Structural Insights into the Design of Synthetic Nanobody Libraries
Abstract
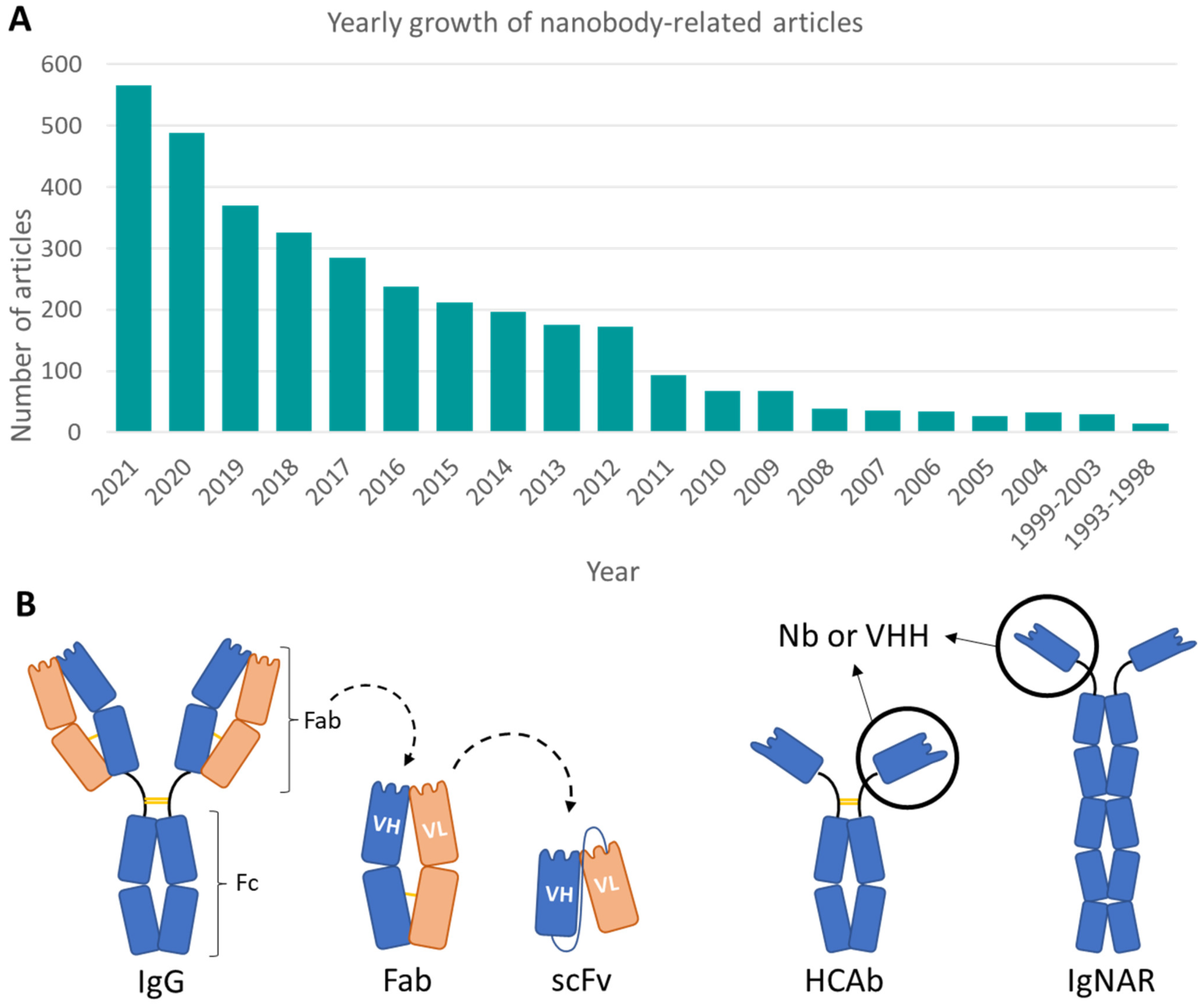
:1. Introduction
2. Structural Bases for the Design of Synthetic Nb Libraries
2.1. Key Features for Framework Selection
2.2. Design of the Hypervariable Loops
3. Advantages and Limitations of Synthetic Libraries
3.1. Advantages
3.2. Limitations
4. Selected Examples of Synthetic Nb Libraries
4.1. Moutel et al., (2016)
4.2. McMahon et al., (2018)
4.3. Zimmermann et al., (2018)
4.4. Chen et al., (2021)
4.5. Other Synthetic Libraries
4.6. Impact of Synthetic Nb Libraries
5. General Discussion
5.1. Theoretical vs. Experimental Variability
5.2. Concluding Remarks
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hamers-Casterman, C.; Atarhouch, T.; Muyldermans, S.; Robinson, G.; Hammers, C.; Songa, E.B.; Bendahman, N.; Hammers, R. Naturally Occurring Antibodies Devoid of Light Chains. Nature 1993, 363, 446–448. [Google Scholar] [CrossRef]
- Morrison, C. Nanobody Approval Gives Domain Antibodies a Boost. Nature reviews. Drug Discov. 2019, 18, 485–487. [Google Scholar] [CrossRef] [PubMed]
- Raybould, M.I.J.; Kovaltsuk, A.; Marks, C.; Deane, C.M. CoV-AbDab: The Coronavirus Antibody Database. Bioinformatics 2021, 37, 734–735. [Google Scholar] [CrossRef] [PubMed]
- Zare, H.; Aghamollaei, H.; Hosseindokht, M.; Heiat, M.; Razei, A.; Bakherad, H. Nanobodies, the Potent Agents to Detect and Treat the Coronavirus Infections: A Systematic Review. Mol. Cell. Probes 2021, 55, 101692. [Google Scholar] [CrossRef]
- Muyldermans, S. Nanobodies: Natural Single-Domain Antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef] [Green Version]
- Steeland, S.; Vandenbroucke, R.E.; Libert, C. Nanobodies as Therapeutics: Big Opportunities for Small Antibodies. Drug Discov. Today 2016, 21, 1076–1113. [Google Scholar] [CrossRef]
- Muyldermans, S. A Guide to: Generation and Design of Nanobodies. FEBS J. 2021, 288, 2084. [Google Scholar] [CrossRef]
- Goldman, E.R.; Andersen, G.P.; Liu, J.L.; Delehanty, J.B.; Sherwood, L.J.; Osborn, L.E.; Cummins, L.B.; Hayhurst, A. Facile Generation of Heat-Stable Antiviral and Antitoxin Single Domain Antibodies from a Semisynthetic Llama Library. Anal. Chem. 2006, 78, 8245–8255. [Google Scholar] [CrossRef] [Green Version]
- Könning, D.; Rhiel, L.; Empting, M.; Grzeschik, J.; Sellmann, C.; Schröter, C.; Zielonka, S.; Dickgießer, S.; Pirzer, T.; Yanakieva, D.; et al. Semi-Synthetic VNAR Libraries Screened against Therapeutic Antibodies Primarily Deliver Anti-Idiotypic Binders. Sci. Rep. 2017, 7, 9676. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.L.; Anderson, G.P.; Goldman, E.R. Isolation of Anti-Toxin Single Domain Antibodies from a Semi-Synthetic Spiny Dogfish Shark Display Library. BMC Biotechnol. 2007, 7, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grzeschik, J.; Könning, D.; Hinz, S.C.; Krah, S.; Schröter, C.; Empting, M.; Kolmar, H.; Zielonka, S. Generation of Semi-Synthetic Shark IgNAR Single-Domain Antibody Libraries. Methods Mol. Biol. 2018, 1701, 147–167. [Google Scholar] [CrossRef] [PubMed]
