Exploration of Flavonoids as Lead Compounds against Ewing Sarcoma through Molecular Docking, Pharmacogenomics Analysis, and Molecular Dynamics Simulations
Abstract
:1. Introduction
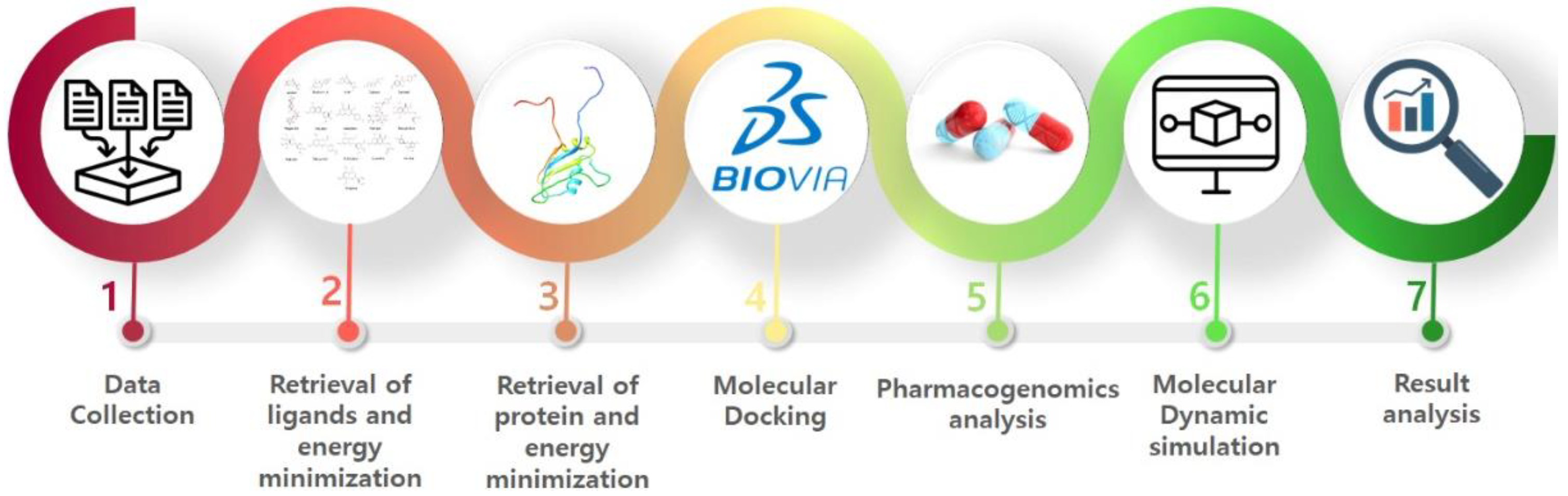
2. In Silico Methodology
2.1. Protein Structure Retrieval
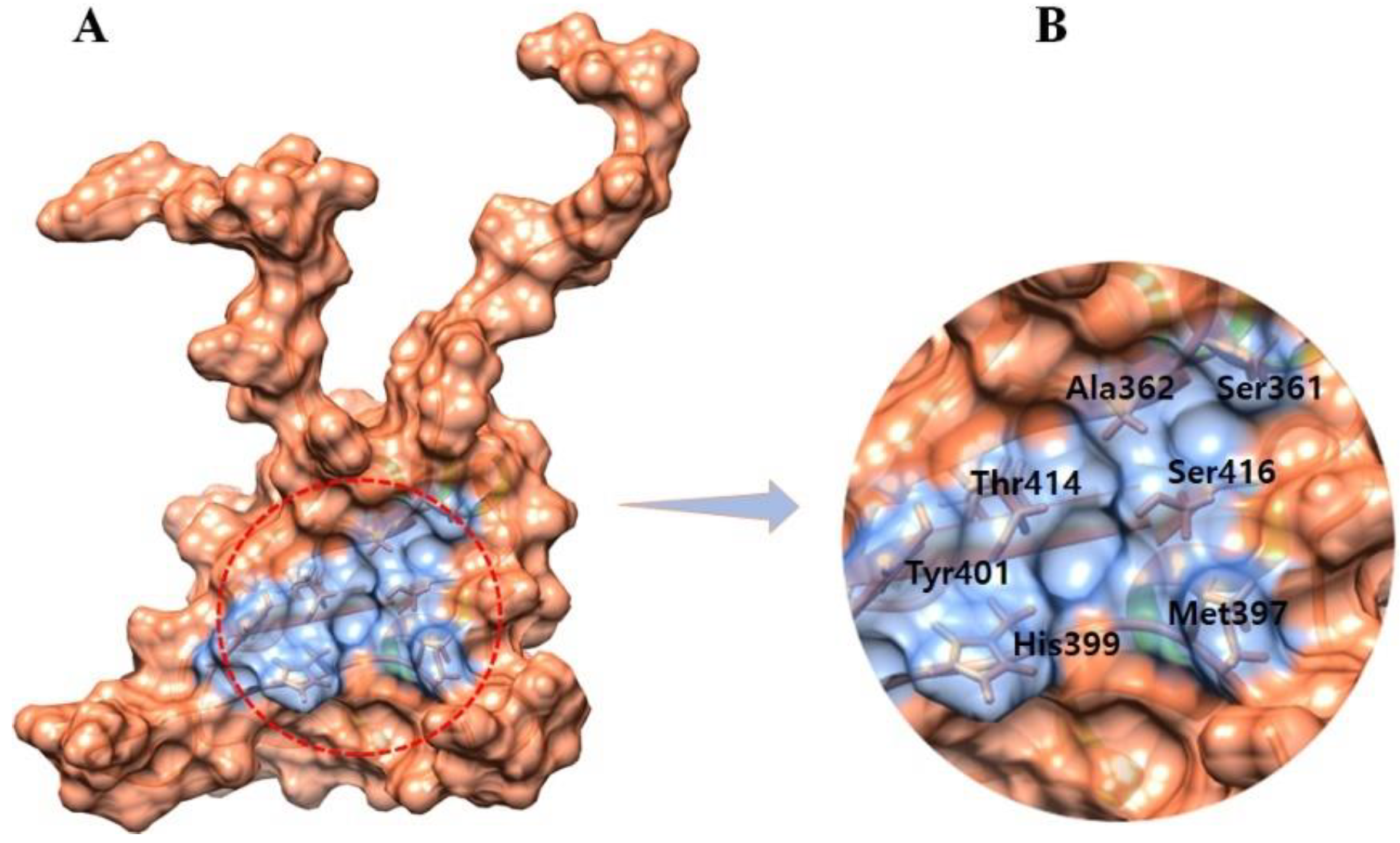
