Revisiting Proteasome Inhibitors: Molecular Underpinnings of Their Development, Mechanisms of Resistance and Strategies to Overcome Anti-Cancer Drug Resistance
Abstract
:1. Introduction
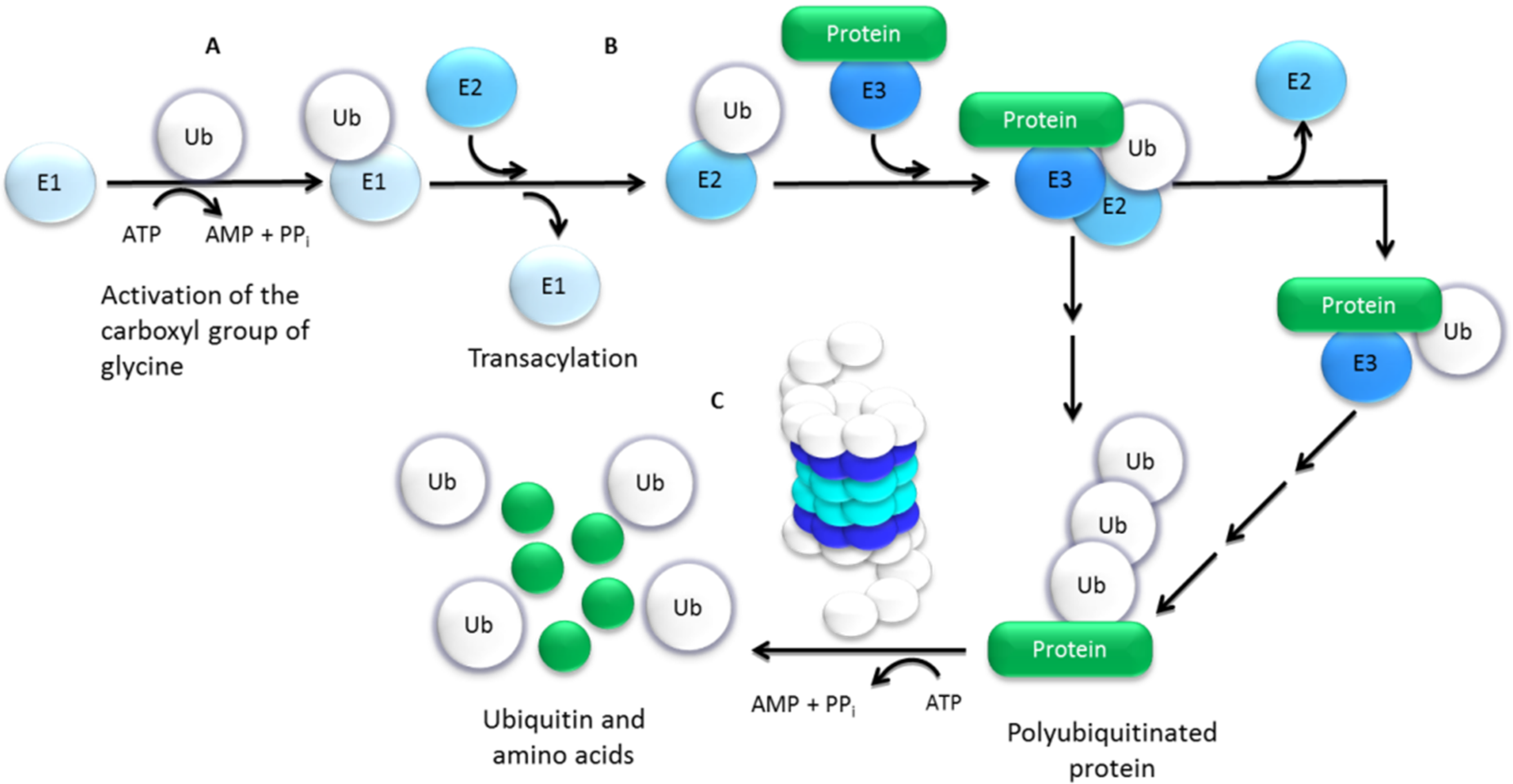
2. Ubiquitin–Proteasome Pathway (UPP)
The 20S Proteasome Core Particle
3. Proteasome Inhibitors (PIs) in Cancer Therapy
3.1. Aldehydes
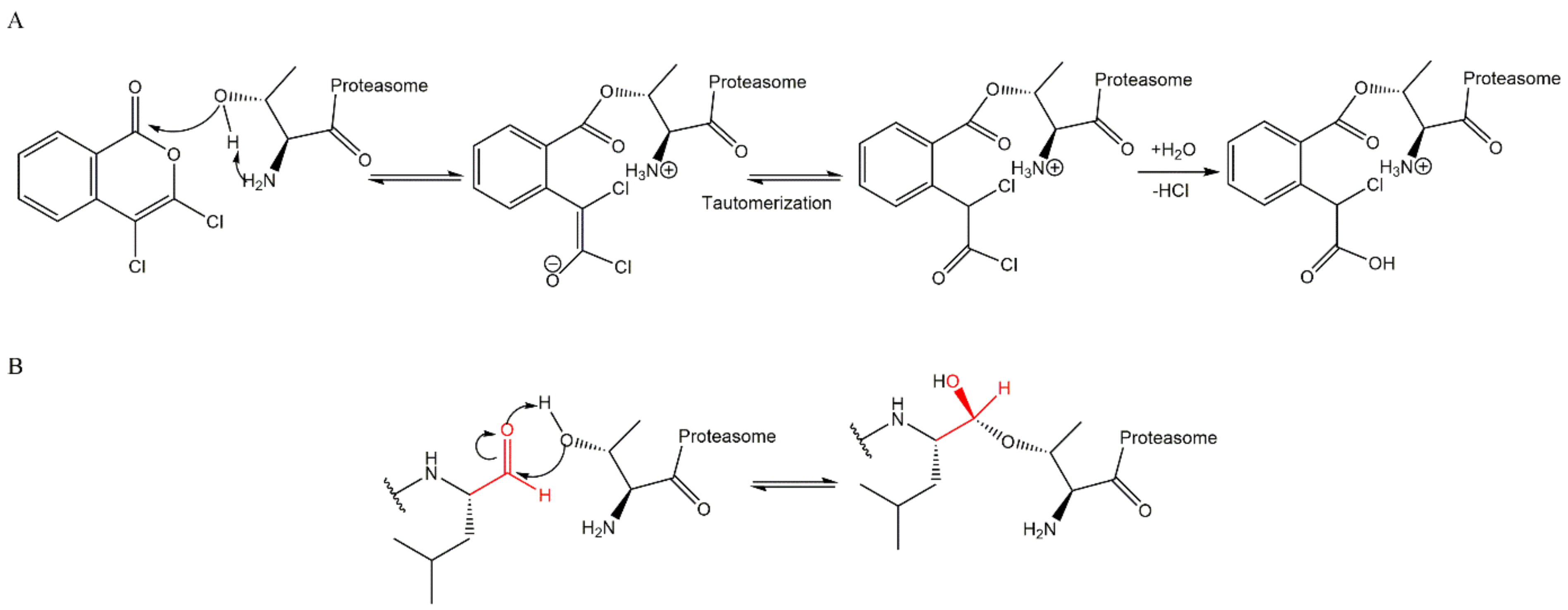
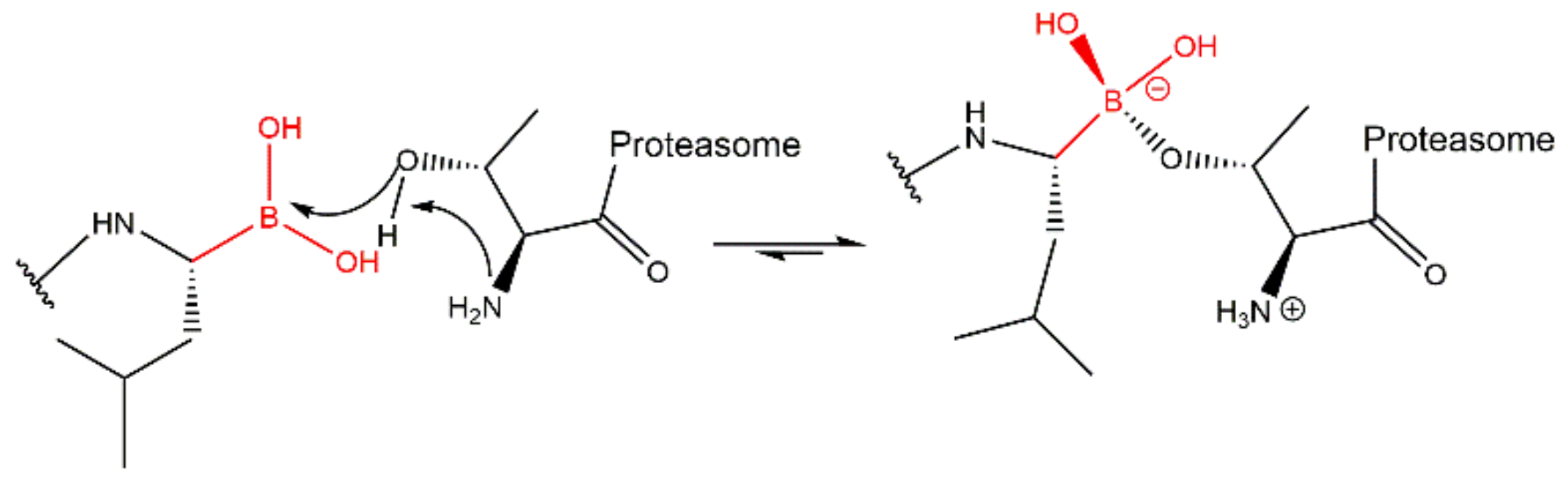
3.2. Boronates
3.3. α’,β’-Epoxyketones
3.4. Non-Covalent Macrocyclics
3.5. α-Ketoaldehydes and α-Ketoamides
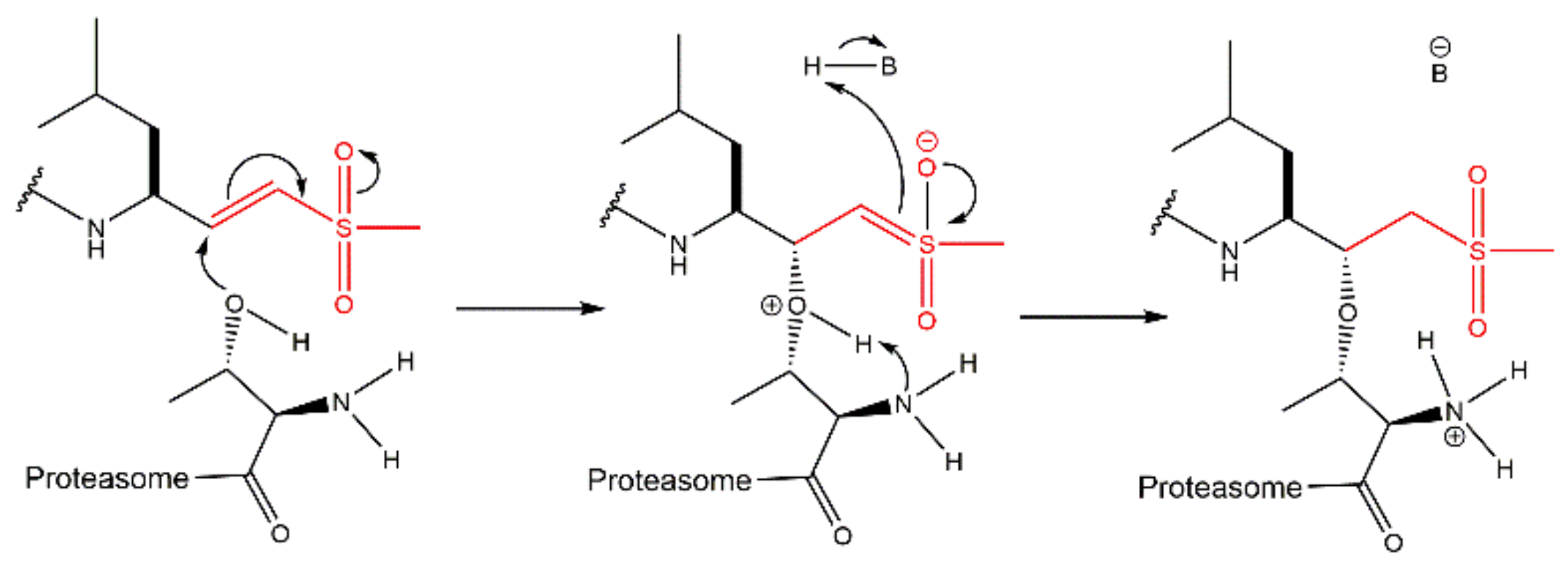
3.6. Peptide Vinyl Derivatives
3.7. β-Lactones
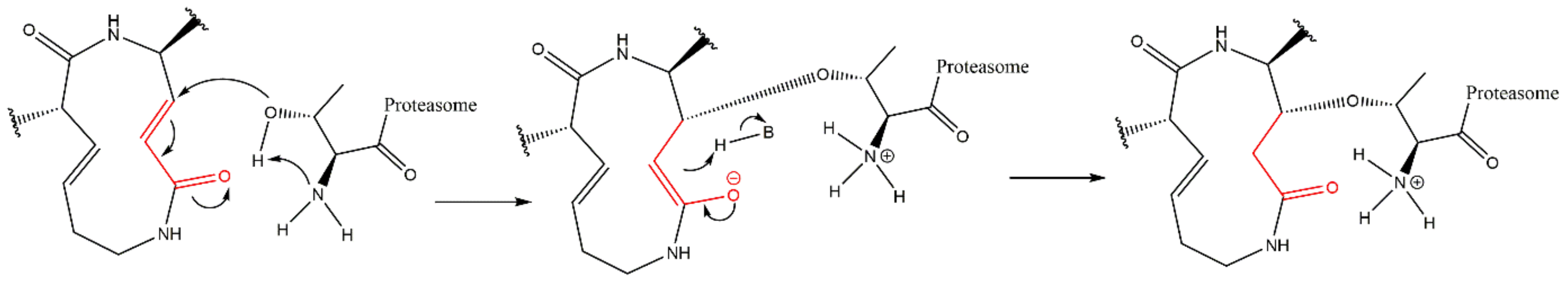
3.8. Syrbactins
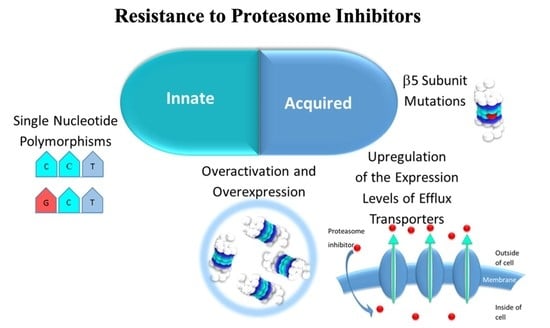
4. Resistance Mechanisms to Proteasome Inhibitors (PIs)
4.1. Innate Resistance
4.1.1. Proteasome Overactivation and Overexpression
4.1.2. Single Nucleotide Polymorphisms (SNPs) of the Genes Encoding the Three Catalytic Subunits β of the 20S CP
4.2. Acquired Resistance
4.2.1. 20S CP β Subunits Overexpression
4.2.2. β5. Subunit Mutation
4.2.3. Alteration of the Expression Levels of Transporters which Mediate Efflux of 20S CP Inhibitors
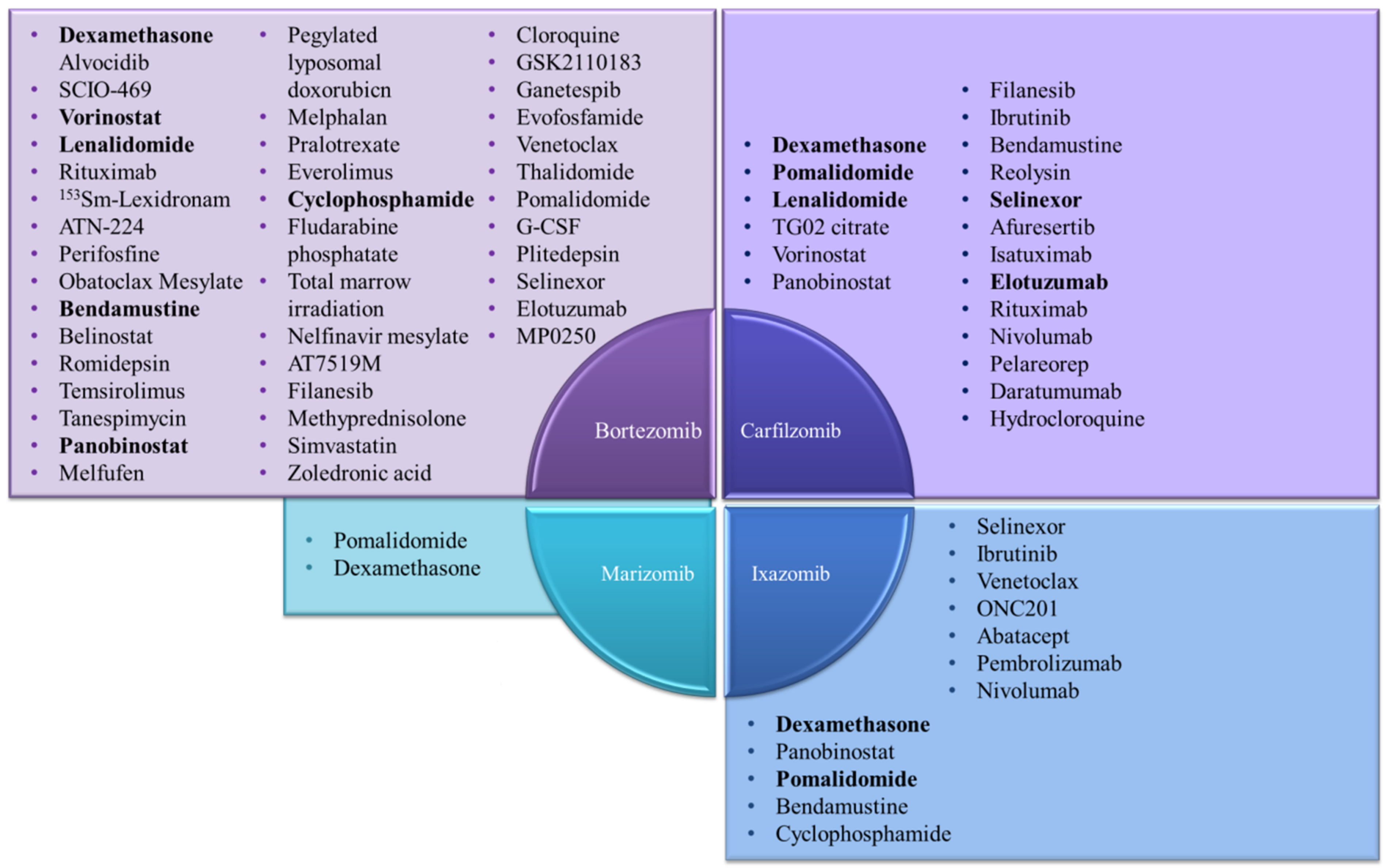
5. Combination Therapy to Overcome Resistance to Proteasome Inhibitors (PIs)
6. Final Considerations and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef] [PubMed]
- Obrist, F.; Manic, G.; Kroemer, G.; Vitale, I.; Galluzzi, L. Trial Watch: Proteasomal inhibitors for anticancer therapy. Mol. Cell. Oncol. 2015, 2, e974463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manasanch, E.E.; Orlowski, R.Z. Proteasome inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 2017, 14, 417–433. [Google Scholar] [CrossRef] [PubMed]
- EMA Ninlaro®: EPAR—Product Information. Available online: https://www.ema.europa.eu/en/documents/product-information/ninlaro-epar-product-information_en.pdf (accessed on 5 April 2021).
- EMA Kyprolis®: EPAR—Product Information. Available online: https://www.ema.europa.eu/en/documents/product-information/kyprolis-epar-product-information_en.pdf (accessed on 5 April 2021).
- FDA Highlights of Prescribing Information Velcade® (Bortezomib). Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2019/206927s000lbl.pdf (accessed on 5 April 2021).
- FDA Highlights of Prescribing Information Ninlaro® (Ixazomib). Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2021/208462s010lbl.pdf (accessed on 5 April 2021).
- FDA Highlights of Prescribing Information Kyprolis® (Carfilzomib). Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/202714s030lbl.pdf (accessed on 5 April 2021).
- EMA Velcade: EPAR—Product Information. Available online: https://www.ema.europa.eu/en/documents/product-information/velcade-epar-product-information_en.pdf (accessed on 5 April 2021).
- Cancer Stat Facts: Myeloma. Available online: https://seer.cancer.gov/statfacts/html/mulmy.html (accessed on 24 February 2022).
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2020. [Google Scholar]
- Kuehl, W.M.; Bergsagel, P.L. Molecular pathogenesis of multiple myeloma and its premalignant precursor. J. Clin. Investig. 2012, 122, 3456–3463. [Google Scholar] [CrossRef] [PubMed]
- Offidani, M.; Corvatta, L.; Morè, S.; Olivieri, A. Novel experimental drugs for treatment of multiple myeloma. J. Exp. Pharmacol. 2021, 13, 245–264. [Google Scholar] [CrossRef]
- Goldman-Mazur, S.; Kumar, S.K. Current approaches to management of high-risk multiple myeloma. Am. J. Hematol. 2021. [Google Scholar] [CrossRef]
- Roué, G.; Sola, B. Management of drug resistance in mantle cell lymphoma. Cancers 2020, 12, 1565. [Google Scholar] [CrossRef]
- Pérez-Galán, P.; Dreyling, M.; Wiestner, A. Mantle cell lymphoma: Biology, pathogenesis, and the molecular basis of treatment in the genomic era. Blood 2011, 117, 26–38. [Google Scholar] [CrossRef] [Green Version]
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef]
- Ciechanover, A.; Brundin, P. The ubiquitin proteasome system in neurodegenerative diseases. Neuron 2003, 40, 427–446. [Google Scholar] [CrossRef] [Green Version]