- Moutel, S.; Bery, N.; Bernard, V.; Keller, L.; Lemesre, E.; de Marco, A.; Ligat, L.; Rain, J.C.; Favre, G.; Olichon, A.; et al. NaLi-H1: A Universal Synthetic Library of Humanized Nanobodies Providing Highly Functional Antibodies and Intrabodies. eLife 2016, 5, 1–31. [Google Scholar] [CrossRef] [PubMed]
- McMahon, C.; Baier, A.S.; Pascolutti, R.; Wegrecki, M.; Zheng, S.; Ong, J.X.; Erlandson, S.C.; Hilger, D.; Rasmussen, S.G.F.; Ring, A.M.; et al. Yeast Surface Display Platform for Rapid Discovery of Conformationally Selective Nanobodies. Nat. Struct. Mol. Biol. 2018, 25, 289–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, I.; Egloff, P.; Hutter, C.A.J.; Arnold, F.M.; Stohler, P.; Bocquet, N.; Hug, M.N.; Huber, S.; Siegrist, M.; Hetemann, L.; et al. Synthetic Single Domain Antibodies for the Conformational Trapping of Membrane Proteins. eLife 2018, 7, e34317. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Gentili, M.; Hacohen, N.; Regev, A. A Cell-Free Nanobody Engineering Platform Rapidly Generates SARS-CoV-2 Neutralizing Nanobodies. Nat. Commun. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- Yan, J.; Li, G.; Hu, Y.; Ou, W.; Wan, Y. Construction of a Synthetic Phage-Displayed Nanobody Library with CDR3 Regions Randomized by Trinucleotide Cassettes for Diagnostic Applications. J. Transl. Med. 2014, 12, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Sevy, A.M.; Chen, M.T.; Castor, M.; Sylvia, T.; Krishnamurthy, H.; Ishchenko, A.; Hsieh, C.M. Structure- and Sequence-Based Design of Synthetic Single-Domain Antibody Libraries. Protein Eng. Des. Sel. PEDS 2020, 33, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, Y.; Su, W.; Li, S. Construction of Synthetic Nanobody Library in Mammalian Cells by DsDNA-Based Strategies. ChemBioChem 2021, 22, 2957–2965. [Google Scholar] [CrossRef] [PubMed]
- Wagner, H.J.; Wehrle, S.; Weiss, E.; Cavallari, M.; Weber, W. A Two-Step Approach for the Design and Generation of Nanobodies. Int. J. Mol. Sci. 2018, 19, 3444. [Google Scholar] [CrossRef] [Green Version]
- Chi, X.; Liu, X.; Wang, C.; Zhang, X.; Li, X.; Hou, J.; Ren, L.; Jin, Q.; Wang, J.; Yang, W. Humanized Single Domain Antibodies Neutralize SARS-CoV-2 by Targeting the Spike Receptor Binding Domain. Nat. Commun. 2020, 11, 1–7. [Google Scholar] [CrossRef]
- Ferrari, D.; Garrapa, V.; Locatelli, M.; Bolchi, A. A Novel Nanobody Scaffold Optimized for Bacterial Expression and Suitable for the Construction of Ribosome Display Libraries. Mol. Biotechnol. 2020, 62, 43–55. [Google Scholar] [CrossRef]
- Stefan, M.A.; Light, Y.K.; Schwedler, J.L.; McIlroy, P.R.; Courtney, C.M.; Saada, E.A.; Thatcher, C.E.; Phillips, A.M.; Bourguet, F.A.; Mageeney, C.M.; et al. Development of Potent and Effective Synthetic SARS-CoV-2 Neutralizing Nanobodies. mAbs 2021, 13, 442911. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Meng, W.; Guo, H.; Pan, W.; Liu, J.; Peng, T.; Chen, L.; Chen, C.Y. Potent Neutralization of Influenza A Virus by a Single-Domain Antibody Blocking M2 Ion Channel Protein. PLoS ONE 2011, 6, e28309. [Google Scholar] [CrossRef]
- Saerens, D.; Pellis, M.; Loris, R.; Pardon, E.; Dumoulin, M.; Matagne, A.; Wyns, L.; Muyldermans, S.; Conrath, K. Identification of a Universal VHH Framework to Graft Non-Canonical Antigen-Binding Loops of Camel Single-Domain Antibodies. J. Mol. Biol. 2005, 352, 597–607. [Google Scholar] [CrossRef]