2.2. Selection of the Binding Pocket
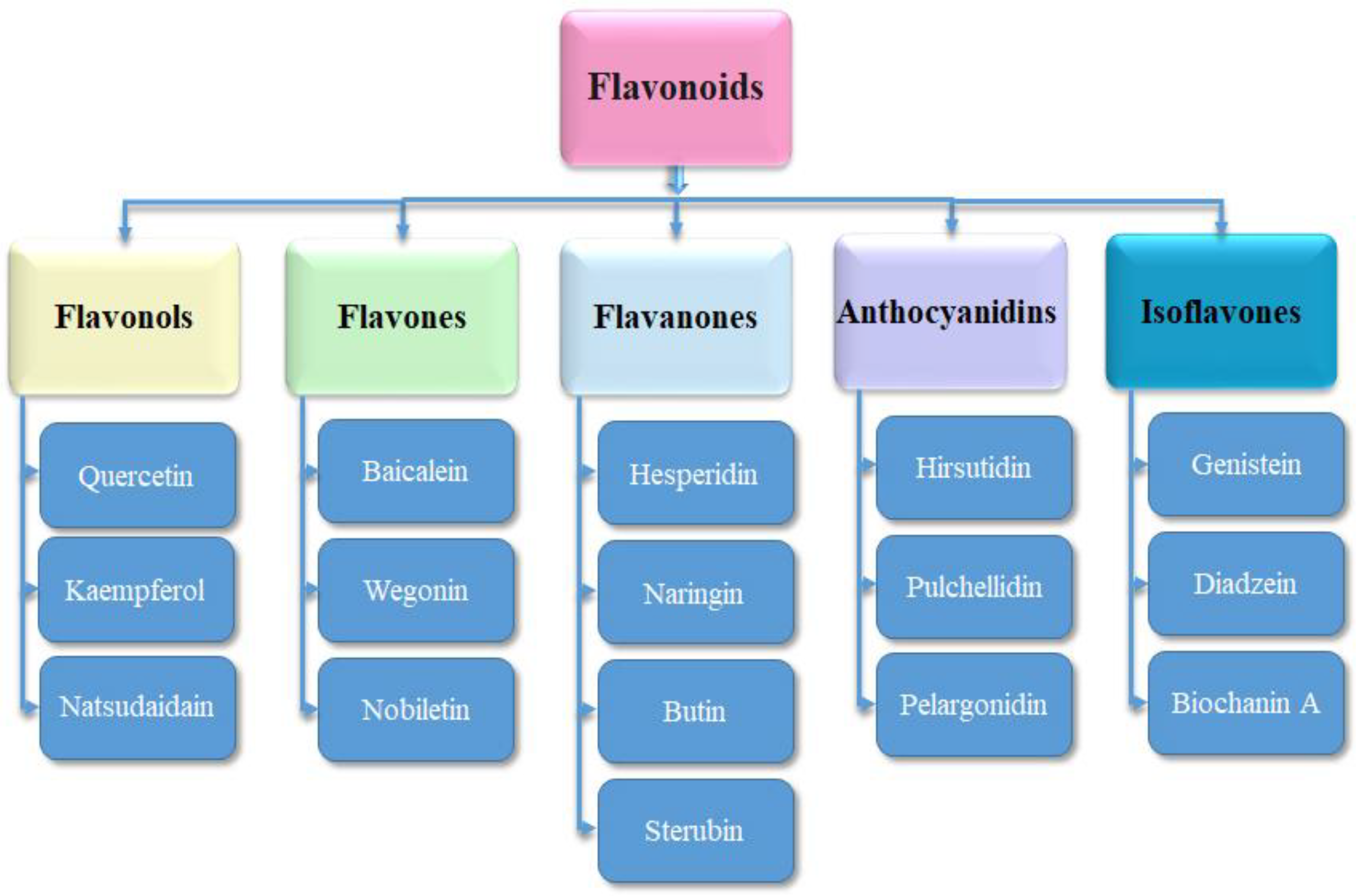
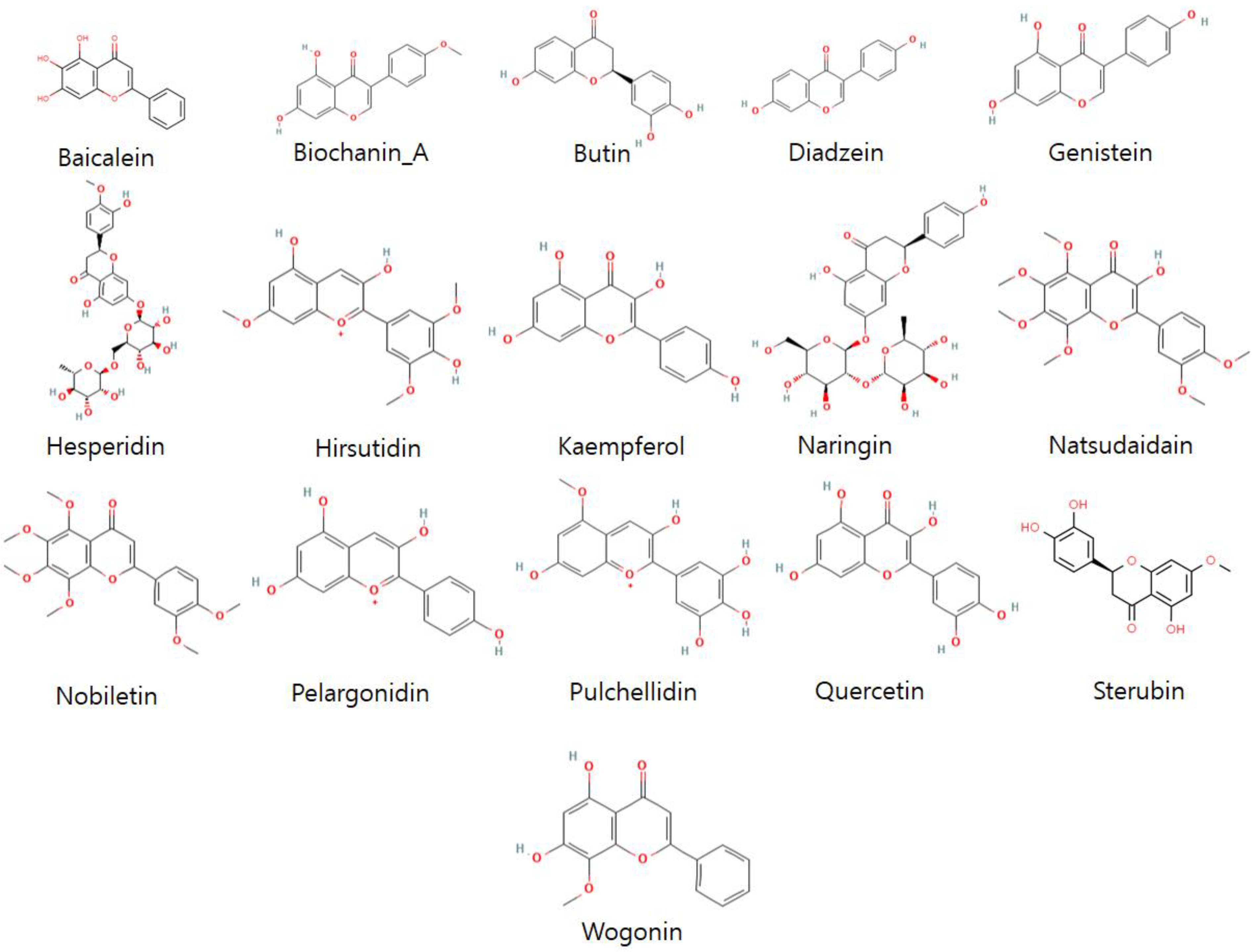
2.3. Ligand’s Preparation