- Thompson, S.; Loftus, L.; Ashley, M.; Meller, R. Ubiquitin–proteasome system as a modulator of cell fate. Curr. Opin. Pharmacol. 2008, 8, 90–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhat, K.P.; Greer, S.F. Proteolytic and non-proteolytic roles of ubiquitin and the ubiquitin proteasome system in transcriptional regulation. Biochim. Biophys. Acta Gene Regul. Mech. 2011, 1809, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, A.L.; Ciechanover, A. Targeting proteins for destruction by the ubiquitin system: Implications for human pathobiology. Annu. Rev. Pharmacol. Toxicol. 2009, 49, 73–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.M.; Yu, Y.; Cheng, Y. Structure characterization of the 26S proteasome. Biochim. Biophys. Acta Gene Regul. Mech. 2011, 1809, 67–79. [Google Scholar] [CrossRef] [Green Version]
- Orlowski, N.; Wilk, S. A multicatalytical protease complex from pituitary that forms enkephalin and enkephalin containing peptides. Biochem. Biophys. Res. Commun. 1981, 101, 814–822. [Google Scholar] [CrossRef]
- Young, P.; Deveraux, Q.; Beal, R.E.; Pickart, C.M.; Rechsteiner, M. Characterization of two polyubiquitin binding sites in the 26 S protease subunit 5a. J. Biol. Chem. 1998, 273, 5461–5467. [Google Scholar] [CrossRef] [Green Version]
- Yao, T.; Cohen, R.E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 2002, 419, 403–407. [Google Scholar] [CrossRef]
- Harer, S.L.; Bhatia, M.S.; Bhatia, N.M. Proteasome inhibitors mechanism; source for design of newer therapeutic agents. J. Antibiot. 2012, 65, 279–288. [Google Scholar] [CrossRef]
- Kisselev, A.F.; Goldberg, A.L. Proteasome inhibitors: From research tools to drug candidates. Chem. Biol. 2001, 8, 739–758. [Google Scholar] [CrossRef] [Green Version]
- Borissenko, L.; Groll, M. 20S proteasome and its inhibitors: Crystallographic knowledge for drug development. Chem. Rev. 2007, 107, 687–717. [Google Scholar] [CrossRef]
- Huber, E.M.; Groll, M. Inhibitors for the immuno- and constitutive proteasome: Current and future trends in drug development. Angew. Chemie Int. Ed. 2012, 51, 8708–8720. [Google Scholar] [CrossRef] [PubMed]
- Zwickl, P.; Baumeister, W. (Eds.) The Proteasome—Ubiquitin Protein Degradation Pathway; Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2002; Volume 268, ISBN 978-3-642-63971-5. [Google Scholar]
- Guedes, R.A.; Serra, P.; Salvador, J.; Guedes, R. Computational approaches for the discovery of human proteasome inhibitors: An overview. Molecules 2016, 21, 927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, B.S.; Eustáquio, A.S.; McGlinchey, R.P. Advances in and applications of proteasome inhibitors. Curr. Opin. Chem. Biol. 2008, 12, 434–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, J. (Ed.) Proteasome Inhibitors in Cancer Therapy; Humana Press: Totowa, NJ, USA, 2004; ISBN 978-1-61737-452-4. [Google Scholar]
- Dou, Q.; Zonder, J. Overview of proteasome inhibitor-based anti-cancer therapies: Perspective on bortezomib and second generation proteasome inhibitors versus future generation inhibitors of ubiquitin-proteasome system. Curr. Cancer Drug Targets 2014, 14, 517–536. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Frezza, M.; Schmitt, S.; Kanwar, J.; Dou, Q.P. Bortezomib as the first proteasome inhibitor anticancer drug: Current status and future perspectives. Curr. Cancer Drug Targets 2011, 11, 239–253. [Google Scholar] [CrossRef] [Green Version]
- Crawford, L.J.; Walker, B.; Irvine, A.E. Proteasome inhibitors in cancer therapy. J. Cell Commun. Signal. 2011, 5, 101–110. [Google Scholar] [CrossRef] [Green Version]
- Achanta, G. A boronic-chalcone derivative exhibits potent anti-cancer activity through inhibition of the proteasome. Mol. Pharmacol. 2006, 579, 3932–3940. [Google Scholar] [CrossRef] [Green Version]
- Han, L.-Q.; Yuan, X.; Wu, X.-Y.; Li, R.-D.; Xu, B.; Cheng, Q.; Liu, Z.-M.; Zhou, T.-Y.; An, H.-Y.; Wang, X.; et al. Urea-containing peptide boronic acids as potent proteasome inhibitors. Eur. J. Med. Chem. 2017, 125, 925–939. [Google Scholar] [CrossRef]
- Ruschak, A.M.; Slassi, M.; Kay, L.E.; Schimmer, A.D. Novel proteasome inhibitors to overcome bortezomib resistance. JNCI J. Natl. Cancer Inst. 2011, 103, 1007–1017. [Google Scholar] [CrossRef] [Green Version]
- ClinicalTrials.gov. Phase I Study of the Proteosome Inhibitor CEP 18770 in Patients with Solid Tumours or Non-Hodgkin’s Lymphomas. Available online: https://www.clinicaltrials.gov/ct2/show/study/NCT00572637?term=delanzomib&rank=3 (accessed on 28 February 2017).