- Conrath, K.E.; Lauwereys, M.; Galleni, M.; Matagne, A.; Frère, J.-M.; Kinne, J.; Wyns, L.; Muyldermans, S. β-Lactamase Inhibitors Derived from Single-Domain Antibody Fragments Elicited in the Camelidae. Antimicrob. Agents Chemother. 2001, 45, 2807. [Google Scholar] [CrossRef] [Green Version]
- Turner, K.B.; Zabetakis, D.; Goldman, E.R.; Anderson, G.P. Enhanced Stabilization of a Stable Single Domain Antibody for SEB Toxin by Random Mutagenesis and Stringent Selection. Protein Eng. Des. Sel. 2014, 27, 89–95. [Google Scholar] [CrossRef] [Green Version]
- Goldman, E.R.; Liu, J.L.; Zabetakis, D.; Anderson, G.P. Enhancing Stability of Camelid and Shark Single Domain Antibodies: An Overview. Front. Immunol. 2017, 8, 1. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.L.; Shriver-Lake, L.C.; Anderson, G.P.; Zabetakis, D.; Goldman, E.R. Selection, Characterization, and Thermal Stabilization of Llama Single Domain Antibodies towards Ebola Virus Glycoprotein. Microb. Cell Factories 2017, 16, 223. [Google Scholar] [CrossRef] [Green Version]
- Zabetakis, D.; Shriver-Lake, L.C.; Olson, M.A.; Goldman, E.R.; Anderson, G.P. Experimental Evaluation of Single-Domain Antibodies Predicted by Molecular Dynamics Simulations to Have Elevated Thermal Stability. Protein Sci. 2019, 28, 1909–1912. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.L.; Goldman, E.R.; Zabetakis, D.; Walper, S.A.; Turner, K.B.; Shriver-Lake, L.C.; Anderson, G.P. Enhanced Production of a Single Domain Antibody with an Engineered Stabilizing Extra Disulfide Bond. Microb Cell Fact 2015, 14, 158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fink, A.L. Protein Aggregation: Folding Aggregates, Inclusion Bodies and Amyloid. Fold. Des. 1998, 3, R9–R23. [Google Scholar] [CrossRef] [Green Version]
- Merlini, G.; Bellotti, V.; Andreola, A.; Palladini, G.; Obici, L.; Casarini, S.; Perfetti, V. Protein Aggregation. Clin. Chem. Lab. Med. 2001, 39, 1065–1075. [Google Scholar] [CrossRef]
- Lindner, A.B.; Demarez, A. Protein Aggregation as a Paradigm of Aging. Biochim. Biophys. Acta 2009, 1790, 980–996. [Google Scholar] [CrossRef]
- Hussack, G.; Hirama, T.; Ding, W.; Mackenzie, R.; Tanha, J. Engineered Single-Domain Antibodies with High Protease Resistance and Thermal Stability. PLoS ONE 2011, 6, 28218. [Google Scholar] [CrossRef] [Green Version]
- Dumoulin, M.; Conrath, K.; van Meirhaeghe, A.; Meersman, F.; Heremans, K.; Frenken, L.G.J.; Muyldermans, S.; Wyns, L.; Matagne, A. Single-Domain Antibody Fragments with High Conformational Stability. Protein Sci. 2002, 11, 500–515. [Google Scholar] [CrossRef]
- Mendoza, M.N.; Jian, M.; King, M.T.; Brooks, C.L. Role of a Noncanonical Disulfide Bond in the Stability, Affinity, and Flexibility of a VHH Specific for the Listeria Virulence Factor InlB. Protein Sci. 2020, 29, 990–1003. [Google Scholar] [CrossRef]
- Hagihara, Y.; Mine, S.; Uegaki, K. Stabilization of an Immunoglobulin Fold Domain by an Engineered Disulfide Bond at the Buried Hydrophobic Region. J. Biol. Chem. 2007, 282, 36489–36495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saerens, D.; Conrath, K.; Govaert, J.; Muyldermans, S. Disulfide Bond Introduction for General Stabilization of Immunoglobulin Heavy-Chain Variable Domains. J. Mol. Biol. 2008, 377, 478–488. [Google Scholar] [CrossRef]
