Diterpenoids from Blumea balsamifera and Their Anti-Inflammatory Activities
Abstract
:1. Introduction
2. Results and Discussion
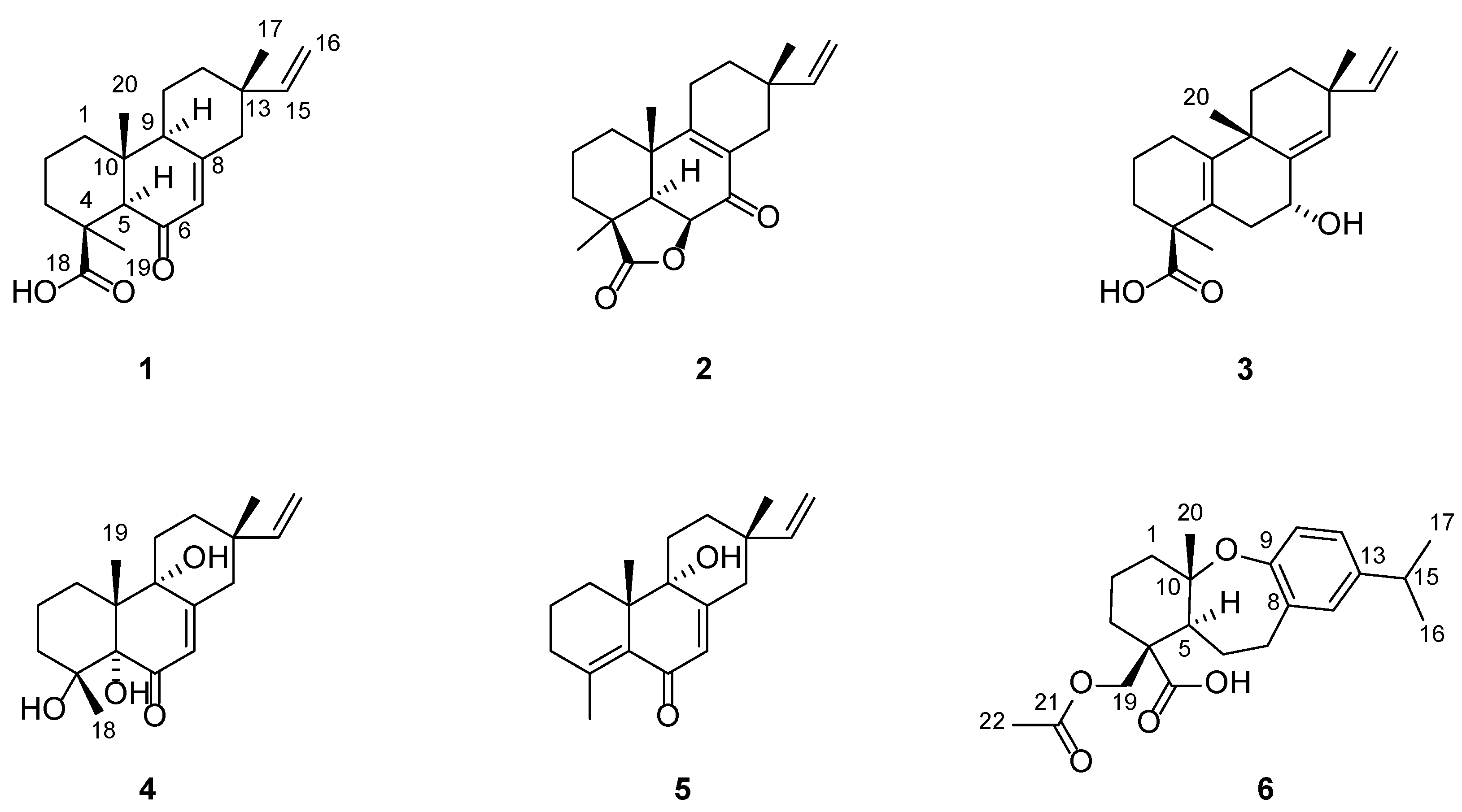
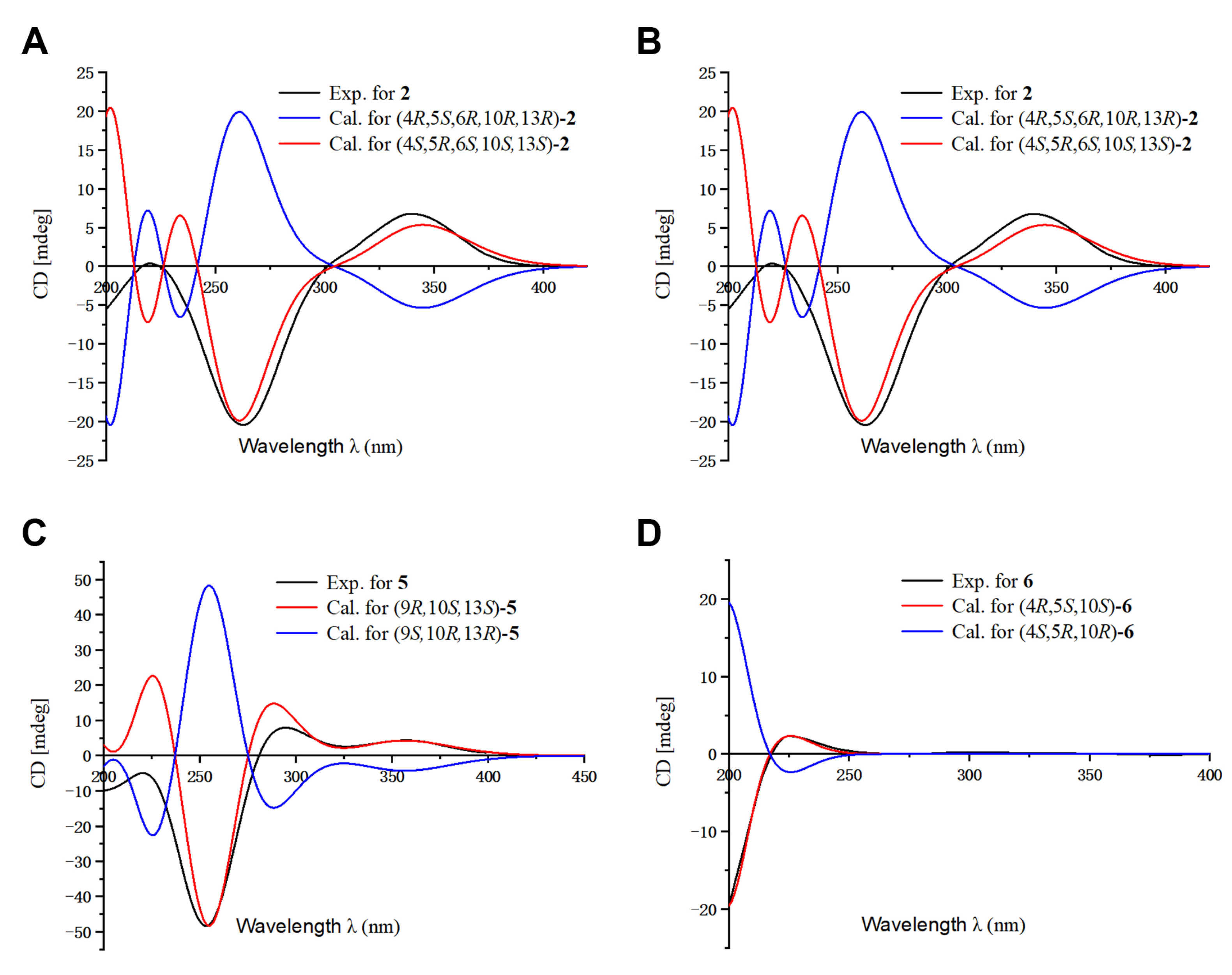
2.1. Structure Elucidation of the Compounds
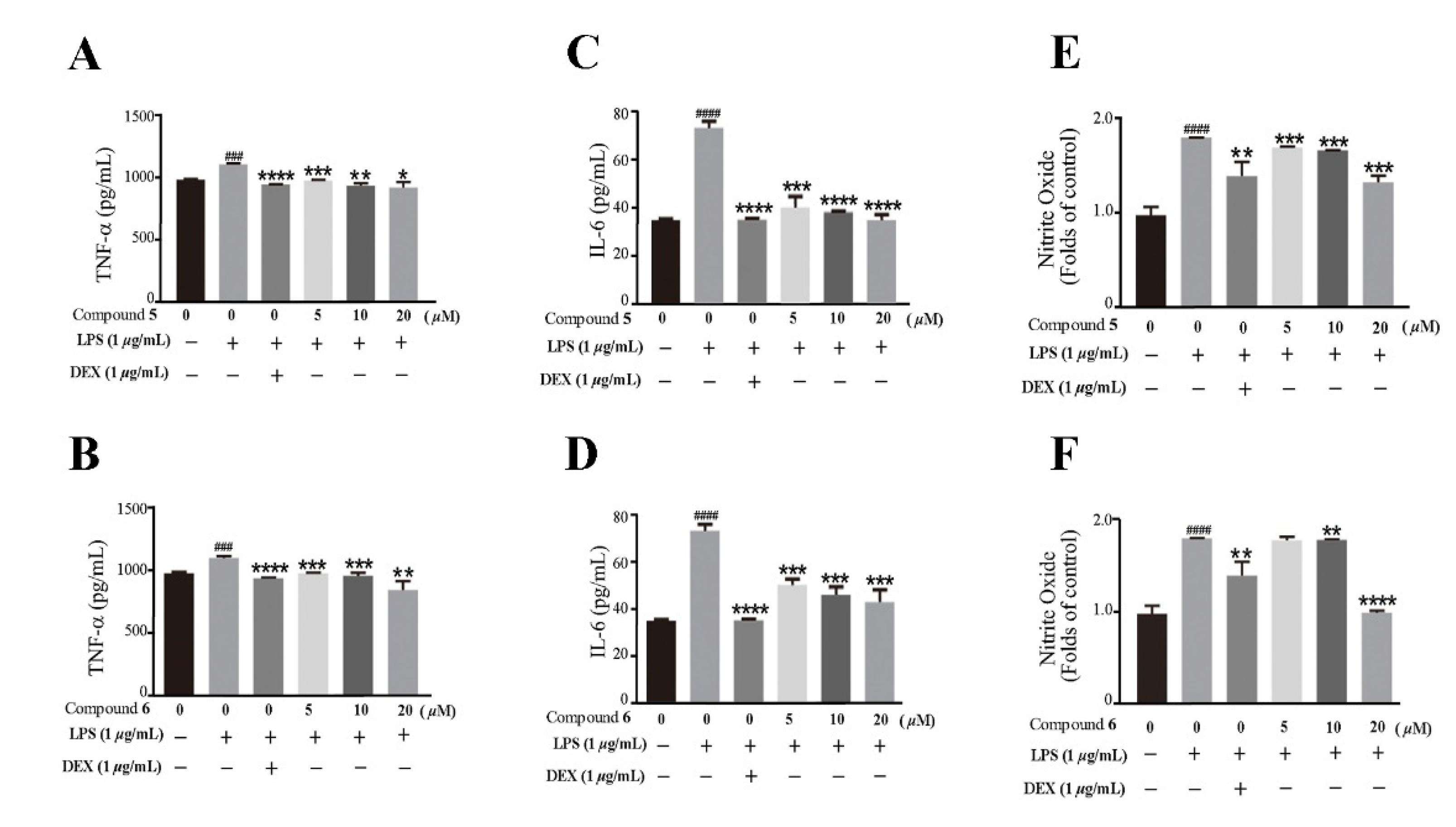
2.2. Biological Evaluation
3. Experimental Section
3.1. General Procedures
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Compound Characterization Data
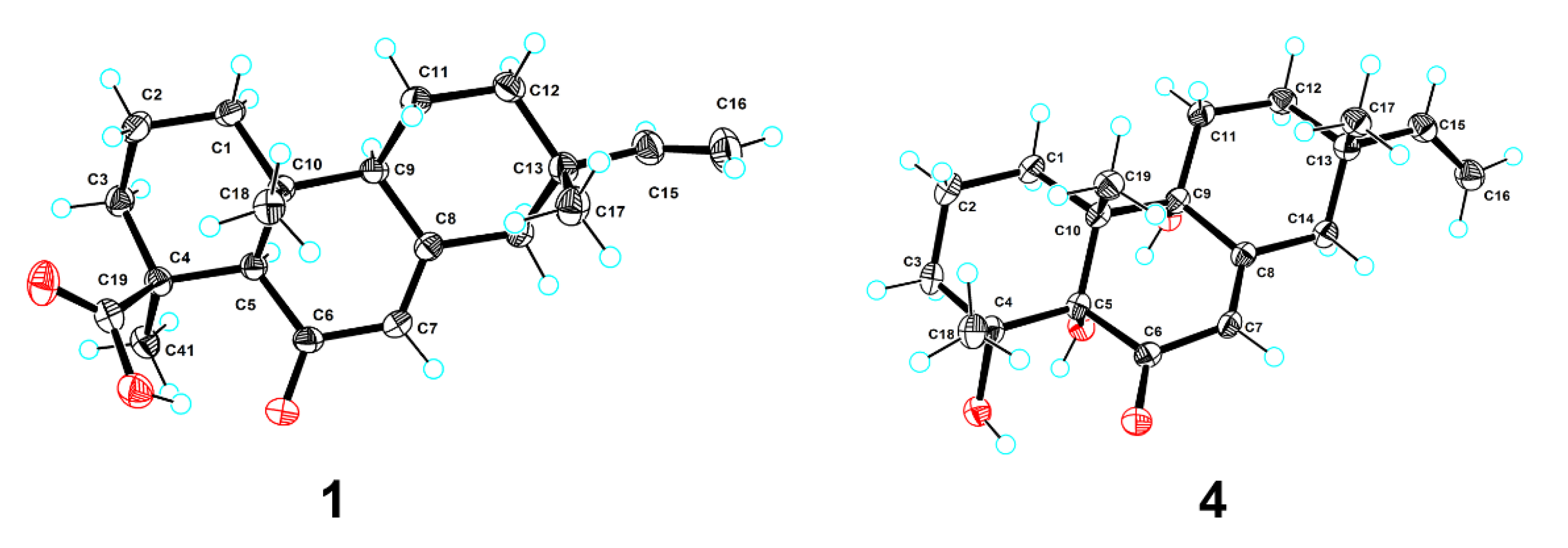
3.5. Crystal Structure Determination of 1 and 4
3.6. ECD Calculations
3.7. NMR Calculations of 5
3.8. Anti-Inflammatory Activity
3.8.1. Cell Culture
3.8.2. Cell Viability Assay
3.8.3. ELISA of TNF-α and IL-6
3.8.4. Determination of Nitrite Oxide
3.8.5. Western Blot
3.8.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Pang, Y.; Wang, D.; Fan, Z.; Chen, X.; Yu, F.; Hu, X.; Wang, K.; Yuan, L. Blumea balsamifera—A phytochemical and pharmacological review. Molecules 2014, 19, 9453–9477. [Google Scholar] [CrossRef] [Green Version]
- Fan, Z.W.; Pang, Y.X.; Wang, K.; Yu, F.L.; Wang, D.; Yang, Q.; Ma, Q.S.; Li, X.T.; Zou, J.; Zhang, W.Q.; et al. Blumea balsamifera oil for the acceleration of healing of burn injuries. Molecules 2015, 20, 17166–17179. [Google Scholar] [CrossRef] [Green Version]
- Cordero, C.S.; Ligsay, A.; Alejandro, G.J.D. Ethnobotanical documentation of medicinal plants used by the Ati tribe in Malay, Aklan, Philippines. J. Complement. Med. Res. 2020, 11, 170–198. [Google Scholar] [CrossRef]