- Gallerani, E.; Zucchetti, M.; Brunelli, D.; Marangon, E.; Noberasco, C.; Hess, D.; Delmonte, A.; Martinelli, G.; Böhm, S.; Driessen, C.; et al. A first in human phase I study of the proteasome inhibitor CEP-18770 in patients with advanced solid tumours and multiple myeloma. Eur. J. Cancer 2013, 49, 290–296. [Google Scholar] [CrossRef]
- ClinicalTrials.gov. Study to Determine the Maximum Tolerated Dose and Evaluate the Efficacy and Safety of CEP-18770 (Delanzomib) in Patients with Relapsed Multiple Myeloma Refractory to the Most Recent Therapy. Available online: https://www.clinicaltrials.gov/ct2/show/NCT01023880?term=delanzomib&rank=1 (accessed on 28 February 2017).
- Vogl, D.T.; Martin, T.G.; Vij, R.; Hari, P.; Mikhael, J.R.; Siegel, D.; Wu, K.L.; Delforge, M.; Gasparetto, C. Phase I/II study of the novel proteasome inhibitor delanzomib (CEP-18770) for relapsed and refractory multiple myeloma. Leuk. Lymphoma 2017, 58, 1872–1879. [Google Scholar] [CrossRef] [PubMed]
- ClinicalTrials.gov. Delanzomib (CEP-18770) in Combination with Lenalidomide and Dexamethasone in Relapsed or Refractory Multiple Myeloma. Available online: https://www.clinicaltrials.gov/ct2/show/NCT01348919?term=delanzomib&rank=2 (accessed on 28 February 2017).
- ClinicalTrials.gov. A Study of Oprozomib, Melphalan, and Prednisone in Transplant Ineligible Patients with Newly Diagnosed Multiple Myeloma. Available online: https://clinicaltrials.gov/ct2/show/study/NCT02072863?cond=oprozomib&rank=6 (accessed on 5 October 2017).
- ClinicalTrials.gov. Phase 1 Study of Oprozomib Administered Orally in Patients with Advanced Refractory or Recurrent Solid Tumors. Available online: https://clinicaltrials.gov/ct2/show/study/NCT01129349?cond=oprozomib&rank=5 (accessed on 5 October 2017).
- ClinicalTrials.gov. Study of Oprozomib and Dexamethasone, in Combination with Lenalidomide or Oral Cyclophosphamide in Patients With Newly Diagnosed Multiple Myeloma. Available online: https://clinicaltrials.gov/ct2/show/NCT01881789?recrs=d&cond=oprozomib&rank=1 (accessed on 5 October 2017).
- ClinicalTrials.gov. Phase 1b/2, Multicenter, Open-label Study of Oprozomib and Dexamethasone in Patients with Relapsed and/or Refractory Multiple Myeloma. Available online: https://clinicaltrials.gov/ct2/show/NCT01832727?recrs=d&cond=oprozomib&rank=2 (accessed on 5 October 2017).
- ClinicalTrials.gov. Open-label Study of the Safety and Activity of Oprozomib in Patients with Hematologic Malignancies. Available online: https://clinicaltrials.gov/ct2/show/NCT01416428?recrs=d&cond=oprozomib&rank=3 (accessed on 5 October 2017).
- ClinicalTrials.gov. A Study of Oprozomib, Pomalidomide, and Dexamethasone in Subjects with Primary Refractory or Relapsed and Refractory Multiple Myeloma. Available online: https://clinicaltrials.gov/ct2/show/NCT01999335?recrs=d&cond=oprozomib&rank=4 (accessed on 5 October 2017).
- ClinicalTrials.gov. A Phase 1 Study of Oprozomib to Assess Food Effect, Drug-Drug Interaction with Midazolam, and Safety and Tolerability in Patients with Advanced Malignancies. Available online: https://clinicaltrials.gov/ct2/show/NCT02244112?recrs=d&cond=oprozomib&rank=5 (accessed on 5 October 2017).
- ClinicalTrials.gov. Phase 1b Study Evaluating OPomD in Relapsed or Refractory Multiple Myeloma. Available online: https://www.clinicaltrials.gov/ct2/show/NCT02939183?term=Oprozomib&rank=9 (accessed on 28 February 2017).
- Basmadjian, C.; Zhao, Q.; Bentouhami, E.; Djehal, A.; Nebigil, C.G.; Johnson, R.A.; Serova, M.; de Gramont, A.; Faivre, S.; Raymond, E.; et al. Cancer wars: Natural products strike back. Front. Chem. 2014, 2, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groll, M.; Nazif, T.; Huber, R.; Bogyo, M. Probing structural determinants distal to the site of hydrolysis that control substrate specificity of the 20S proteasome. Chem. Biol. 2002, 9, 655–662. [Google Scholar] [CrossRef] [Green Version]
- Gräwert, M.A.; Gallastegui, N.; Stein, M.; Schmidt, B.; Kloetzel, P.-M.; Huber, R.; Groll, M. Elucidation of the α-keto-aldehyde binding mechanism: A lead structure motif for proteasome inhibition. Angew. Chemie Int. Ed. 2011, 50, 542–544. [Google Scholar] [CrossRef]
- Stein, M.L.; Cui, H.; Beck, P.; Dubiella, C.; Voss, C.; Krüger, A.; Schmidt, B.; Groll, M. Systematic comparison of peptidic proteasome inhibitors highlights the α-ketoamide electrophile as an auspicious reversible lead motif. Angew. Chemie Int. Ed. 2014, 53, 1679–1683. [Google Scholar] [CrossRef]
- Schrader, J.; Henneberg, F.; Mata, R.A.; Tittmann, K.; Schneider, T.R.; Stark, H.; Bourenkov, G.; Chari, A. The inhibition mechanism of human 20 S proteasomes enables next-generation inhibitor design. Science 2016, 353, 594–598. [Google Scholar] [CrossRef] [Green Version]
- Koguchi, Y.; Kohno, J.; Nishio, M.; Takahashi, K.; Okuda, T.; Ohnuki, T.; Komatsubara, S. TMC-95A, B, C, and D, novel proteasome inhibitors produced by Apiospora Montagnei Sacc. TC 1093. taxonomy, production, isolation, and biological activities. J. Antibiot. 2000, 53, 105–109. [Google Scholar] [CrossRef] [Green Version]
- Kohno, J.; Koguchi, Y.; Nishio, M.; Nakao, K.; Kuroda, M.; Shimizu, R.; Ohnuki, T.; Komatsubara, S. Structures of TMC-95A-D: Novel proteasome inhibitors from Apiospora montagnei Sacc. TC 1093. J. Org. Chem. 2000, 65, 990–995. [Google Scholar] [CrossRef]
- Groll, M.; Koguchi, Y.; Huber, R.; Kohno, J. Crystal structure of the 20 S proteasome:TMC-95A complex: A non-covalent proteasome inhibitor. J. Mol. Biol. 2001, 311, 543–548. [Google Scholar] [CrossRef]
- Génin, E.; Reboud-Ravaux, M.; Vidal, J. Proteasome inhibitors: Recent advances and new perspectives in medicinal chemistry. Curr. Top. Med. Chem. 2010, 10, 232–256. [Google Scholar] [CrossRef] [Green Version]
- Nazif, T.; Bogyo, M. Global analysis of proteasomal substrate specificity using positional-scanning libraries of covalent inhibitors. Proc. Natl. Acad. Sci. USA 2001, 98, 2967–2972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masters, E.I. Structural Studies of the Proteasome and Implications for Proteasome Activation. Ph.D. Thesis, Department of Biochemistry, University of Utah, Salt Lake City, UT, USA, 2008. [Google Scholar]
- Marastoni, M.; Baldisserotto, A.; Cellini, S.; Gavioli, R.; Tomatis, R. Peptidyl vinyl ester derivatives: New class of selective inhibitors of proteasome trypsin-like activity. J. Med. Chem. 2005, 48, 5038–5042. [Google Scholar] [CrossRef] [PubMed]
- Marastoni, M.; Baldisserotto, A.; Trapella, C.; Gavioli, R.; Tomatis, R. Synthesis and biological evaluation of new vinyl ester pseudotripeptide proteasome inhibitors. Eur. J. Med. Chem. 2006, 41, 978–984. [Google Scholar] [CrossRef] [PubMed]
- Marastoni, M.; Baldisserotto, A.; Canella, A.; Gavioli, R.; De Risi, C.; Pollini, G.P.; Tomatis, R. Arecoline tripeptide inhibitors of proteasome. J. Med. Chem. 2004, 47, 1587–1590. [Google Scholar] [CrossRef]
- Baldisserotto, A.; Destro, F.; Vertuani, G.; Marastoni, M.; Gavioli, R.; Tomatis, R. N-Terminal-prolonged vinyl ester-based peptides as selective proteasome β1 subunit inhibitors. Bioorg. Med. Chem. 2009, 17, 5535–5540. [Google Scholar] [CrossRef]
- Baldisserotto, A.; Marastoni, M.; Fiorini, S.; Pretto, L.; Ferretti, V.; Gavioli, R.; Tomatis, R. Vinyl ester-based cyclic peptide proteasome inhibitors. Bioorg. Med. Chem. Lett. 2008, 18, 1849–1854. [Google Scholar] [CrossRef]
- Baldisserotto, A.; Marastoni, M.; Gavioli, R.; Tomatis, R. New cyclic peptide proteasome inhibitors. Bioorg. Med. Chem. Lett. 2009, 19, 1966–1969. [Google Scholar] [CrossRef]
- Franceschini, C.; Trapella, C.; Sforza, F.; Gavioli, R.; Marastoni, M. Synthesis and biological properties of C-terminal vinyl ketone pseudotripeptides. J. Enzyme Inhib. Med. Chem. 2013, 28, 560–564. [Google Scholar] [CrossRef]
- Baldisserotto, A.; Ferretti, V.; Destro, F.; Franceschini, C.; Marastoni, M.; Gavioli, R.; Tomatis, R. α,β-unsaturated N -acylpyrrole peptidyl derivatives: New proteasome inhibitors. J. Med. Chem. 2010, 53, 6511–6515. [Google Scholar] [CrossRef]
- Asai, A.; Hasegawa, A.; Ochiai, K.; Yamashita, Y.; Mizukami, T. Belactosin A, a Novel antitumor antibiotic acting on cyclin/CDK mediated cell cycle regulation, produced by Streptomyces sp. J. Antibiot. 2000, 53, 81–83. [Google Scholar] [CrossRef] [Green Version]
- Asai, A.; Tsujita, T.; Sharma, S.V.; Yamashita, Y.; Akinaga, S.; Funakoshi, M.; Kobayashi, H.; Mizukami, T. A new structural class of proteasome inhibitors identified by microbial screening using yeast-based assay. Biochem. Pharmacol. 2004, 67, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, S.; Unno, Y.; List, A.; Mizuno, A.; Tanaka, M.; Sasaki, T.; Arisawa, M.; Asai, A.; Groll, M.; Shuto, S. Potent proteasome inhibitors derived from the unnatural cis-cyclopropane isomer of Belactosin A: Synthesis, biological activity, and mode of action. J. Med. Chem. 2013, 56, 3689–3700. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Watanabe, M.; Ban, H.S.; Nabeyama, W.; Asai, A. Synthesis and biological evaluation of boron peptide analogues of Belactosin C as proteasome inhibitors. Bioorg. Med. Chem. Lett. 2009, 19, 3220–3224. [Google Scholar] [CrossRef]
- COMP Marizomib for the Treatment of Plasma Cell Myeloma. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/Orphan_designation/2014/09/WC500171952.pdf (accessed on 6 April 2017).