- Zabetakis, D.; Olson, M.A.; Anderson, G.P.; Legler, P.M.; Goldman, E.R. Evaluation of Disulfide Bond Position to Enhance the Thermal Stability of a Highly Stable Single Domain Antibody. PLoS ONE 2014, 9, e115405. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.W.; Messer, A. Intrabody Applications in Neurological Disorders: Progress and Future Prospects. Mol. Ther. 2005, 12, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Marasco, W.A. Intrabodies: Turning the Humoral Immune System Outside in for Intracellular Immunization. Gene Ther. 1997, 4, 11–15. [Google Scholar] [CrossRef] [Green Version]
- Lo, A.S.Y.; Zhu, Q.; Marasco, W.A. Intracellular Antibodies (Intrabodies) and Their Therapeutic Potential. Handb. Exp. Pharmacol. 2008, 181, 343–373. [Google Scholar] [CrossRef]
- Slastnikova, T.A.; Ulasov, A.V.; Rosenkranz, A.A.; Sobolev, A.S. Targeted Intracellular Delivery of Antibodies: The State of the Art. Front. Pharmacol. 2018, 9, 1208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Audenhove, I.; Gettemans, J. Nanobodies as Versatile Tools to Understand, Diagnose, Visualize and Treat Cancer. EBioMedicine 2016, 8, 40–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asaadi, Y.; Jouneghani, F.F.; Janani, S.; Rahbarizadeh, F. A Comprehensive Comparison between Camelid Nanobodies and Single Chain Variable Fragments. Biomark. Res. 2021, 9, 1–20. [Google Scholar] [CrossRef]
- de Marco, A. Recombinant Expression of Nanobodies and Nanobody-Derived Immunoreagents. Protein Expr. Purif. 2020, 172, 105645. [Google Scholar] [CrossRef]
- Reiter, Y.; Schuck, P.; Boyd, L.F.; Plaksin, D. An Antibody Single-Domain Phage Display Library of a Native Heavy Chain Variable Region: Isolation of Functional Single-Domain VH Molecules with a Unique Interface. J. Mol. Biol. 1999, 290, 685–698. [Google Scholar] [CrossRef]
- Kunz, P.; Flock, T.; Soler, N.; Zaiss, M.; Vincke, C.; Sterckx, Y.; Kastelic, D.; Muyldermans, S.; Hoheisel, J.D. Exploiting Sequence and Stability Information for Directing Nanobody Stability Engineering. Biochim. Biophys. Acta-Gen. Subj. 2017, 1861, 2196–2205. [Google Scholar] [CrossRef]
- Zabetakis, D.; Anderson, G.P.; Bayya, N.; Goldman, E.R. Contributions of the Complementarity Determining Regions to the Thermal Stability of a Single-Domain Antibody. PLoS ONE 2013, 8, e77678. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, T.; Hoshikawa, K.; Ezura, K.; Okazawa, R.; Fujita, S.; Takaoka, M.; Mason, H.S.; Ezura, H.; Miura, K. Improvement of the Transient Expression System for Production of Recombinant Proteins in Plants. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Kaur, J.J.; Kumar, A.; Kaur, J.J. Strategies for Optimization of Heterologous Protein Expression in E. coli: Roadblocks and Reinforcements. Int. J. Biol. Macromol. 2018, 106, 803–822. [Google Scholar] [CrossRef]