- Chinese Pharmacopeia Commission. Pharmacopoeia of the People’s Republic of China; China Medical Science Press: Beijing, China, 2020; p. 90. [Google Scholar]
- Chen, M.; Jin, H.Z.; Zhang, W.D.; Yan, S.K.; Shen, Y.H. Chemical constituents of plants from the genus Blumea. Chem. Biodivers. 2009, 6, 809–817. [Google Scholar] [CrossRef]
- Pang, Y.; Zhang, Y.; Huang, L.; Xu, L.; Wang, K.; Wang, D.; Guan, L.; Zhang, Y.; Yu, F.; Chen, Z.; et al. Effects and mechanisms of total flavonoids from Blumea balsamifera (L.) DC. on skin wound in rats. Int. J. Mol. Sci. 2017, 18, 2766. [Google Scholar] [CrossRef] [Green Version]
- Norikura, T.; Kojima-Yuasa, A.; Shimizu, M.; Huang, X.; Xu, S.; Kametani, S.; Rho, S.-N.; Kennedy, D.O.; Matsui-Yuasa, I. Anticancer activities and mechanisms of Blumea balsamifera extract in hepatocellular carcinoma cells. Am. J. Chin. Med. 2008, 36, 411–424. [Google Scholar] [CrossRef]
- Yang, H.; Gao, Y.; Long, L.; Cai, Y.; Liao, J.; Peng, J.; Wang, L. Antibacterial effect of Blumea balsamifera (L.) DC. essential oil against Staphylococcus aureus. Arch. Microbiol. 2021, 203, 3981–3988. [Google Scholar] [CrossRef]
- Liao, J.; Xie, X.; Wang, W.; Gao, Y.; Cai, Y.; Peng, J.; Li, T.; Yi, Q.; He, C.; Wang, L. Anti-inflammatory activity of essential oil from leaves of Blumea balsamifera (L.) DC through inhibiting TLR4/NF-kB signaling pathways and NLRP3 inflammasome activation in LPS-induced RAW 264.7 macrophage cells. J. Essent. Oil-Bear. Plants 2021, 24, 160–176. [Google Scholar] [CrossRef]