2.4. Molecular Docking
2.5. Pharmacogenomics Analysis
2.6. Molecular Dynamics Simulations
3. Results and Discussion
3.1. Structural Analysis of the EWS Protein
3.2. The Binding Pocket Analysis
3.3. Ligand’s Preparation
3.4. Molecular Docking Analysis
3.5. Pharmacogenomics Analysis
3.6. Hydrogen Bond Interaction Analysis
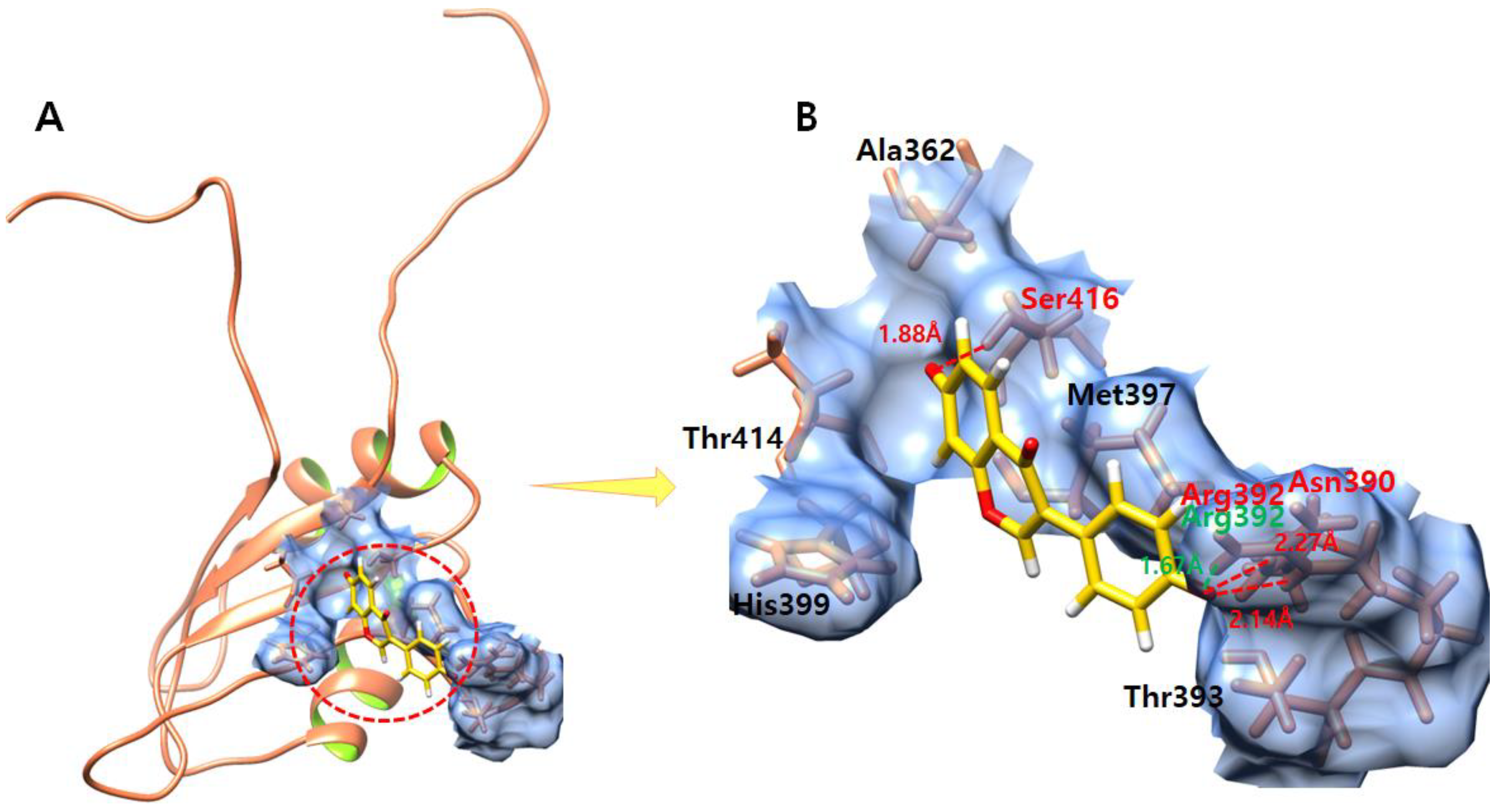
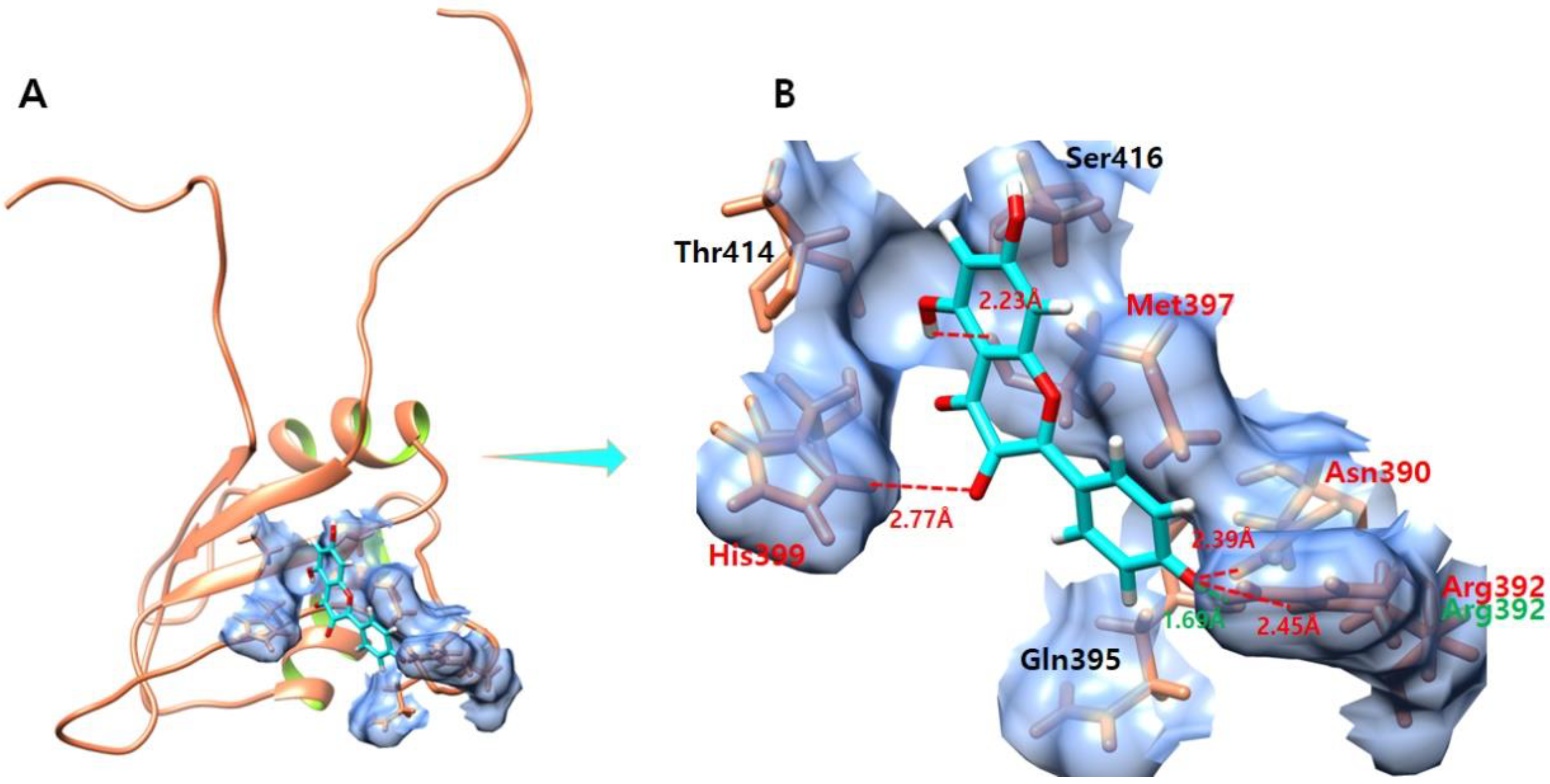
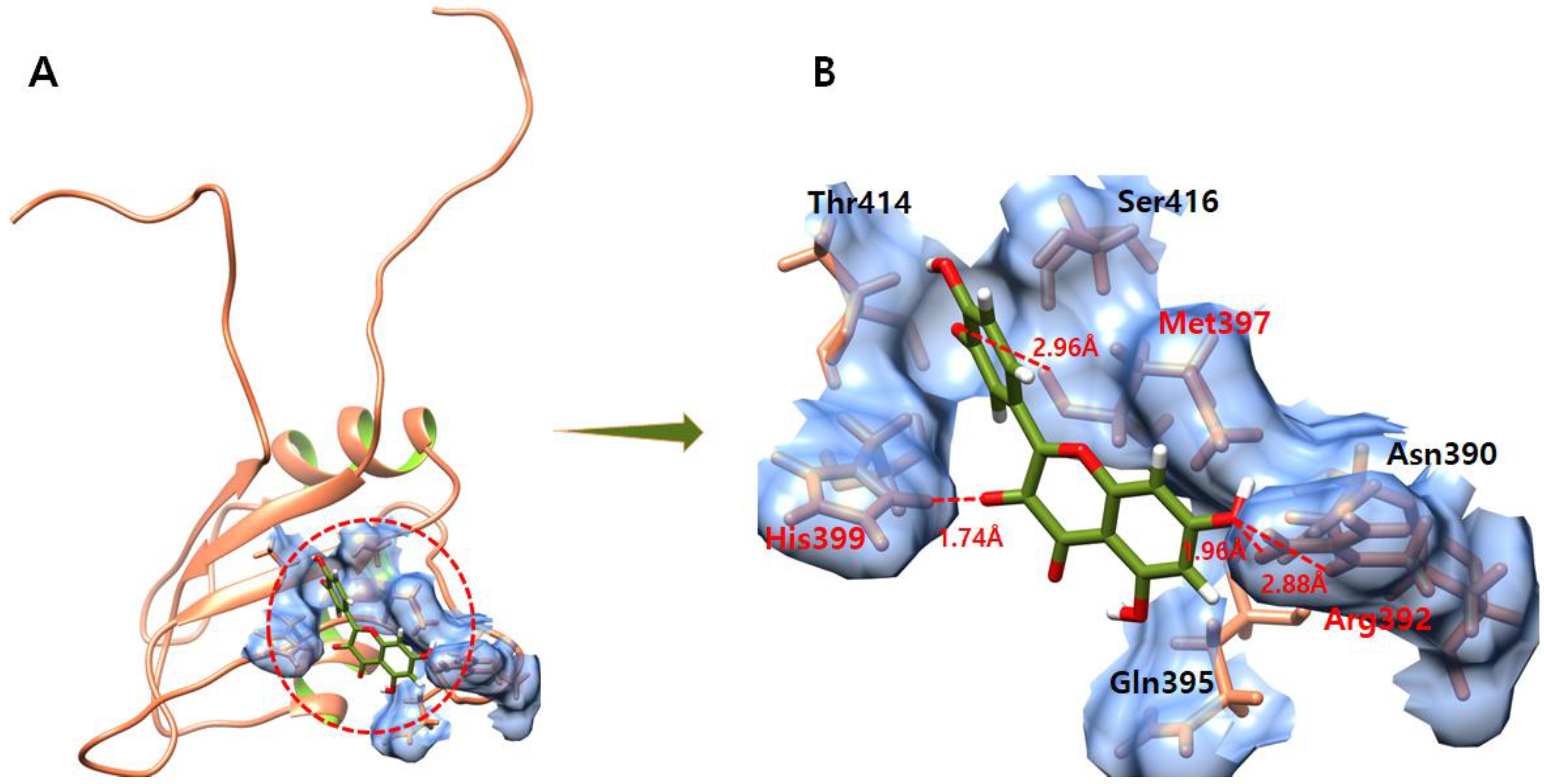
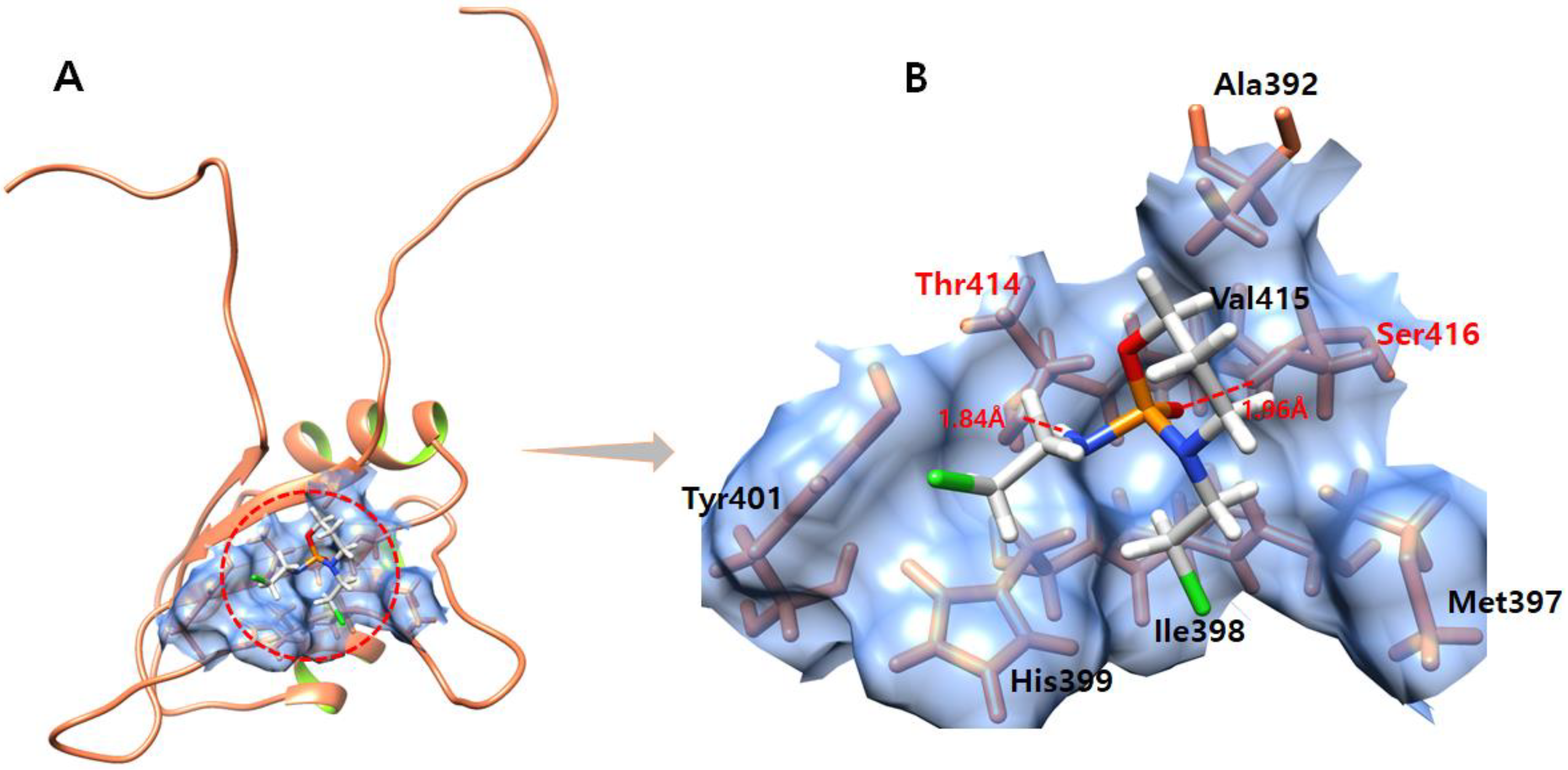