- Tripathi, N.K.; Shrivastava, A. Recent Developments in Bioprocessing of Recombinant Proteins: Expression Hosts and Process Development. Front. Bioeng. Biotechnol. 2019, 7, 420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vincke, C.; Loris, R.; Saerens, D.; Martinez-Rodriguez, S.; Muyldermans, S.; Conrath, K. General Strategy to Humanize a Camelid Single-Domain Antibody and Identification of a Universal Humanized Nanobody Scaffold Cé Cile Vincke. J. Biol. Chem. 2008, 284, 3273–3284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanck-Silva, G.; Fatori Trevizan, L.N.; Petrilli, R.; de Lima, F.T.; Eloy, J.O.; Chorilli, M. A Critical Review of Properties and Analytical/Bioanalytical Methods for Characterization of Cetuximab. Crit. Rev. Anal. Chem. 2019, 50, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Banisadr, A.; Safdari, Y.; Kianmehr, A.; Pourafshar, M. Production of a Germline-Humanized Cetuximab ScFv and Evaluation of Its Activity in Recognizing EGFR- Overexpressing Cancer Cells. Hum. Vaccines Immunother. 2017, 14, 856–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, L.S.; Colwell, L.J. Analysis of Nanobody Paratopes Reveals Greater Diversity than Classical Antibodies. Protein Engineering. Des. Sel. 2018, 31, 267–275. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, L.S.; Colwell, L.J. Comparative Analysis of Nanobody Sequence and Structure Data. Proteins Struct. Funct. Bioinform. 2018, 86, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Vattekatte, A.M.; Shinada, N.K.; Narwani, T.J.; Noël, F.; Bertrand, O.; Meyniel, J.P.; Malpertuy, A.; Gelly, J.C.; Cadet, F.; de Brevern, A.G. Discrete Analysis of Camelid Variable Domains: Sequences, Structures, and in-Silico Structure Prediction. PeerJ 2020, 2020, e8408. [Google Scholar] [CrossRef]
- Kunz, P.; Zinner, K.; Mücke, N.; Bartoschik, T.; Muyldermans, S.; Hoheisel, J.D. The Structural Basis of Nanobody Unfolding Reversibility and Thermoresistance. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Erreni, M.; Schorn, T.; D’autilia, F.; Doni, A. Nanobodies as Versatile Tool for Multiscale Imaging Modalities. Biomolecules 2020, 10, 1695. [Google Scholar] [CrossRef]
- Sabir, J.S.M.; Atef, A.; El-Domyati, F.M.; Edris, S.; Hajrah, N.; Alzohairy, A.M.; Bahieldin, A. Construction of Naïve Camelids VHH Repertoire in Phage Display-Based Library. Comptes Rendus Biol. 2014, 337, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, Q.; Sun, Z.; Wang, Y.; Su, B.; Zhang, C.; Cao, H.; Liu, X. Nanobody Affinity Improvement: Directed Evolution of the Anti-Ochratoxin A Single Domain Antibody. Int. J. Biol. Macromol. 2020, 151, 312–321. [Google Scholar] [CrossRef]
- Rothbauer, U.; Zolghadr, K.; Tillib, S.; Nowak, D.; Schermelleh, L.; Gahl, A.; Backmann, N.; Conrath, K.; Muyldermans, S.; Cardoso, M.C.; et al. Targeting and Tracing Antigens in Live Cells with Fluorescent Nanobodies. Nat. Methods 2006, 3, 887–889. [Google Scholar] [CrossRef] [PubMed]
- Kirchhofer, A.; Helma, J.; Schmidthals, K.; Frauer, C.; Cui, S.; Karcher, A.; Pellis, M.; Muyldermans, S.; Casas-Delucchi, C.S.; Cardoso, M.C.; et al. Modulation of Protein Properties in Living Cells Using Nanobodies. Nat. Struct. Mol. Biol. 2009, 17, 133–138. [Google Scholar] [CrossRef]