- COMP Marizomib for the Treatment of Glioma. Available online: https://www.ema.europa.eu/en/documents/orphan-designation/eu/3/18/2119-public-summary-opinion-orphan-designation-marizomib-treatment-glioma_en.pdf (accessed on 21 February 2020).
- Triphase Accelerator Corporation Marizomib. Available online: http://triphaseco.com/marizomib/ (accessed on 2 October 2017).
- Triphase Accelerator Corporation Triphase Receives FDA Orphan Drug Designation for Marizomib in Multiple Myeloma. Available online: http://triphaseco.com/wp-content/uploads/2014/02/20140226_Triphase_orphan_drug_news-release_final.pdf (accessed on 4 October 2018).
- Triphase Accelerator Corporation Triphase Accelerator Corporation Announces FDA Orphan Drug Designation for Marizomib in Malignant Glioma. Available online: http://triphaseco.com/wp-content/uploads/2015/11/MRZ-Orphan-Press-Release-11192015_FINAL.pdf (accessed on 4 October 2018).
- ClinicalTrials.gov. Phase 1 Trial of Marizomib Alone and in Combination with Panobinostat for Children with Diffuse Intrinsic Pontine Glioma. Available online: https://www.clinicaltrials.gov/ct2/show/NCT04341311?cond=marizomib&draw=3&rank=4 (accessed on 5 April 2021).
- ClinicalTrials.gov. Phase 2 Clinical Trial of NPI-0052 in Patients with Relapsed or Relapsed/Refractory Multiple Myeloma. Available online: https://clinicaltrials.gov/ct2/show/NCT00461045?term=marizomib&draw=2&rank=7 (accessed on 21 February 2020).
- ClinicalTrials.gov. ABI-009 (Nab-Rapamycin) in Recurrent High Grade Glioma and Newly Diagnosed Glioblastoma. Available online: https://clinicaltrials.gov/ct2/show/NCT03463265?term=marizomib&draw=2&rank=10 (accessed on 20 February 2020).
- ClinicalTrials.gov. Phase 1 Clinical Trial of NPI-0052 in Patients with Advanced Malignancies. Available online: https://clinicaltrials.gov/ct2/show/NCT00629473?cond=marizomib&rank=5 (accessed on 6 October 2017).
- ClinicalTrials.gov. NPI-0052 and Vorinostat in Patients with Non-small Cell Lung Cancer, Pancreatic Cancer, Melanoma or Lymphoma. Available online: https://clinicaltrials.gov/ct2/show/NCT00667082?term=marizomib&draw=2&rank=3 (accessed on 21 February 2020).
- ClinicalTrials.gov. Combination Study of Pomalidomide, Marizomib, and Low-Dose Dexamethasone in Relapsed and Refractory Multiple Myeloma. Available online: https://www.clinicaltrials.gov/ct2/show/NCT02103335?term=Marizomib&rank=3 (accessed on 28 February 2017).
- ClinicalTrials.gov. Stage 1: Marizomib + Bevacizumab in WHO Gr IV GBM; Stage 2: Marizomib Alone; Stage 3: Combination of Marizomib and Bevacizumab. Available online: https://clinicaltrials.gov/ct2/show/NCT02330562?cond=marizomib&rank=3 (accessed on 6 October 2017).
- ClinicalTrials.gov. Study of Marizomib With Temozolomide and Radiotherapy in Patients with Newly Diagnosed Brain Cancer. Available online: https://clinicaltrials.gov/ct2/show/NCT02903069?cond=marizomib&rank=1 (accessed on 6 October 2017).
- ClinicalTrials.gov. A Phase III Trial of With Marizomib in Patients with Newly Diagnosed Glioblastoma. Available online: https://www.clinicaltrials.gov/ct2/show/NCT03345095?term=NPI-0052&draw=2&rank=9 (accessed on 5 April 2021).
- ClinicalTrials.gov. Marizomib for Recurrent Low-Grade and Anaplastic Supratentorial, Infratentorial, and Spinal Cord Ependymoma. Available online: https://clinicaltrials.gov/ct2/show/NCT03727841?term=marizomib&draw=2&rank=2 (accessed on 20 February 2020).
- ClinicalTrials.gov. Phase 1 Clinical Trial of NPI-0052 in Patients with Advanced Solid Tumor Malignancies or Refractory Lymphoma. Available online: https://clinicaltrials.gov/ct2/show/NCT00396864?cond=marizomib&rank=7 (accessed on 6 October 2017).
- Harrison, S.J.; Mainwaring, P.; Price, T.; Millward, M.J.; Padrik, P.; Underhill, C.R.; Cannell, P.K.; Reich, S.D.; Trikha, M.; Spencer, A. Phase I clinical trial of marizomib (NPI-0052) in patients with advanced malignancies including multiple myeloma: Study NPI-0052-102 final results. Clin. Cancer Res. 2016, 22, 4559–4566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millward, M.; Price, T.; Townsend, A.; Sweeney, C.; Spencer, A.; Sukumaran, S.; Longenecker, A.; Lee, L.; Lay, A.; Sharma, G.; et al. Phase 1 clinical trial of the novel proteasome inhibitor marizomib with the histone deacetylase inhibitor vorinostat in patients with melanoma, pancreatic and lung cancer based on in vitro assessments of the combination. Invest. New Drugs 2012, 30, 2303–2317. [Google Scholar] [CrossRef]
- Spencer, A.; Laubach, J.P.; Zonder, J.A.; Badros, A.Z.; Harrison, S.; Khot, A.; Chauhan, D.; Anderson, K.C.; Reich, S.D.; Trikha, M.; et al. Phase 1, multicenter, open-label, combination study (NPI-0052-107; NCT02103335) of pomalidomide (POM), marizomib (MRZ, NPI-0052), and low-dose dexamethasone (LD-DEX) in patients with relapsed and refractory multiple myeloma. Blood 2015, 126, 4220. [Google Scholar] [CrossRef]
- Spencer, A.; Harrison, S.; Laubach, J.P.; Zonder, J.; Badros, A.Z.; Bergin, K.; Khot, A.; Zimmerman, T.; Anderson, K.C.; MacLaren, A.; et al. Pmd-107: Marizomib, pomalidomide and low dose-dexamethasone combination study in relapsed/refractory multiple myeloma (NCT02103335): Full enrollment results from a phase-1 multicenter, open label study. Blood 2016, 128, 3326. [Google Scholar] [CrossRef]
- Bota, D.A.; Mason, W.; Kesari, S.; Magge, R.; Winograd, B.; Elias, I.; Reich, S.D.; Levin, N.; Trikha, M.; Desjardins, A. Marizomib alone or in combination with bevacizumab in patients with recurrent glioblastoma: Phase I/II clinical trial data. Neuro-Oncol. Adv. 2021, 3, 1–10. [Google Scholar] [CrossRef]
- Mason, W.P.; Kesari, S.; Stupp, R.; Aregawi, D.G.; Piccioni, D.E.; Roth, P.; Desjardins, A.; Reich, S.D.; Casadebaig, M.-L.; Elias, I.; et al. Full enrollment results from an extended phase I, multicenter, open label study of marizomib (MRZ) with temozolomide (TMZ) and radiotherapy (RT) in newly diagnosed glioblastoma (GBM). J. Clin. Oncol. 2019, 37, 2021. [Google Scholar] [CrossRef]
- ClinicalTrials.gov. Marizomib Central Nervous System (CNS). Available online: https://clinicaltrials.gov/ct2/show/NCT05050305?term=marizomib&draw=1&rank=4 (accessed on 22 February 2022).
- Clerc, J.; Groll, M.; Illich, D.J.; Bachmann, A.S.; Huber, R.; Schellenberg, B.; Dudler, R.; Kaiser, M. Synthetic and structural studies on syringolin A and B reveal critical determinants of selectivity and potency of proteasome inhibition. Proc. Natl. Acad. Sci. USA 2009, 106, 6507–6512. [Google Scholar] [CrossRef] [Green Version]
- Clerc, J.; Schellenberg, B.; Groll, M.; Bachmann, A.S.; Huber, R.; Dudler, R.; Kaiser, M. Convergent synthesis and biological evaluation of syringolin A and derivatives as eukaryotic 20S proteasome inhibitors. Eur. J. Org. Chem. 2010, 2010, 3991–4003. [Google Scholar] [CrossRef]
- Kisselev, A.F.; van der Linden, W.A.; Overkleeft, H.S. Proteasome inhibitors: An expanding army attacking a unique target. Chem. Biol. 2012, 19, 99–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krahn, D.; Ottmann, C.; Kaiser, M. The chemistry and biology of syringolins, glidobactins and cepafungins (syrbactins). Nat. Prod. Rep. 2011, 28, 1854. [Google Scholar] [CrossRef] [PubMed]
- Archer, C.R.; Groll, M.; Stein, M.L.; Schellenberg, B.; Clerc, J.; Kaiser, M.; Kondratyuk, T.P.; Pezzuto, J.M.; Dudler, R.; Bachmann, A.S. Activity enhancement of the synthetic syrbactin proteasome inhibitor hybrid and biological evaluation in tumor cells. Biochemistry 2012, 51, 6880–6888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richardson, P.G.; Mitsiades, C.; Hideshima, T.; Anderson, K.C. Bortezomib: Proteasome inhibition as an effective anticancer therapy. Annu. Rev. Med. 2006, 57, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Orlowski, R.Z.; Kuhn, D.J. Proteasome inhibitors in cancer therapy: Lessons from the first decade. Clin. Cancer Res. 2008, 14, 1649–1657. [Google Scholar] [CrossRef] [Green Version]
- Dou, Q.P. (Ed.) Resistance to Proteasome Inhibitors in Cancer; Resistance to Targeted Anti-Cancer Therapeutics; Springer International Publishing: Los Angeles, CA, USA, 2014; ISBN 978-3-319-06751-3. [Google Scholar] [CrossRef]
- Suzuki, E.; Demo, S.; Deu, E.; Keats, J.; Arastu-Kapur, S.; Bergsagel, P.L.; Bennett, M.K.; Kirk, C.J. Molecular mechanisms of bortezomib resistant adenocarcinoma cells. PLoS ONE 2011, 6, e27996. [Google Scholar] [CrossRef] [Green Version]
- Verbrugge, S.E.; Al, M.; Assaraf, Y.G.; Niewerth, D.; Van Meerloo, J.; Cloos, J.; Van Der Veer, M.; Scheffer, G.L.; Peters, G.J.; Chan, E.T.; et al. Overcoming bortezomib resistance in human B cells by anti-CD20/rituximab-mediated complement-dependent cytotoxicity and epoxyketone-based irreversible proteasome inhibitors. Exp. Hematol. Oncol. 2013, 2, 1. [Google Scholar] [CrossRef] [Green Version]
- Rückrich, T.; Kraus, M.; Gogel, J.; Beck, A.; Ovaa, H.; Verdoes, M.; Overkleeft, H.S.; Kalbacher, H.; Driessen, C. Characterization of the ubiquitin–proteasome system in bortezomib-adapted cells. Leukemia 2009, 23, 1098–1105. [Google Scholar] [CrossRef] [Green Version]
- Vij, R.; Siegel, D.S.; Jagannath, S.; Jakubowiak, A.J.; Stewart, A.K.; McDonagh, K.; Bahlis, N.; Belch, A.; Kunkel, L.A.; Wear, S.; et al. An open-label, single-arm, phase 2 study of single-agent carfilzomib in patients with relapsed and/or refractory multiple myeloma who have been previously treated with bortezomib. Br. J. Haematol. 2012, 158, 739–748. [Google Scholar] [CrossRef] [Green Version]
- Siegel, D.S.; Martin, T.; Wang, M.; Vij, R.; Jakubowiak, A.J.; Lonial, S.; Trudel, S.; Kukreti, V.; Bahlis, N.; Alsina, M.; et al. A phase 2 study of single-agent carfilzomib (PX-171-003-A1) in patients with relapsed and refractory multiple myeloma. Blood 2012, 120, 2817–2825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berenson, J.R.; Hilger, J.D.; Yellin, O.; Dichmann, R.; Patel-Donnelly, D.; Boccia, R.V.; Bessudo, A.; Stampleman, L.; Gravenor, D.; Eshaghian, S.; et al. Replacement of bortezomib with carfilzomib for multiple myeloma patients progressing from bortezomib combination therapy. Leukemia 2014, 28, 1529–1536. [Google Scholar] [CrossRef] [PubMed]
- ClinicalTrials.gov. Ixazomib as a Replacement for Carfilzomib and Bortezomib for Multiple Myeloma Patients. Available online: https://clinicaltrials.gov/ct2/show/NCT02206425?term=bortezomib&draw=4&rank=669 (accessed on 7 October 2018).