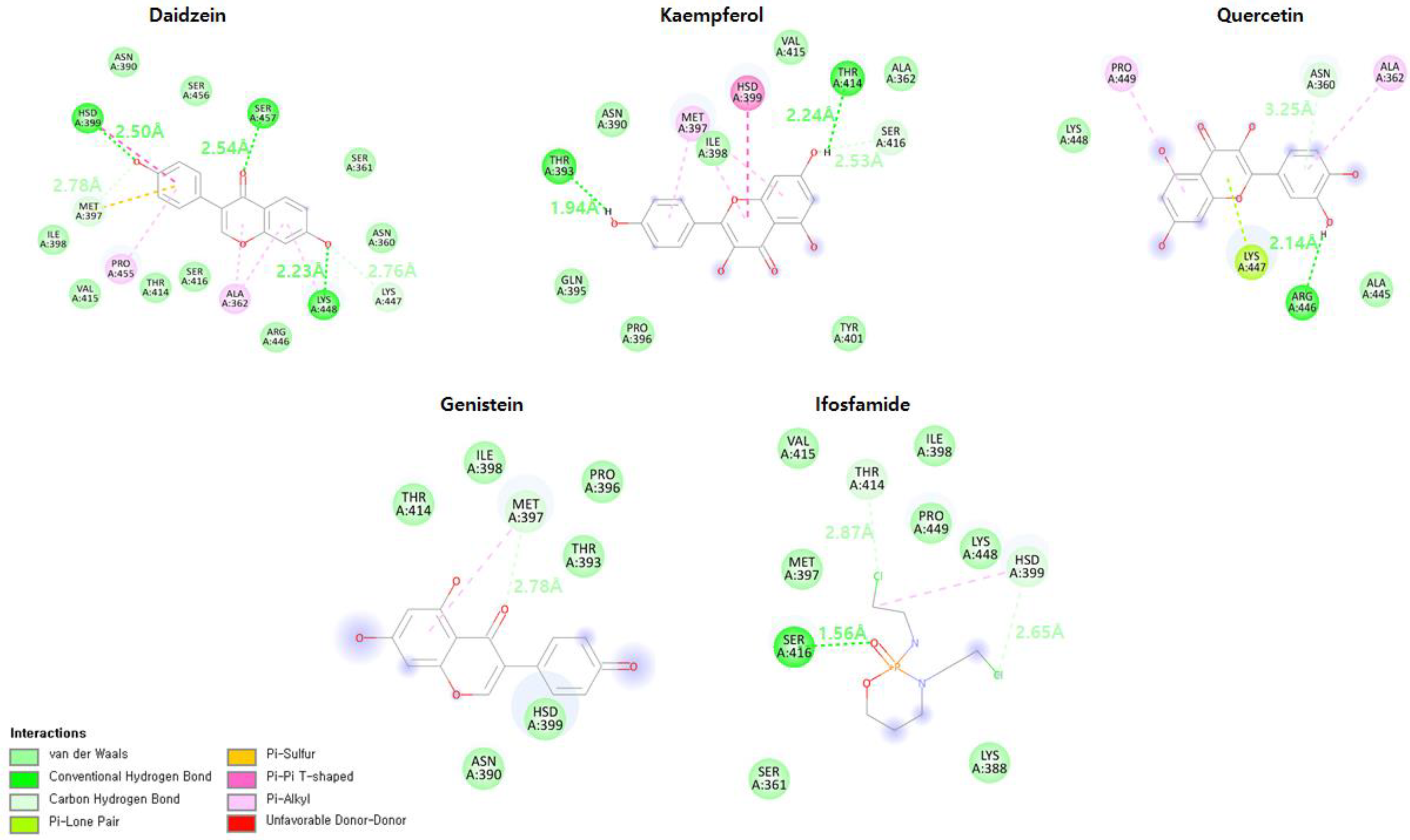
3.7. Daidzein
3.8. Kaempferol
3.9. Genistein
3.10. Quercetin
3.11. Ifosfamide
3.12. Molecular Dynamics Simulations
