith AP, Lindeque JZ, van der Walt MM. Untargeted Metabolomics Reveals the Potential Antidepressant Activity of a Novel Adenosine Receptor Antagonist. Molecules. 2022; 27(7):2094. https://doi.org/10.3390/molecules27072094
Chicago/Turabian StyleSmith, Arnold Petrus, Jeremie Zander Lindeque, and Mietha Magdalena van der Walt. 2022. "Untargeted Metabolomics Reveals the Potential Antidepressant Activity of a Novel Adenosine Receptor Antagonist" Molecules 27, no. 7: 2094. https://doi.org/10.3390/molecules27072094
APA StyleSmith, A. P., Lindeque, J. Z., & van der Walt, M. M. (2022). Untargeted Metabolomics Reveals the Potential Antidepressant Activity of a Novel Adenosine Receptor Antagonist. Molecules, 27(7), 2094. https://doi.org/10.3390/molecules27072094