- Berenson, J.R.; Cohen, A.; Spektor, T.M.; Bitran, J.D.; Chen, G.Q.; Moezi, M.M.; Bessudo, A.; Ye, J.Z.; Hager, S.J.; Moss, R.A.; et al. Replacement of ixazomib for relapsed/refractory multiple myeloma patients refractory to a bortezomib or carfilzomib-containing combination therapy. J. Clin. Oncol. 2017, 35, 8013. [Google Scholar] [CrossRef]
- Levin, N.; Spencer, A.; Harrison, S.J.; Chauhan, D.; Burrows, F.J.; Anderson, K.C.; Reich, S.D.; Richardson, P.G.; Trikha, M. Marizomib irreversibly inhibits proteasome to overcome compensatory hyperactivation in multiple myeloma and solid tumour patients. Br. J. Haematol. 2016, 174, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Oerlemans, R.; Franke, N.E.; Assaraf, Y.G.; Cloos, J.; van Zantwijk, I.; Berkers, C.R.; Scheffer, G.L.; Debipersad, K.; Vojtekova, K.; Lemos, C.; et al. Molecular basis of bortezomib resistance: Proteasome subunit 5 (PSMB5) gene mutation and overexpression of PSMB5 protein. Blood 2008, 112, 2489–2499. [Google Scholar] [CrossRef] [Green Version]
- Chauhan, D.; Li, G.; Shringarpure, R.; Podar, K.; Ohtake, Y.; Hideshima, T.; Anderson, K.C. Blockade of Hsp27 overcomes Bortezomib/proteasome inhibitor PS-341 resistance in lymphoma cells. Cancer Res. 2003, 63, 6174–6177. [Google Scholar]
- Kuhn, D.J.; Berkova, Z.; Jones, R.J.; Woessner, R.; Bjorklund, C.C.; Ma, W.; Davis, R.E.; Lin, P.; Wang, H.; Madden, T.L.; et al. Targeting the insulin-like growth factor-1 receptor to overcome bortezomib resistance in preclinical models of multiple myeloma. Blood 2012, 120, 3260–3270. [Google Scholar] [CrossRef] [Green Version]
- Cohen, Y.C.; Zada, M.; Wang, S.-Y.; Bornstein, C.; David, E.; Moshe, A.; Li, B.; Shlomi-Loubaton, S.; Gatt, M.E.; Gur, C.; et al. Identification of resistance pathways and therapeutic targets in relapsed multiple myeloma patients through single-cell sequencing. Nat. Med. 2021, 27, 491–503. [Google Scholar] [CrossRef]
- De Wilt, L.H.A.M.; Jansen, G.; Assaraf, Y.G.; van Meerloo, J.; Cloos, J.; Schimmer, A.D.; Chan, E.T.; Kirk, C.J.; Peters, G.J.; Kruyt, F.A.E. Proteasome-based mechanisms of intrinsic and acquired bortezomib resistance in non-small cell lung cancer. Biochem. Pharmacol. 2012, 83, 207–217. [Google Scholar] [CrossRef]
- Lippert, T.H.; Ruoff, H.-J.; Volm, M. Intrinsic and Acquired Drug Resistance in Malignant Tumors The main reason for therapeutic failure. Arzneimittel-forsch. Drug Res. 2008, 58, 261–264. [Google Scholar] [CrossRef]
- Wang, L.; Kumar, S.; Fridley, B.L.; Kalari, K.R.; Moon, I.; Pelleymounter, L.L.; Hildebrandt, M.A.T.; Batzler, A.; Eckloff, B.W.; Wieben, E.D.; et al. Proteasome β subunit pharmacogenomics: Gene resequencing and functional genomics. Clin. Cancer Res. 2008, 14, 3503–3513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Wood, T.E.; Sprangers, R.; Jansen, G.; Franke, N.E.; Mao, X.; Wang, X.; Zhang, Y.; Verbrugge, S.E.; Adomat, H.; et al. Effect of noncompetitive proteasome inhibition on bortezomib resistance. JNCI J. Natl. Cancer Inst. 2010, 102, 1069–1082. [Google Scholar] [CrossRef] [PubMed]
- Shuqing, L.; Jianmin, Y.; Chongmei, H.; Hui, C.; Wang, J. Upregulated expression of the PSMB5 gene may contribute to drug resistance in patient with multiple myeloma when treated with bortezomib-based regimen. Exp. Hematol. 2011, 39, 1117–1118. [Google Scholar] [CrossRef] [PubMed]
- Niewerth, D.; Franke, N.E.; Jansen, G.; Assaraf, Y.G.; van Meerloo, J.; Kirk, C.J.; Degenhardt, J.; Anderl, J.; Schimmer, A.D.; Zweegman, S.; et al. Higher ratio immune versus constitutive proteasome level as novel indicator of sensitivity of pediatric acute leukemia cells to proteasome inhibitors. Haematologica 2013, 98, 1896–1904. [Google Scholar] [CrossRef]
- Niewerth, D.; Kaspers, G.J.L.; Jansen, G.; Van Meerloo, J.; Zweegman, S.; Jenkins, G.; Whitlock, J.A.; Hunger, S.P.; Lu, X.; Alonzo, T.A.; et al. Proteasome subunit expression analysis and chemosensitivity in relapsed paediatric acute leukaemia patients receiving bortezomib-containing chemotherapy. J. Hematol. Oncol. 2016, 9, 82. [Google Scholar] [CrossRef] [Green Version]
- Lü, S.; Wang, J. The resistance mechanisms of proteasome inhibitor bortezomib. Biomark. Res. 2013, 1, 13. [Google Scholar] [CrossRef] [Green Version]
- Lichter, D.I.; Danaee, H.; Pickard, M.D.; Tayber, O.; Sintchak, M.; Shi, H.; Richardson, P.G.; Cavenagh, J.; Bladé, J.; Façon, T.; et al. Sequence analysis of β-subunit genes of the 20S proteasome in patients with relapsed multiple myeloma treated with bortezomib or dexamethasone. Blood 2012, 120, 4513–4516. [Google Scholar] [CrossRef]
- Coiffier, B.; Li, W.; Henitz, E.D.; Karkera, J.D.; Favis, R.; Gaffney, D.; Shapiro, A.; Theocharous, P.; Elsayed, Y.A.; Van De Velde, H.; et al. Prespecified candidate biomarkers identify follicular lymphoma patients who achieved longer progression-free survival with bortezomib-rituximab versus rituximab. Clin. Cancer Res. 2013, 19, 2551–2561. [Google Scholar] [CrossRef] [Green Version]
- Varga, G.; Mikala, G.; Kiss, K.P.; Kosóczki, É.; Szabó, E.; Meggyesi, N.; Balassa, K.; Kövy, P.; Tegze, B.; Szombath, G.; et al. Proteasome Subunit Beta Type 1 P11A Polymorphism Is a New Prognostic Marker in Multiple Myeloma. Clin. Lymphoma Myeloma Leuk. 2017, 17, 734–742. [Google Scholar] [CrossRef]
- Huber, E.M.; Heinemeyer, W.; Groll, M. Bortezomib-resistant mutant proteasomes: Structural and biochemical evaluation with carfilzomib and ONX 0914. Structure 2015, 23, 407–417. [Google Scholar] [CrossRef] [Green Version]
- Kale, A.J.; Moore, B.S. Molecular mechanisms of acquired proteasome inhibitor resistance. J. Med. Chem. 2012, 55, 10317–10327. [Google Scholar] [CrossRef] [PubMed]
- Niewerth, D.; Jansen, G.; Assaraf, Y.G.; Zweegman, S.; Kaspers, G.J.L.; Cloos, J. Molecular basis of resistance to proteasome inhibitors in hematological malignancies. Drug Resist. Updat. 2015, 18, 18–35. [Google Scholar] [CrossRef]
- Niewerth, D.; Kaspers, G.J.; Assaraf, Y.G.; Van Meerloo, J.; Kirk, C.J.; Anderl, J.; Blank, J.L.; Van De Ven, P.M.; Zweegman, S.; Jansen, G.; et al. Interferon-γ-induced upregulation of immunoproteasome subunit assembly overcomes bortezomib resistance in human hematological cell lines. J. Hematol. Oncol. 2014, 7, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niewerth, D.; van Meerloo, J.; Jansen, G.; Assaraf, Y.G.; Hendrickx, T.C.; Kirk, C.J.; Anderl, J.L.; Zweegman, S.; Kaspers, G.J.L.; Cloos, J. Anti-leukemic activity and mechanisms underlying resistance to the novel immunoproteasome inhibitor PR-924. Biochem. Pharmacol. 2014, 89, 43–51. [Google Scholar] [CrossRef]
- Wu, Y.-X.; Yang, J.-H.; Saitsu, H. Bortezomib-resistance is associated with increased levels of proteasome subunits and apoptosis-avoidance. Oncotarget 2016, 7, 77622–77634. [Google Scholar] [CrossRef] [Green Version]
- Ding, W.-X.; Ni, H.-M.; Gao, W.; Yoshimori, T.; Stolz, D.B.; Ron, D.; Yin, X.-M. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am. J. Pathol. 2007, 171, 513–524. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, D.; Berges, C.; Opelz, G.; Daniel, V.; Naujokat, C. Increased expression and altered subunit composition of proteasomes induced by continuous proteasome inhibition establish apoptosis resistance and hyperproliferation of Burkitt lymphoma cells. J. Cell. Biochem. 2008, 103, 270–283. [Google Scholar] [CrossRef]
- Lü, S.; Chen, Z.; Yang, J.; Chen, L.; Gong, S.; Zhou, H.; Guo, L.; Wang, J. Overexpression of the PSMB5 gene contributes to bortezomib resistance in T-lymphoblastic lymphoma/leukemia cells derived from Jurkat line. Exp. Hematol. 2008, 36, 1278–1284. [Google Scholar] [CrossRef]
- Franke, N.E.; Niewerth, D.; Assaraf, Y.G.; van Meerloo, J.; Vojtekova, K.; van Zantwijk, C.H.; Zweegman, S.; Chan, E.T.; Kirk, C.J.; Geerke, D.P.; et al. Impaired bortezomib binding to mutant β5 subunit of the proteasome is the underlying basis for bortezomib resistance in leukemia cells. Leukemia 2012, 26, 757–768. [Google Scholar] [CrossRef] [Green Version]
- Verbrugge, S.E.; Assaraf, Y.G.; Dijkmans, B.A.C.; Scheffer, G.L.; Al, M.; den Uyl, D.; Oerlemans, R.; Chan, E.T.; Kirk, C.J.; Peters, G.J.; et al. Inactivating PSMB5 mutations and P-glycoprotein (multidrug resistance-associated protein/ATP-binding cassette B1) mediate resistance to proteasome inhibitors: Ex vivo efficacy of (immuno)proteasome inhibitors in mononuclear blood cells from patients with rheumatoid arthritis. J. Pharmacol. Exp. Ther. 2012, 341, 174–182. [Google Scholar] [CrossRef] [Green Version]
- Lü, S.; Yang, J.; Chen, Z.; Gong, S.; Zhou, H.; Xu, X.; Wang, J. Different mutants of PSMB5 confer varying bortezomib resistance in T lymphoblastic lymphoma/leukemia cells derived from the Jurkat cell line. Exp. Hematol. 2009, 37, 831–837. [Google Scholar] [CrossRef] [PubMed]
- Ri, M.; Iida, S.; Nakashima, T.; Miyazaki, H.; Mori, F.; Ito, A.; Inagaki, A.; Kusumoto, S.; Ishida, T.; Komatsu, H.; et al. Bortezomib-resistant myeloma cell lines: A role for mutated PSMB5 in preventing the accumulation of unfolded proteins and fatal ER stress. Leukemia 2010, 24, 1506–1512. [Google Scholar] [CrossRef]
- Niewerth, D.; Jansen, G.; Riethoff, L.F.V.; van Meerloo, J.; Kale, A.J.; Moore, B.S.; Assaraf, Y.G.; Anderl, J.L.; Zweegman, S.; Kaspers, G.J.L.; et al. Antileukemic activity and mechanism of drug resistance to the marine Salinispora tropica proteasome inhibitor salinosporamide A (Marizomib). Mol. Pharmacol. 2014, 86, 12–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groll, M.; Berkers, C.R.; Ploegh, H.L.; Ovaa, H. Crystal Structure of the Boronic Acid-Based Proteasome Inhibitor Bortezomib in Complex with the Yeast 20S Proteasome. Structure 2006, 14, 451–456. [Google Scholar] [CrossRef] [Green Version]
- Harshbarger, W.; Miller, C.; Diedrich, C.; Sacchettini, J.; Correspondence, J.S.; Sacchettini, J. Crystal structure of the human 20S proteasome in complex with carfilzomib. Structure 2015, 23, 418–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Politou, M.; Karadimitris, A.; Terpos, E.; Kotsianidis, I.; Apperley, J.F.; Rahemtulla, A. No evidence of mutations of the PSMB5 (beta-5 subunit of proteasome) in a case of myeloma with clinical resistance to Bortezomib. Leuk. Res. 2006, 30, 240–241. [Google Scholar] [CrossRef]
- Balsas, P.; Galán-Malo, P.; Marzo, I.; Naval, J. Bortezomib resistance in a myeloma cell line is associated to PSMβ5 overexpression and polyploidy. Leuk. Res. 2012, 36, 212–218. [Google Scholar] [CrossRef]