- Wang, Y.H.; Yu, X.Y. Biological activities and chemical compositions of volatile oil and essential oil from the leaves of Blumea balsamifera. J. Essent. Oil-Bear. Plants 2019, 21, 1511–1531. [Google Scholar] [CrossRef]
- Xiong, Y.; Yi, P.; Li, Y.; Gao, R.; Chen, J.; Hu, Z.; Lou, H.; Du, C.; Zhang, J.; Zhang, Y.; et al. New sesquiterpeniod esters from Blumea balsamifera (L.) DC. and their anti-influenza virus activity. Nat. Prod. Res. 2020, 36, 1151–1160. [Google Scholar] [CrossRef]
- Yuan, Y.; Pang, Y.X.; Yuan, C. Antibacterial Constituents of Ethyl Acetate Extract from Blumea balsamifera (L.) DC. Chin. J. Trop. Crops 2018, 39, 1195–1199. [Google Scholar] [CrossRef]
- Cai, Y.; Liao, J.; Peng, J.; Gao, Y.; Wang, W.; Yi, Q.; Wang, L. Screening of anti-inflammatory substances from Blumea balsamifera (L.) DC.oil and their effects on inflammatory factors. Nat. Prod. Res. Dev. 2021, 33, 402–409. [Google Scholar] [CrossRef]
- Li, X.; Wu, H.; Yang, Y.; Wang, X.; Xu, F. Comparison of anti-inflammatory effects of Blumea balsamifera from different areas of Guizhou. Lishizhen Med. Mater. Med. Res. 2018, 29, 200–201. [Google Scholar] [CrossRef]
- Wang, Y.; Peng, X.; He, Y.; Sun, X.; Ma, X.; Wang, G.; Lan, Y.; Li, Y. Spectrum-effect Relationship of Anti-inflammatory Active Components from Nonvolatile Fraction of Blumea balsamifera Based on Gray Correlation Analysis. Chin. J. Exp. Tradit. Med. Formulae 2019, 25, 180–186. [Google Scholar] [CrossRef]
- Ulubelen, A.; Öksüz, S.; Topcu, G.; Gören, A.C.; Bozok-Johansson, C.; Çelik, C.; Kökdil, G.; Voelter, W. A new antibacterial diterpene from the roots of Salvia caespitosa. Nat. Prod. Lett. 2001, 15, 307–314. [Google Scholar] [CrossRef]