- Rasmussen, S.G.F.; Choi, H.J.; Fung, J.J.; Pardon, E.; Casarosa, P.; Chae, P.S.; Devree, B.T.; Rosenbaum, D.M.; Thian, F.S.; Kobilka, T.S.; et al. Structure of a Nanobody-Stabilized Active State of the Β2 Adrenoceptor. Nature 2011, 469, 175–180. [Google Scholar] [CrossRef] [Green Version]
- De Genst, E.; Silence, K.; Decanniere, K.; Conrath, K.; Loris, R.; Kinne, J.; Muyldermans, S.; Wyns, L. Molecular Basis for the Preferential Cleft Recognition by Dromedary Heavy-Chain Antibodies. Proc. Natl. Acad. Sci. USA 2006, 103, 4586–4591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Leung, G.P.H. Equilibrative Nucleoside Transporters 1 and 4: Which One Is a Better Target for Cardioprotection Against Ischemia-Reperfusion Injury? J. Cardiovasc. Pharmacol. 2015, 65, 517–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, I.; Egloff, P.; Hutter, C.A.J.J.; Kuhn, B.T.; Bräuer, P.; Newstead, S.; Dawson, R.J.P.P.; Geertsma, E.R.; Seeger, M.A. Generation of Synthetic Nanobodies against Delicate Proteins. Nat. Protoc. 2020, 15, 1707–1741. [Google Scholar] [CrossRef]
- Hansen, J.; Baum, A.; Pascal, K.E.; Russo, V.; Giordano, S.; Wloga, E.; Fulton, B.O.; Yan, Y.; Koon, K.; Patel, K.; et al. Studies in Humanized Mice and Convalescent Humans Yield a SARS-CoV-2 Antibody Cocktail. Science 2020, 369, 1010–1014. [Google Scholar] [CrossRef]
- Scheid, J.F.; Barnes, C.O.; Eraslan, B.; Hudak, A.; Keeffe, J.R.; Cosimi, L.A.; Brown, E.M.; Muecksch, F.; Weisblum, Y.; Zhang, S.; et al. B Cell Genomics behind Cross-Neutralization of SARS-CoV-2 Variants and SARS-CoV. Cell 2021, 184, 3205–3221.e24. [Google Scholar] [CrossRef]
- Jang, H.I.; Wilson, P.G.; Sau, M.; Chawla, U.; Rodgers, D.W.; Galperin, E. Single-Domain Antibodies for Functional Targeting of the Signaling Scaffold Shoc2. Mol. Immunol. 2020, 118, 110–116. [Google Scholar] [CrossRef]
- Walter, J.D.; Hutter, C.A.J.; Garaeva, A.A.; Scherer, M.; Zimmermann, I.; Wyss, M.; Rheinberger, J.; Ruedin, Y.; Earp, J.C.; Egloff, P.; et al. Biparatopic Sybody Constructs Neutralize SARS-CoV-2 Variants of Concern and Mitigate Emergence of Drug Resistance. bioRxiv 2021, 2020, 2020-11. [Google Scholar] [CrossRef]
- Hong, C.; Byrne, N.J.; Zamlynny, B.; Tummala, S.; Xiao, L.; Shipman, J.M.; Partridge, A.T.; Minnick, C.; Breslin, M.J.; Rudd, M.T.; et al. Structures of Active-State Orexin Receptor 2 Rationalize Peptide and Small-Molecule Agonist Recognition and Receptor Activation. Nat. Commun. 2021, 12, 1–11. [Google Scholar] [CrossRef]
- Li, T.; Cai, H.; Yao, H.; Zhou, B.; Zhang, N.; van Vlissingen, M.F.; Kuiken, T.; Han, W.; GeurtsvanKessel, C.H.; Gong, Y.; et al. A Synthetic Nanobody Targeting RBD Protects Hamsters from SARS-CoV-2 Infection. Nat. Commun. 2021, 12, 1–13. [Google Scholar] [CrossRef]
- Schoof, M.; Faust, B.; Saunders, R.A.; Sangwan, S.; Rezelj, V.; Hoppe, N.; Boone, M.; Billesbølle, C.B.; Puchades, C.; Azumaya, C.M.; et al. An Ultrapotent Synthetic Nanobody Neutralizes SARS-CoV-2 by Stabilizing Inactive Spike. Science 2021, 370, 1473–1479. [Google Scholar] [CrossRef]
- Fan, C.; Fan, M.; Orlando, B.J.; Fastman, N.M.; Zhang, J.; Xu, Y.; Chambers, M.G.; Xu, X.; Perry, K.; Liao, M.; et al. X-Ray and Cryo-EM Structures of the Mitochondrial Calcium Uniporter. Nature 2018, 559, 575–579. [Google Scholar] [CrossRef]