- Mandery, K.; Glaeser, H.; Fromm, M.F. Interaction of innovative small molecule drugs used for cancer therapy with drug transporters. Br. J. Pharmacol. 2012, 165, 345–362. [Google Scholar] [CrossRef] [Green Version]
- Lü, S.; Chen, Z.; Yang, J.; Chen, L.; Zhou, H.; Xu, X.; Li, J.; Han, F.; Wang, J. The effects of proteasome inhibitor bortezomib on a P-gp positive leukemia cell line K562/A02. Int. J. Lab. Hematol. 2010, 32, e123–e131. [Google Scholar] [CrossRef]
- Minderman, H.; Zhou, Y.; O’Loughlin, K.L.; Baer, M.R.; O’Loughlin, K.L.; Baer, M.R. Bortezomib activity and in vitro interactions with anthracyclines and cytarabine in acute myeloid leukemia cells are independent of multidrug resistance mechanisms and p53 status. Cancer Chemother. Pharmacol. 2007, 60, 245–255. [Google Scholar] [CrossRef]
- Clemens, J.; Seckinger, A.; Hose, D.; Theile, D.; Longo, M.; Haefeli, W.E.; Burhenne, J.; Weiss, J. Cellular uptake kinetics of bortezomib in relation to efficacy in myeloma cells and the influence of drug transporters. Cancer Chemother. Pharmacol. 2015, 75, 281–291. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, R.; Ooi, M.G.; Meiller, J.; Jakubikova, J.; Klippel, S.; Delmore, J.; Richardson, P.; Anderson, K.; Clynes, M.; Mitsiades, C.S.; et al. The interaction of bortezomib with multidrug transporters: Implications for therapeutic applications in advanced multiple myeloma and other neoplasias. Cancer Chemother. Pharmacol. 2013, 71, 1357–1368. [Google Scholar] [CrossRef] [PubMed]
- Besse, A.; Stolze, S.C.; Rasche, L.; Weinhold, N.; Morgan, G.J.; Kraus, M.; Bader, J.; Overkleeft, H.S.; Besse, L.; Driessen, C. Carfilzomib resistance due to ABCB1/MDR1 overexpression is overcome by nelfinavir and lopinavir in multiple myeloma. Leukemia 2017, 32, 391–401. [Google Scholar] [CrossRef] [Green Version]
- Ao, L.; Wu, Y.; Kim, D.; Jang, E.R.; Kim, K.; Lee, D.-M.; Kim, K.B.; Lee, W. Development of peptide-based reversing agents for p-glycoprotein-mediated resistance to carfilzomib. Mol. Pharm. 2012, 9, 2197–2205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutman, D.; Morales, A.A.; Boise, L.H. Acquisition of a multidrug-resistant phenotype with a proteasome inhibitor in multiple myeloma. Leukemia 2009, 23, 2181–2183. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, D.J.; Chen, Q.; Voorhees, P.M.; Strader, J.S.; Shenk, K.D.; Sun, C.M.; Demo, S.D.; Bennett, M.K.; van Leeuwen, F.W.B.; Chanan-Khan, A.A.; et al. Potent activity of carfilzomib, a novel, irreversible inhibitor of the ubiquitin-proteasome pathway, against preclinical models of multiple myeloma. Blood 2007, 110, 3281–3290. [Google Scholar] [CrossRef] [Green Version]
- Rumpold, H.; Salvador, C.; Wolf, A.M.; Tilg, H.; Gastl, G.; Wolf, D. Knockdown of PgP resensitizes leukemic cells to proteasome inhibitors. Biochem. Biophys. Res. Commun. 2007, 361, 549–554. [Google Scholar] [CrossRef]
- Clemens, J.; Welti, L.; Schäfer, J.; Seckinger, A.; Burhenne, J.; Theile, D.; Weiss, J. Bortezomib, carfilzomib and ixazomib do not mediate relevant transporter-based drug-drug interactions. Oncol. Lett. 2017. [Google Scholar] [CrossRef] [Green Version]
- Hawley, T.S.; Riz, I.; Yang, W.; Wakabayashi, Y.; DePalma, L.; Chang, Y.-T.; Peng, W.; Zhu, J.; Hawley, R.G. Identification of an ABCB1 (P-glycoprotein)-positive carfilzomib-resistant myeloma subpopulation by the pluripotent stem cell fluorescent dye CDy1. Am. J. Hematol. 2013, 88, 265–272. [Google Scholar] [CrossRef] [Green Version]
- Hideshima, T.; Richardson, P.G.; Anderson, K.C. Mechanism of action of proteasome inhibitors and deacetylase inhibitors and the biological basis of synergy in multiple myeloma. Mol. Cancer Ther. 2011, 10, 2034–2042. [Google Scholar] [CrossRef] [Green Version]
- Chhabra, S. Saurabh novel proteasome inhibitors and histone deacetylase inhibitors: Progress in myeloma therapeutics. Pharmaceuticals 2017, 10, 40. [Google Scholar] [CrossRef] [PubMed]
- Catley, L.; Weisberg, E.; Kiziltepe, T.; Tai, Y.-T.; Hideshima, T.; Neri, P.; Tassone, P.; Atadja, P.; Chauhan, D.; Munshi, N.C.; et al. Aggresome induction by proteasome inhibitor bortezomib and alpha-tubulin hyperacetylation by tubulin deacetylase (TDAC) inhibitor LBH589 are synergistic in myeloma cells. Blood 2006, 108, 3441–3449. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, P.; Santagata, S.; Lindquist, S. Methods and Compositions Relating to Proteasome Inhibitor Resistance. U.S. Patent 2018/0353445, 2017. [Google Scholar]
- Dou, Q.; Yang, H.; Heath, E.I. Treatments and Diagnostics for Cancers. U.S. Patent 2018/0280412, 2017. [Google Scholar]
- Roden, R.B.; Anchoori, R.K.; Balasubramanyam, K. Novel Bis-Benzylidine Piperidone Proteasome Inhibitor with Anticancer Activity. Patent WO 2014/182744, 2014. [Google Scholar]
- Chiriva-Internati, M.; Cobos, E.; John, C. Galectin-3C Combination Therapy for Human Cancer. U.S. Patent 2016/0250289, 2012. [Google Scholar]
- Gaczynska, M.E.; Osmulski, P.A. Rapamycin Analogs Targeting Proteasome Function in the Treatment of Cancer. U.S. Patent 2015/328192, 2014. [Google Scholar]
- Keegan, M.; Grant, S. Combination Therapy. U.S. Patent 2009/0105200, 2008. [Google Scholar]
- Anderson, K.C.; Hideshima, T.; Mitsiades, C.S.; Mitsiades, N. Methods and Compositions for Treating Cancer using Proteasome Inhibitors. U.S. Patent 2012/0214757, 2004. [Google Scholar]
- Dai, C.; Tang, Z. Methods of Treating Cancer by Administering a MEK Inhibitor in Combination with a Proteasome Inhibitor. Patent WO 2017/161093, 2017. [Google Scholar]
- Dolloff, N.G.; Robinson, R.M. Caffeic Acid Derivatives and Uses Thereof. Patent WO 2017/161093, 2017. [Google Scholar]
- FDA. Highlights of Prescribing Information XPovio® (selinexor). Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/212306s001lbl.pdf (accessed on 4 April 2021).
- Karyopharm Therapeutics Inc U.S. Food and Drug Administration Accepts Karyopharm’s Supplemental New Drug Application for XPOVIO® (Selinexor) as a Treatment for Patients with Relapsed or Refractory Diffuse Large B-Cell Lymphoma. Available online: https://www.karyopharm.com/products/ (accessed on 21 February 2020).
- Bahlis, N.J.; Sutherland, H.; White, D.; Sebag, M.; Lentzsch, S.; Kotb, R.; Venner, C.P.; Gasparetto, C.; Del Col, A.; Neri, P.; et al. Selinexor plus low-dose bortezomib and dexamethasone for patients with relapsed or refractory multiple myeloma. Blood 2018, 132, 2546–2554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakubowiak, A.J.; Jasielec, J.K.; Rosenbaum, C.A.; Cole, C.E.; Chari, A.; Mikhael, J.; Nam, J.; McIver, A.; Severson, E.; Stephens, L.A.; et al. Phase 1 study of selinexor plus carfilzomib and dexamethasone for the treatment of relapsed/refractory multiple myeloma. Br. J. Haematol. 2019, 186, 549–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ClinicalTrials.gov. Selinexor, Carfilzomib, and Dexamethasone in Treating Patients with Relapsed or Refractory Multiple Myeloma (SINE). Available online: https://clinicaltrials.gov/ct2/show/NCT02199665 (accessed on 21 February 2020).
- ClinicalTrials.gov. Selinexor, Carfilzomib, and Dexamethasone Versus Placebo, Carfilzomib, and Dexamethasone in Multiple Myeloma. Available online: https://clinicaltrials.gov/ct2/show/NCT02628704 (accessed on 21 February 2020).
- ClinicalTrials.gov. Selinexor and Backbone Treatments of Multiple Myeloma Patients (STOMP). Available online: https://clinicaltrials.gov/ct2/show/NCT02343042 (accessed on 21 February 2020).
- ClinicalTrials.gov. A Study to Test the Combination of Selinexor (KPT-330), Ixazomib, and Dexamethasone in Patients With Myeloma. Available online: https://clinicaltrials.gov/ct2/show/NCT02831686 (accessed on 21 February 2020).
- ClinicalTrials.gov. A Phase 2b, Open-label, Multi-arm Clinical Trial of Selinexor Plus Low-dose Dexamethasone (Sd) in Patients With Penta-refractory Multiple Myeloma or Selinexor and Bortezomib Plus Low-dose Dexamethasone (SVd) in Patients With Triple-class Refractory Multip. Available online: https://clinicaltrials.gov/ct2/show/NCT04414475?term=selinexor&draw=6&rank=48 (accessed on 11 October 2020).
- ClinicalTrials.gov. A Phase 3 Randomized, Controlled, Open-label Study of Selinexor, Bortezomib, and Dexamethasone (SVd) Versus Bortezomib and Dexamethasone (Vd) in Patients with Relapsed or Refractory Multiple Myeloma (RRMM). Available online: https://clinicaltrials.gov/ct2/show/NCT03110562?term=selinexor&draw=8&rank=69 (accessed on 11 October 2020).
- Grosicki, S.; Simonova, M.; Spicka, I.; Pour, L.; Kriachok, I.; Gavriatopoulou, M.; Pylypenko, H.; Auner, H.W.; Leleu, X.; Doronin, V.; et al. Once-per-week selinexor, bortezomib, and dexamethasone versus twice-per-week bortezomib and dexamethasone in patients with multiple myeloma (BOSTON): A randomised, open-label, phase 3 trial. Lancet 2020, 396, 1563–1573. [Google Scholar] [CrossRef]
- Mateos, M.V.; Gavriatopoulou, M.; Facon, T.; Auner, H.W.; Leleu, X.; Hájek, R.; Dimopoulos, M.A.; Delimpasi, S.; Simonova, M.; Špička, I.; et al. Effect of prior treatments on selinexor, bortezomib, and dexamethasone in previously treated multiple myeloma. J. Hematol. Oncol. 2021, 14, 59. [Google Scholar] [CrossRef]
- Richard, S.; Chari, A.; Delimpasi, S.; Simonova, M.; Spicka, I.; Pour, L.; Kriachok, I.; Dimopoulos, M.A.; Pylypenko, H.; Auner, H.W.; et al. Selinexor, bortezomib, and dexamethasone versus bortezomib and dexamethasone in previously treated multiple myeloma: Outcomes by cytogenetic risk. Am. J. Hematol. 2021, 96, 1120–1130. [Google Scholar] [CrossRef]
- Auner, H.W.; Gavriatopoulou, M.; Delimpasi, S.; Simonova, M.; Spicka, I.; Pour, L.; Dimopoulos, M.A.; Kriachok, I.; Pylypenko, H.; Leleu, X.; et al. Effect of age and frailty on the efficacy and tolerability of once-weekly selinexor, bortezomib, and dexamethasone in previously treated multiple myeloma. Am. J. Hematol. 2021, 96, 708–718. [Google Scholar] [CrossRef]
- ClinicalTrials.gov. An Open-label, Multicenter, Phase 2 Trial of Selinexor (KPT-330), Bortezomib and Low-dose Dexamethasone Plus Daratumumab (SELIBORDARA) for the Treatment of Patients With Refractory or Relapsed and Refractory Multiple Myeloma. Available online: https://clinicaltrials.gov/ct2/show/NCT03589222?term=selinexor&draw=8&rank=63 (accessed on 11 October 2020).