- Hu, C.L.; Xiong, J.; Gao, L.X.; Li, J.; Zeng, H.Q.; Zou, Y.K.; Hu, J.F. Diterpenoids from the shed trunk barks of the endangered plant Pinus dabeshanensis and their PTP1B inhibitory effects. RSC Adv. 2016, 6, 60467–60478. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, L.; Wang, F.; Li, Z.-H.; Dong, Z.-J.; Liu, J.-K. New diterpenes from cultures of the fungus Engleromyces goetzii and their CETP inhibitory activity. Natur. Prod. Bioprosp. 2015, 5, 69–75. [Google Scholar] [CrossRef] [Green Version]
- Ohtsu, H.; Tanaka, R.; Matsunaga, S.; Tokuda, H.; Nishino, H. Anti-tumor-promoting rearranged abietane diterpenes from the leaves of Larix kaempferi. Planta Med. 1999, 65, 664–666. [Google Scholar] [CrossRef]
- Zhang, M.; Chen, M.; Hou, Y.; Fan, C.; Wei, H.; Shi, L.; Ma, G.; Zhang, J. Inflammatory and Cytotoxic Activities of Abietane Terpenoids from Nepeta bracteata Benth. Molecules 2021, 26, 5603. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian, Version 09, Expanding the Limits of Computational Chemistry; Gaussian, Inc.: Wallingford, CT, USA, 2013. [Google Scholar]
- Lodewyk, M.W.; Siebert, M.R.; Tantillo, D.J. Computational prediction of 1H and 13C chemical shifts: A useful tool for natural product, mechanistic, and synthetic organic chemistry. Chem. Rev. 2012, 112, 1839–1862. [Google Scholar] [CrossRef]