- Wingler, L.M.; McMahon, C.; Staus, D.P.; Lefkowitz, R.J.; Kruse, A.C. Distinctive Activation Mechanism for Angiotensin Receptor Revealed by a Synthetic Nanobody. Cell 2019, 176, 479–490.e12. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Wu, X.; Sun, D.; Park, E.; Catipovic, M.A.; Rapoport, T.A.; Gao, N.; Li, L. Structure of the Substrate-Engaged SecA-SecY Protein Translocation Machine. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef]
- Wu, T.; Liu, J.; Liu, M.; Liu, S.; Zhao, S.; Tian, R.; Wei, D.; Liu, Y.; Zhao, Y.; Xiao, H.; et al. A Nanobody-Conjugated DNA Nanoplatform for Targeted Platinum-Drug Delivery. Angew. Chem. Int. Ed. 2019, 58, 14224–14228. [Google Scholar] [CrossRef] [PubMed]
- Cornish-Bowden, A. Nomenclature for Incompletely Specified Bases in Nucleic Acid Sequences: Recommendations 1984. Nucleic Acids Res. 1985, 13, 3021. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Song, H.; Chen, Q.; Yu, J.; Xian, M.; Nian, R.; Feng, D. Recent Advances in the Selection and Identification of Antigen-Specific Nanobodies. Mol. Immunol. 2018, 96, 37–47. [Google Scholar] [CrossRef] [PubMed]

| Name a | Framework Source | CDR Lengths | CDR Randomization Strategy | Library Size/Display System | Reference |
|---|---|---|---|---|---|
| NaLi-H1 | Llama-derived (consensus sequence from immune Nb set) | CDR1: 7 CDR2: 7 CDR3: 9, 12, 15, 18 | CDR1 and 2: fully randomized. Partially recapitulates the natural diversity, with the inclusion of polar amino acids. CDR3: fully randomized | 3 × 109 Phage Display | [12] |
| McMahon et al. | Llama-derived (consensus sequence derived from llama genes) | CDR1: 7 CDR2: 5 CDR3: 7, 11, 15 | CDR1 and 2: 4 and 1 highly variable positions, respectively CDR3: fully randomized, including variable adjacent positions | 1 × 108 Yeast Display | [13] |
| Concave | Dromedary-llama (consensus framework between dromedary-derived 3K1K and llama-derived 3P0G.) | CDR1: 7 CDR2: 6 CDR3: 6 | CDR1 and 2: 5 positions were rationally randomized CDR3: 5 positions were rationally randomized | 9 × 1012 Ribosome Display | [14] |
| Loop | Dromedary-llama (consensus framework between dromedary-derived 3K1K and llama-derived 3P0G.) | CDR1: 7 CDR2: 6 CDR3: 12 | CDR1 and 2: 5 positions were rationally randomized CDR3: 6 positions were rationally randomized | 9 × 1012 Ribosome Display | [14] |
| Convex | Dromedary-derived (from Nb 1ZVH) | CDR1: 7 CDR2: 6 CDR3: 16 | CDR1 and 2: 4 positions were rationally randomized CDR3: 10 positions were rationally randomized | 9 × 1012 Ribosome Display | [14] |
| CeVICA | Consensus framework and reported natural Nbs: A310 and GFP-binding nanobody | CDR1: 7 CDR2: 5 CDR3: 6, 9, 10, 13 | CDRs: All positions were fully randomized, including cysteine | 3.68 × 1011 Ribosome Display | [15] |
| Yan et al. | cAbBCII10 | CDR3: 16 | CDR3: fully randomized | 1.65 × 109 Phage Display | [16] |
| Alp_LowDiv, Hum_LowDiv, Alp_HighDiv, Hum_HighDiv | Alpaca-derived and partially humanized (consensus sequence derived from alpaca genes) | CDR1: 8 * CDR2: 7 * CDR3: 6–18 | CDR1 and 2: All (for Alp_HighDiv, Hum_HighDiv) or selected (for Alp_LowDiv, Hum_LowDiv) positions were randomized CDR3: fully randomized | 1.2 × 109, 1.5 × 109, 0.9 × 109, 1.1 × 109 Yeast Display | [17] |