- ClinicalTrials.gov. A Study of Selinexor, in Combination with Carfilzomib, Daratumumab or Pomalidomide in Patients With Multiple Myeloma. Available online: https://www.clinicaltrials.gov/ct2/show/NCT04661137?term=carfilzomib&strd_s=02%2F15%2F2020&strd_e=02%2F16%2F2021&draw=2&rank=5 (accessed on 16 February 2021).
- ClinicalTrials.gov. Selinexor, Pomalidomide, and Dexamethasone with or Without Carfilzomib for the Treatment of Patients With Relapsed Refractory Multiple Myeloma, The SCOPE Trial. Available online: https://www.clinicaltrials.gov/ct2/show/NCT04764942?term=selinexor&draw=8&rank=63 (accessed on 22 February 2022).
- Mitra, A.K.; Ness, B. Van Transcriptional classification and prediction of drug response (t-cap dr). U.S. Patent 2017/0159130, 2017. [Google Scholar]
- Mulligan, G.; Bryant, B.M.; Morrissey, M.P.; Bolt, A.; Damokosh, A.I. Methods for the Identification, Assessment, and Treatment of Patients with Proteasome Inhibition Therapy. U.S. Patent 2017/023577, 2004. [Google Scholar]
- Bhatia, S.; Krieger, V.; Groll, M.; Osko, J.D.; Reßing, N.; Ahlert, H.; Borkhardt, A.; Kurz, T.; Christianson, D.W.; Hauer, J.; et al. Discovery of the first-in-class dual histone deacetylase–proteasome inhibitor. J. Med. Chem. 2018, 61, 10299–10309. [Google Scholar] [CrossRef]




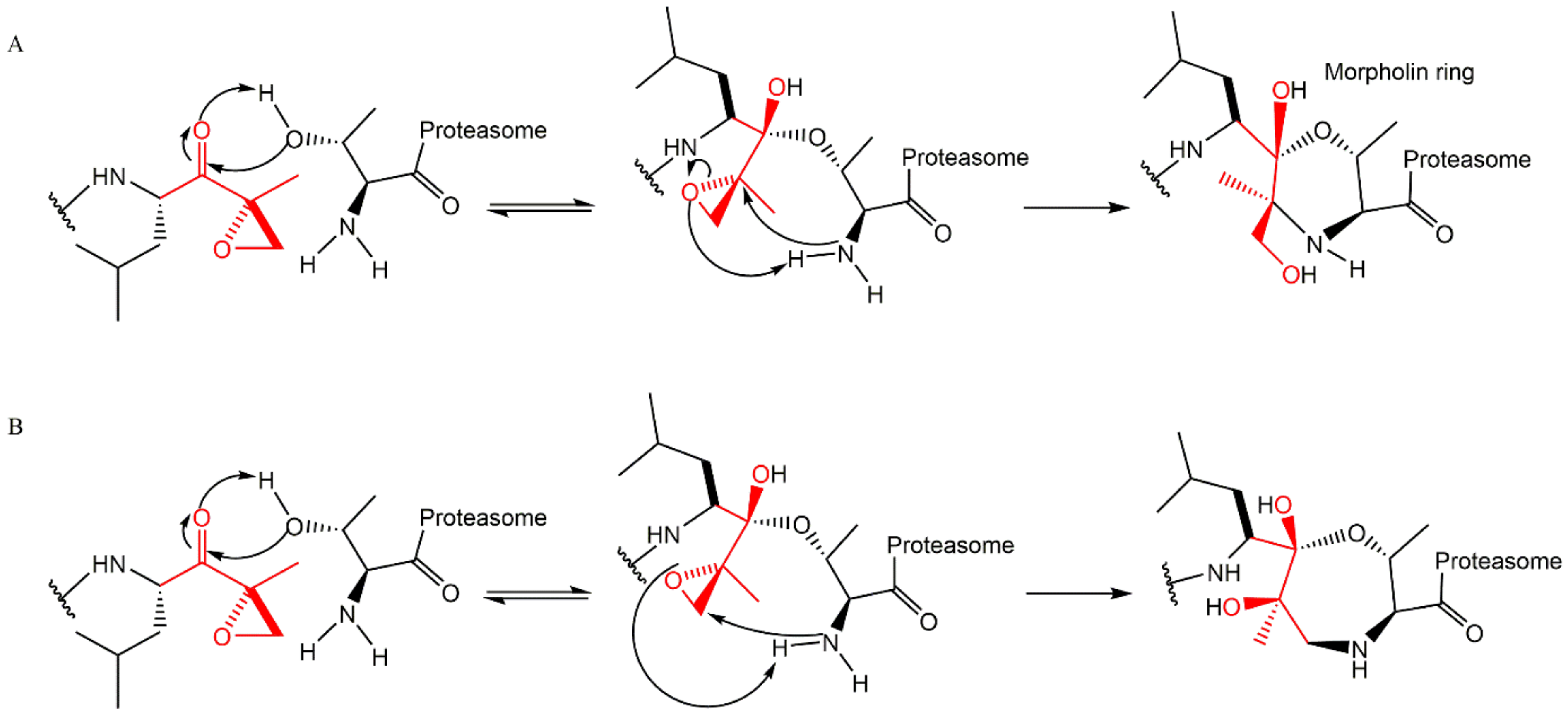
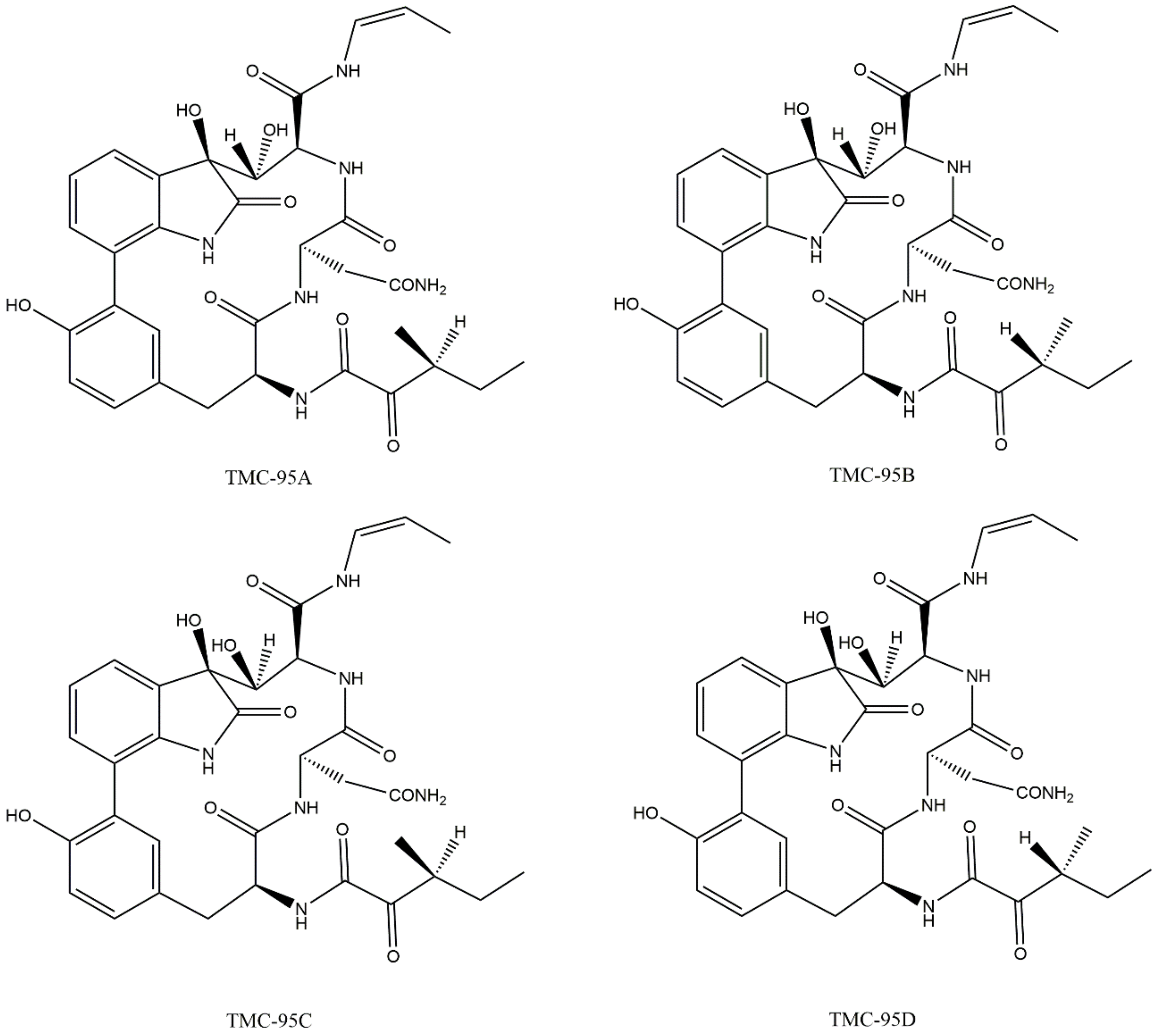
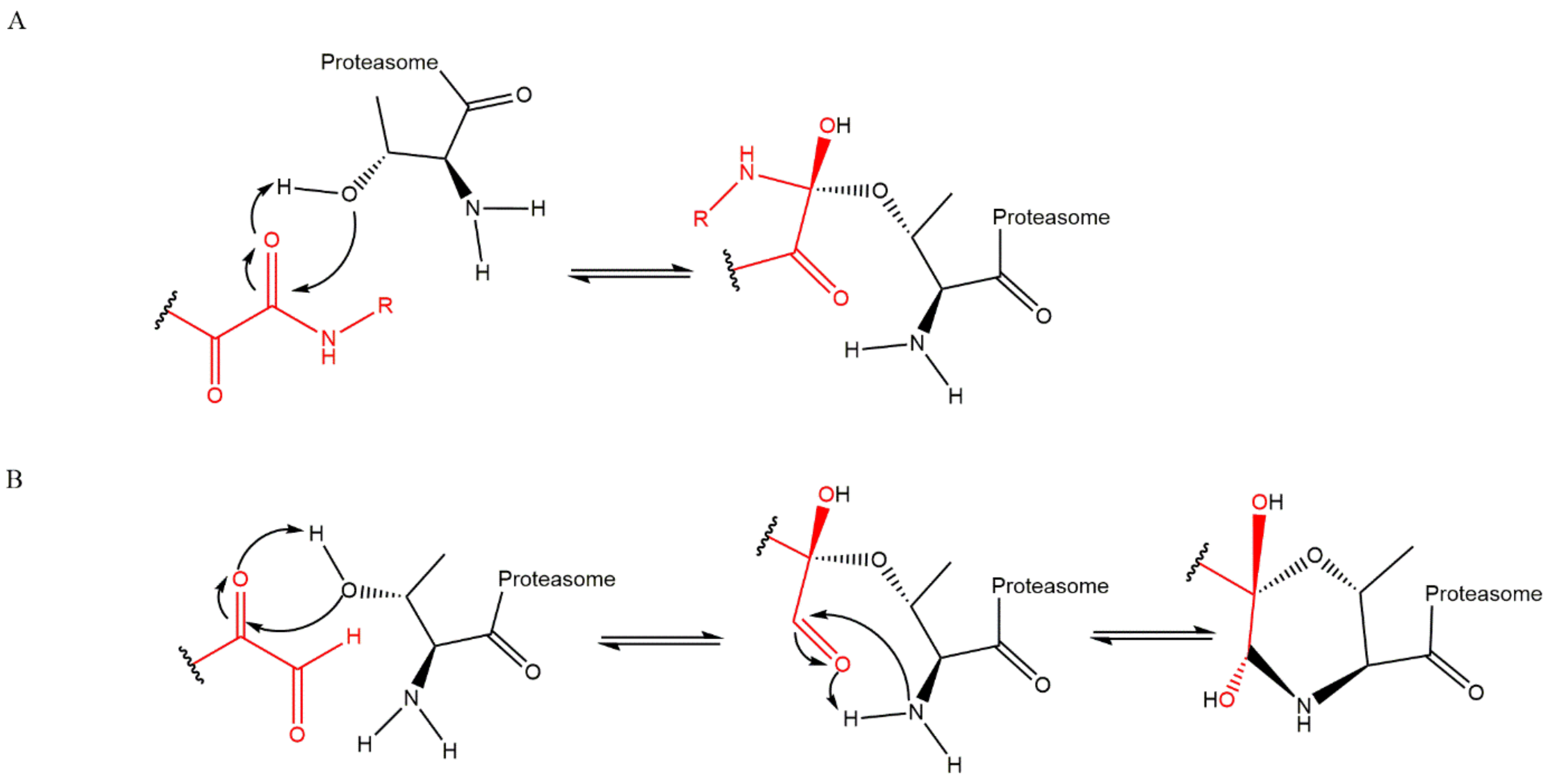
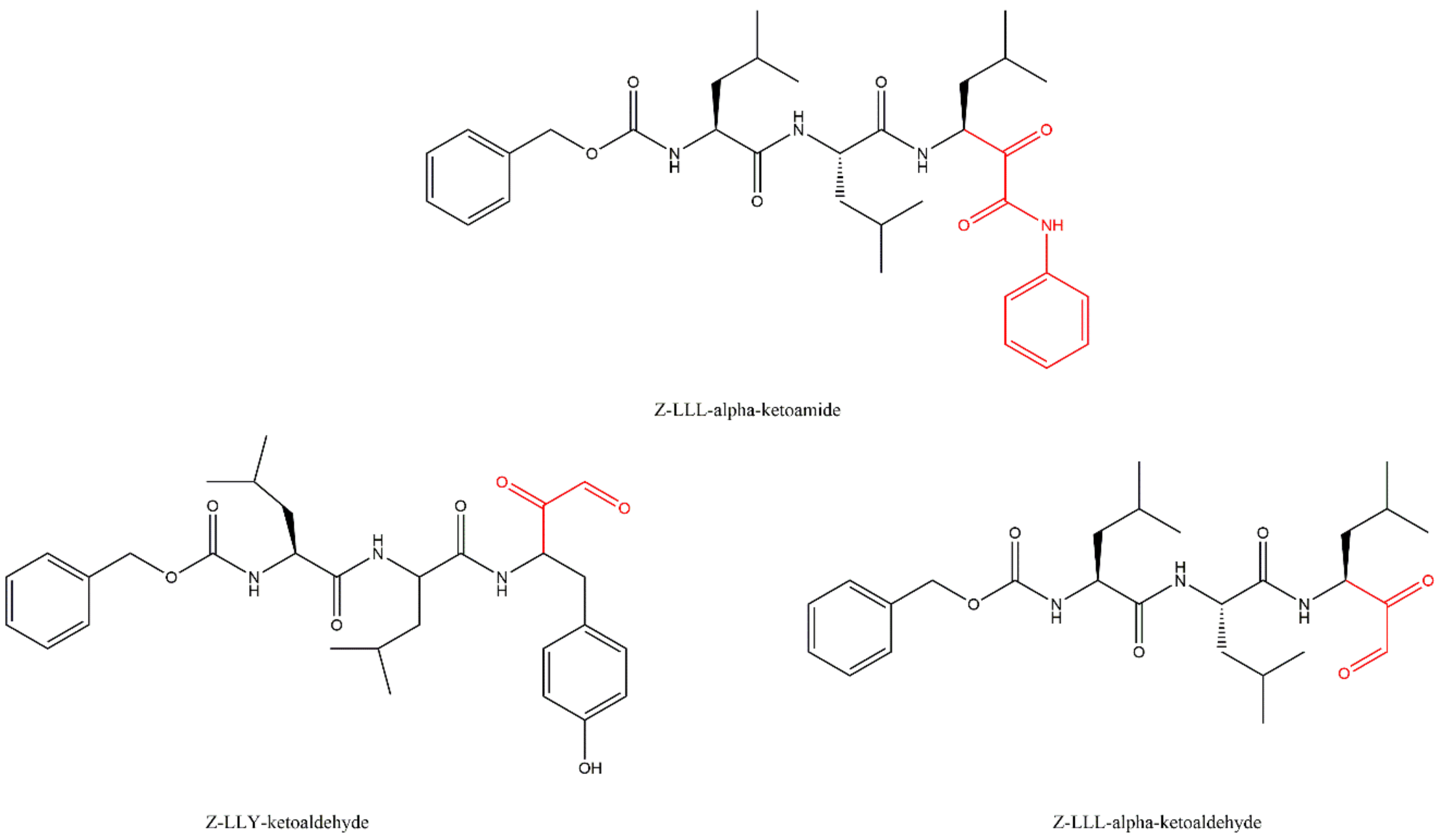
| Bortezomib-Resistant Cell Lines | Observations (Alteration of the Expression Levels of Proteasomal Subunits) | Ref. |
|---|---|---|
| Human Namalwa Burkitt lymphoma | Proteolytic activities increased. Increased expression of the proteolytic subunits α, β1, β2, β4, β5 and β6, indicating that the cells abundantly express and assemble complete 20S complexes. | [138] |
| Jurkat B1 and B5 of lymphoblastic lymphoma/leukemia (highly resistant) | mRNA of the gene encoding the β5 subunit levels overexpressed, amplification of this gene and increased chymotrypsin-like activity. | [139] |
| Jurkat B2 of lymphoblastic lymphoma/leukemia (slightly resistant) | No significant differences were registered in comparison with parental cells. | [139] |
| THP-1 of human myelomonocytic | β5 subunit levels overexpressed up to 60-fold (proportional to the gradually increasing concentrations of bortezomib during the stepwise selection). β1 and β2 subunit’s expression was less than 2-fold increased. | [116] |
| Mutation | Cell Line | Resistance | Observations | Refs. |
|---|---|---|---|---|
| Ala49Thr | THP-1 of human myelomonocytic | Bortezomib | Cross-resistance to carfilzomib and oprozomib. The siRNA-guided silencing of β5 subunit gene expression restored bortezomib sensitivity. | [116,140,141] |
| Jurkat B of lymphoblastic lymphoma/leukemia | Bortezomib | Chymotrypsin-like activity did not differ significantly between mutated and non-mutated cells. | [142] | |
| KMS-11 and OPM-2 of multiple myeloma | Bortezomib and MG132 | The artificial introduction of the mutation in the cell line KMS-11 induced a less prominent resistance. | [143] | |
| 8226 of multiple myeloma | Bortezomib | [140] | ||
| CCRF-CEM of leukemia | Bortezomib | Cross-resistance to marizomib. | [140,144] | |
| H460 e A549 of non-small cell lung cancer human | Bortezomib | Cross-resistance to carfilzomib and MG132. | [120] | |
| Ala49Val | Jurkat B of lymphoblastic lymphoma/leukemia | Bortezomib | Chymotrypsin-like activity did not differ significantly between mutated and non-mutated cells. | [142] |
| CCRF-CEM of leukemia | Bortezomib | [140] | ||
| Ala50Val | Jurkat B of lymphoblastic lymphoma/leukemia | Bortezomib | [142] | |
| Arg24Cys (*) | HT-29 of adenocarcinoma | Bortezomib | Proteasome activity and sensitivity to MG262 did not change. | [107] |
| Cys52Phe (*) | CCRF-CEM of leukemia | Bortezomib | Cross-resistance to marizomib. | [140,144] |
| SW1573 of non-small cell lung cancer human | Bortezomib | Cross-resistance to carfilzomib and MG132. | [120] | |
| Cys63Phe | HT-29 of adenocarcinoma | Bortezomib | [107] | |
| Met45Ile | THP-1 of human myelomonocytic | Bortezomib | Cross-resistance to carfilzomib and oprozomib. | [140,141] |
| Met45Val | THP-1 of human myelomonocytic | Bortezomib | [140] | |
| H549 of non-small cell lung cancer human | Bortezomib | Cross-resistance to carfilzomib and MG132. | [120] | |
| CCRF-CEM of leukemia | Marizomib | Cross-resistance to bortezomib. | [144] | |
| Thr21Ala | 8226 of multiple myeloma | Bortezomib | [140] |
| Cell Line | Resistance to Inhibitor | Observations | Ref. |
|---|---|---|---|