- Grimblat, N.; Zanardi, M.M.; Sarotti, A.M. Beyond DP4: An improved probability for the stereochemical assignment of isomeric compounds using quantum chemical calculations of NMR shifts. J. Org. Chem. 2015, 80, 12526–12534. [Google Scholar] [CrossRef] [PubMed]



| No. | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δH | δC | δH | δC | δH | δC | |
| 1 | Ha: 1.93, m Hb: 1.33, dt (13.3, 4.2) | 39.6, t | 1.62, overlap | 29.5, t | Ha: 2.04, m Hb: 1.90, m | 24.5, t |
| 2 | Ha: 1.87, m Hb: 1.53, overlap | 18.9, t | Ha: 1.79, m Hb: 1.62, overlap | 17.8, t | 1.64, m | 20.1, t |
| 3 | Ha: 2.33, d-like (13.3) Hb: 0.96, td (13.3, 3.3) | 39.3, t | Ha: 2.21, m Hb: 1.52, m | 27.8, t | Ha: 2.08, m Hb: 1.45, m | 36.4, t |
| 4 | 43.8, s | 42.3, s | 46.9, s | |||
| 5 | 2.64, s | 64.3, d | 2.17, d (5.8) | 49.0, d | 125.4, s | |
| 6 | 205.9, s | 4.74, d (5.8) | 75.8, d | Ha: 2.41, dd (14.9, 5.5) Hb: 2.02, m | 40.2, t | |
| 7 | 5.92, brs | 125.6, d | 192.1, s | 4.46, ddd (10.6, 5.5, 1.7) | 67.6, d | |
| 8 | 164.7, s | 130.5, s | 145.4, s | |||
| 9 | 2.25, m | 52.3, d | 166.2, s | 40.2, s | ||
| 10 | 41.3, s | 36.3, s | 138.8, s | |||
| 11 | Ha: 1.83, m Hb: 1.53, overlap | 20.9, t | Ha: 2.30, m Hb: 2.29, m | 23.3, t | Ha: 1.58, m Hb: 1.39, m | 31.5, t |
| 12 | Ha: 1.63, m Hb: 1.53, overlap | 35.5, t | Ha: 1.60, overlap Hb: 1.46, ddd, (13.8, 13.4, 7.0) | 33.1, t | 1.55, m | 32.0, t |
| 13 | 37.8, s | 34.6, s | 37.8, s | |||
| 14 | 2.22, brs | 46.2, t | Ha: 2.40, d-like (17.4) Hb: 2.05, d-like (17.4) | 33.7, t | 5.32, brs | 120.3, d |
| 15 | 5.81, dd (17.5, 10.8) | 148.3, d | 5.74, dd, (17.5, 10.7) | 145.9, d | 5.70, dd, (17.3, 10.4) | 146.8, d |