| Zhao et al. | GFP-binding nanobody (cAbGFP4) | CDR1: 9 * CDR2: 8 * CDR3: 7 | All CDRs were fully randomized | ~1 × 106 Mammalian Cells | [18] |
| Wagner et al. | dromedary-derived (3K1K) | CDR1: 9 CDR2: 6 CDR3: 18 | CDR2 and 3: Randomization of selected positions recapitulating natural diversity and enriching with amino acids of the same nature | 9 × 109 Phage Display | [19] |
| Chi et al. | cAbBCII10-derived | CDR1: 8 CDR2: 8 CDR3: 18 | All CDRs were fully randomized | 1.2 × 1010 Phage Display | [20] |
| Ferrari et al. | Llama-derived (consensus framework from annotated sequences) | CDR1: 9 * CDR2: 7 * CDR3: 16 | CDRs: All positions were randomized recapitulating the natural diversity and excluding cysteine | ~1 × 1012 Ribosome Display | [21] |
| Stefan et al. | NaLi-H1 framework derived | CDR3: 9, 12, 15 | All CDRs were fully randomized | 3.18 × 1010 Phage Display | [22] |
| Wei et al. | cAbBCII10 | CDR1: 8 CDR2: 8 CDR3: 9–20 | CDR1 and 2: Partially randomized CDR3: fully randomized | 1 × 1012 Phage Display | [23] |
| Liu et al. | SPSL1 naïve library | CDR3: 13, 16, 18 | CDR3: fully randomized | 1 × 109 Phage Display | [10] |
| Könning et al. | naïve bamboo shark scaffolds | CDR3: 12, 14, 16, 18 | CDR3: fully randomized | 3 × 109 Yeast Display | [9] |
| Library | Antigen | Antigen Features | Pannings | KD (M) | Ref. |
|---|---|---|---|---|---|
| NaLi-H1 | EGFP mCherry tubulin β-actin p53 (83 first aa fragment) HP1α RHO GTPase HER2 | Soluble protein Soluble protein Polymeric protein Polymeric protein Soluble protein Soluble protein Membrane protein/specific conformer Membrane protein/surface antigen | 2–4 | 1.6 × 10−10–4 × 10−9 | [12] |
| McMahon et al. | HSA Human Adiponectine β2AR A2AR | Soluble protein Soluble protein Membrane protein/specific conformer Membrane protein/specific conformer | 4 | 44 × 10−9–430 × 10−9 | [13] |
| Concave, loop, and convex | MBP ABC transporter TM287/288 ENT1 GlyT1 | Soluble protein Membrane protein/specific mutant Membrane protein/specific conformer Membrane protein/specific conformer | 3 | 4.94 × 10−10–2.5 × 10−6 | [14] |
| CeVICA | EGFP RBD | Soluble protein Soluble protein | 3 | 2.18 × 10−9 a | [15] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valdés-Tresanco, M.S.; Molina-Zapata, A.; Pose, A.G.; Moreno, E. Structural Insights into the Design of Synthetic Nanobody Libraries. Molecules 2022, 27, 2198. https://doi.org/10.3390/molecules27072198
Valdés-Tresanco MS, Molina-Zapata A, Pose AG, Moreno E. Structural Insights into the Design of Synthetic Nanobody Libraries. Molecules. 2022; 27(7):2198. https://doi.org/10.3390/molecules27072198
Chicago/Turabian StyleValdés-Tresanco, Mario S., Andrea Molina-Zapata, Alaín González Pose, and Ernesto Moreno. 2022. "Structural Insights into the Design of Synthetic Nanobody Libraries" Molecules 27, no. 7: 2198. https://doi.org/10.3390/molecules27072198
APA StyleValdés-Tresanco, M. S., Molina-Zapata, A., Pose, A. G., & Moreno, E. (2022). Structural Insights into the Design of Synthetic Nanobody Libraries. Molecules, 27(7), 2198. https://doi.org/10.3390/molecules27072198