| AMO of multiple myeloma and ARH77 of plasmoide leukemia | Carfilzomib (resistance induced by continued exposure to inhibitor) | Additionally, the influence of P-gp transporters on other inhibitors was evaluated. The ratio of IC50 values between the ABCB1-containing and the ABCB1-deficient AMO cells was 2.6, 7.8, 3.7, 2, 6.7 and 1.9, respectively to bortezomib, carfilzomib, delanzomib, ixazomib, oprozomib and marizomib. | [154] |
| CEM/VLB of T-cell leukemia with P-gp transporters overexpressed | Carfilzomib and oprozomib | Comparatively with parental CEM cells, the cells were 4.5-fold resistant to bortezomib, 23-fold resistant to oprozomib and 114-fold resistant to carfilzomib. The P-gp inhibitor P121 restored the capacity of inhibitors to inhibit chymotrypsin-like proteasome activity at inhibitory concentrations obtained with parental CEM cells. | [141] |
| H23 of human lung adenocarcinoma and DLD of human colon adenocarcinoma | Carfilzomib (resistance induced by continued exposure to inhibitor) | Cross-resistance to YU-101. No significant differences, compared with cells, were registered for the BCRP and MRP1–3 transporters levels. The P-gp inhibitor verapamil restored carfilzomib sensitivity of cells. | [155] |
| KMS11 of multiple myeloma | Epoxomicin (resistance induced by continued exposure to inhibitor) | Mutations in the gene encoding the β5 subunit of 20S CP were not identified. The verapamil restored epoxomicin sensitivity of cells. | [156] |
| RPMI8226.Dox40 of doxorubicin-resistant multiple myeloma with P-gp transporters overexpressed | Carfilzomib | Pre-treatment with the verapamil partially overcame the resistance to carfilzomib. | [157] |
| Study (Code) | Title | Status | 20S CP inhibitor | Other Drugs |
|---|---|---|---|---|
| NCT00401011 | An Open-Label Phase I/II Study of the Safety and Efficacy of Perifosine and Bortezomib with or without Dexamethasone for Patients with Relapsed or Refractory Multiple Myeloma Previously Treated with Bortezomib | Completed | Bortezomib | Dexamethasone and perifosine |
| NCT01083602 | A Phase II, Multi-center, Single Arm, Open Label Study of Panobinostat in Combination with Bortezomib and Dexamethasone in Patients with Relapsed and Bortezomib-refractory Multiple Myeloma | Completed | Bortezomib | Dexamethasone and panobinostat |
| NCT01794507 | A Phase 1b Study Evaluating the Safety and Pharmacokinetics of ABT-199 in Relapsed or Refractory Multiple Myeloma Subjects Who Are Receiving Bortezomib and Dexamethasone as Their Standard Therapy | Completed | Bortezomib | Dexamethasone and venetoclax |
| NCT02188537 | Nelfinavir as Bortezomib-sensitizing Drug in Patients with Proteasome Inhibitor-nonresponsive Myeloma. A Multicenter Phase II Trial | Completed | Bortezomib | Dexamethasone and nelfinavir |
| NCT04065789 | Safety, Tolerability, and Efficacy of Once Weekly Carfilzomib in Combination with Daratumumab, Lenalidomide, and Dexamethasone, in Transplant-ineligible Multiple Myeloma Patients Non-responsive to a Bortezomib Based Induction | Completed | Carfilzomib | Daratumumab, lenalidomide and dexamethasone |
| NCT04163107 | Combined Carfilzomib and Hydroxychloroquine in Patients with Relapsed/Refractory Multiple Myeloma–a Phase 1 Trial | Active, not recruiting participants | Carfilzomib | Hydroxychloro-quine and dexamethasone |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leonardo-Sousa, C.; Carvalho, A.N.; Guedes, R.A.; Fernandes, P.M.P.; Aniceto, N.; Salvador, J.A.R.; Gama, M.J.; Guedes, R.C. Revisiting Proteasome Inhibitors: Molecular Underpinnings of Their Development, Mechanisms of Resistance and Strategies to Overcome Anti-Cancer Drug Resistance. Molecules 2022, 27, 2201. https://doi.org/10.3390/molecules27072201
Leonardo-Sousa C, Carvalho AN, Guedes RA, Fernandes PMP, Aniceto N, Salvador JAR, Gama MJ, Guedes RC. Revisiting Proteasome Inhibitors: Molecular Underpinnings of Their Development, Mechanisms of Resistance and Strategies to Overcome Anti-Cancer Drug Resistance. Molecules. 2022; 27(7):2201. https://doi.org/10.3390/molecules27072201
Chicago/Turabian StyleLeonardo-Sousa, Carlota, Andreia Neves Carvalho, Romina A. Guedes, Pedro M. P. Fernandes, Natália Aniceto, Jorge A. R. Salvador, Maria João Gama, and Rita C. Guedes. 2022. "Revisiting Proteasome Inhibitors: Molecular Underpinnings of Their Development, Mechanisms of Resistance and Strategies to Overcome Anti-Cancer Drug Resistance" Molecules 27, no. 7: 2201. https://doi.org/10.3390/molecules27072201
APA StyleLeonardo-Sousa, C., Carvalho, A. N., Guedes, R. A., Fernandes, P. M. P., Aniceto, N., Salvador, J. A. R., Gama, M. J., & Guedes, R. C. (2022). Revisiting Proteasome Inhibitors: Molecular Underpinnings of Their Development, Mechanisms of Resistance and Strategies to Overcome Anti-Cancer Drug Resistance. Molecules, 27(7), 2201. https://doi.org/10.3390/molecules27072201