| 16 | Ha: 4.99, dd (17.5, 0.9) Hb: 4.96, dd (10.8, 0.9) | 110.8, t | Ha: 4.95, dd (10.7, 1.1) Hb: 4.89, dd (17.5, 1.1) | 111.6, t | Ha: 4.92, dd (10.4, 1.8) Hb: 4.80, dd (17.3, 1.8) | 112.6, t |
| 17 | 0.91, s | 22.2, q | 0.98, s | 26.3, q | 1.11, s | 29.3, q |
| 18 | 176.6, s | 180.6, s | 182.5, s | |||
| 19 | 1.42, s | 28.5, q | 1.32, s | 24.2, q | 1.26, s | 24.0, q |
| 20 | 0.88, s | 14.8, q | 1.09, s | 27.8, q | 1.22, s | 26.0, q |
| 4-COOH | 12.45, brs | |||||
| No. | 4 | 5 | 6 | |||
|---|---|---|---|---|---|---|
| δH | δC | δH | δC | δH | δC | |
| 1 | Ha: 2.24, dt (13.5, 4.3) Hb: 1.33, overlap | 25.5, t | Ha: 2.13, overlap Hb: 1.48, m | 29.2, t | Ha: 1.92, dd (13.4, 5.6) Hb: 1.63, m | 36.7, t |
| 2 | Ha: 1.70, m Hb: 1.51, m | 18.4, t | Ha: 1.72, m Hb: 1.52, m | 18.5, t | Ha: 1.84, m Hb: 1.76, m | 19.4, t |
| 3 | Ha: 1.79, m Hb: 1.47, m | 34.8, t | Ha: 2.14, m Hb: 2.09, m | 33.9, t | 1.80, overlap | 31.6, t |
| 4 | 74.4, s | 149.4, s | 52.0, s | |||
| 5 | 80.2, s | 133.0, s | 1.97, t-like (5.5) | 53.4, d | ||
| 6 | 198.8, s | 190.8, s | Ha: 1.80, overlap Hb: 1.72, m | 26.2, t | ||
| 7 | 5.73, brd (2.2) * | 123.4, d | 5.83, brd (2.1) * | 128.2, d | 2.62, m | 29.4, t |
| 8 | 163.1, s | 159.4, s | 127.7, s | |||
| 9 | 74.0, s | 73.6, s | 151.6, s | |||
| 10 | 42.9, s | 44.4, s | 86.2, s | |||
| 11 | Ha: 1.86, m Hb: 1.73, m | 27.3, t | Ha: 1.93, dd (13.7, 4.3) Hb: 1.76, m | 25.7, t | 6.66, d (8.1) | 115.4, d |
| 12 | Ha: 1.95, dt (13.0, 3.9) Hb: 1.33, overlap | 31.5, t | Ha: 1.82, dt (13.1, 4.3) Hb: 1.41, m | 31.8, t | 6.93, dd (8.1, 2.3) | 125.2, d |
| 13 | 38.2, s | 37.2, s | 141.5, s | |||
| 14 | Ha: 2.80, dd (14.5,1.2) * Hb: 1.99, dd (14.5, 3.0) * | 43.7, t | Ha: 2.60, dd (15.2, 1.4) * Hb: 2.06, dd (15.2, 3.0) * | 42.9, t | 6.90, d (2.3) | 128.4, d |
| 15 | 5.85, dd, (17.4, 10.7) | 148.8, d | 5.83, dd, (17.4, 10.7) | 148.7, d | 2.81, h (6.9) | 33.4, d |
| 16 | Ha: 5.00, dd (17.4, 1.0) Hb: 4.95, dd (10.7, 1.0) | 110.5, t | Ha: 5.00, dd (17.4, 1.0) Hb: 4.95, dd (10.7, 1.0) | 110.6, t | 1.21, d (6.9) | 24.4, q |
| 17 | 0.95, s | 22.0, q | 0.94, s | 21.9, q | 1.21, d (6.9) | 24.4, q |
| 18 | 1.67, s | 26.2, q | 1.97, s | 22.7, q | 177.1, s | |
| 19 | 1.10, s | 20.1, q | 1.23, s | 25.2, q | Ha: 4.44, d (11.9) Hb: 4.29, d (11.9) | 64.7, t |
| 20 | 1.53, s | 22.3, q | ||||
| 21 | 170.9, s | |||||
| 22 | 2.04, s | 20.8, q | ||||
| 5-OH | 4.82, brs | |||||
| 9-OH | 4.83, d (2.7) | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, X.-L.; Wang, D.-W.; Liu, Y.-Q.; Cheng, Y.-X. Diterpenoids from Blumea balsamifera and Their Anti-Inflammatory Activities. Molecules 2022, 27, 2890. https://doi.org/10.3390/molecules27092890
Huang X-L, Wang D-W, Liu Y-Q, Cheng Y-X. Diterpenoids from Blumea balsamifera and Their Anti-Inflammatory Activities. Molecules. 2022; 27(9):2890. https://doi.org/10.3390/molecules27092890
Chicago/Turabian StyleHuang, Xiao-Ling, Dai-Wei Wang, Ying-Qian Liu, and Yong-Xian Cheng. 2022. "Diterpenoids from Blumea balsamifera and Their Anti-Inflammatory Activities" Molecules 27, no. 9: 2890. https://doi.org/10.3390/molecules27092890