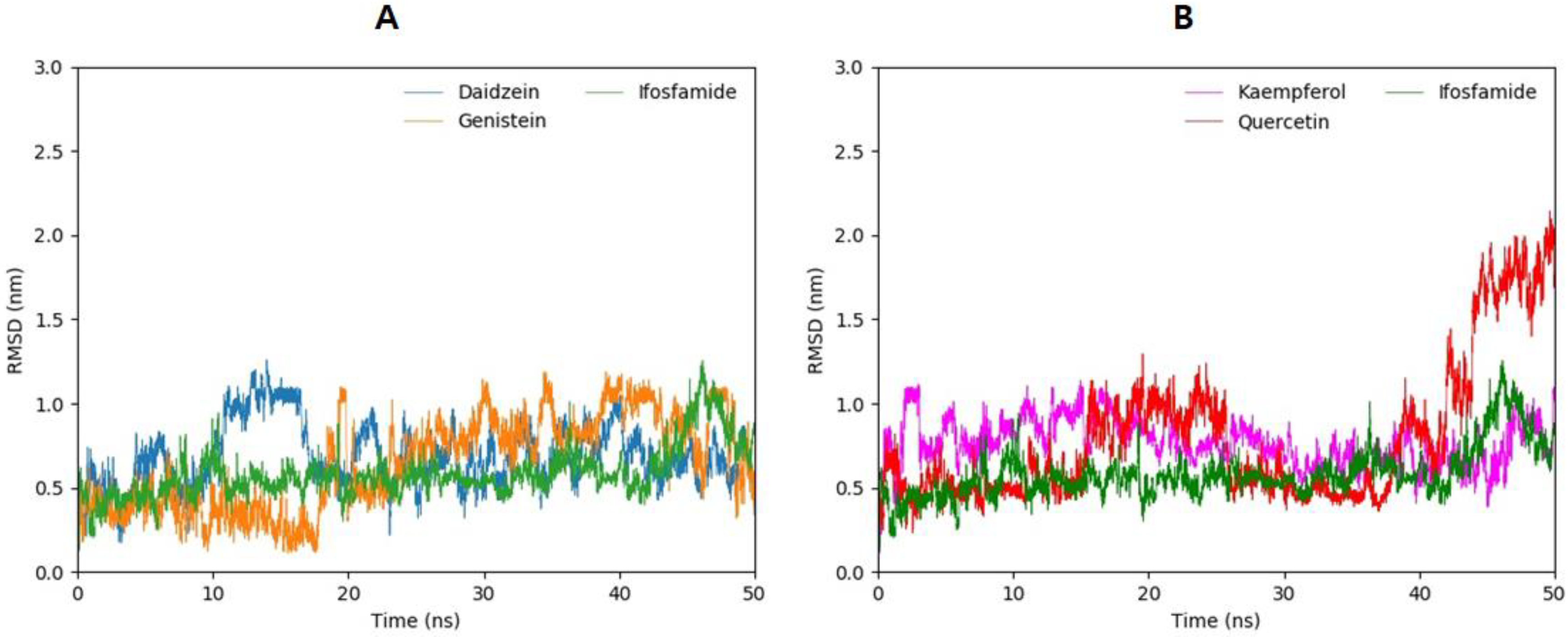
3.13. Root-Mean-Square-Deviation
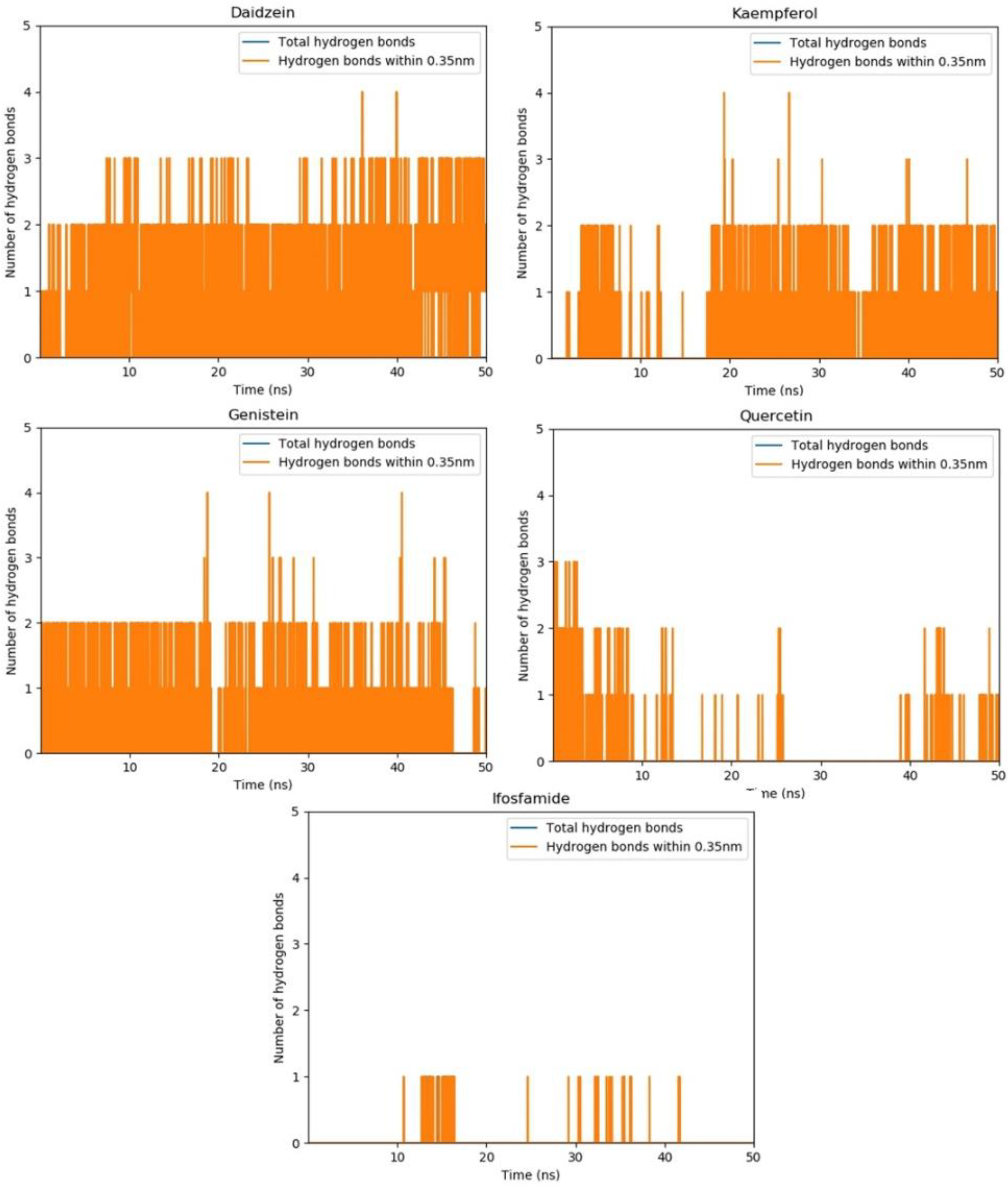
3.14. Hydrogen Bond Plot Analysis
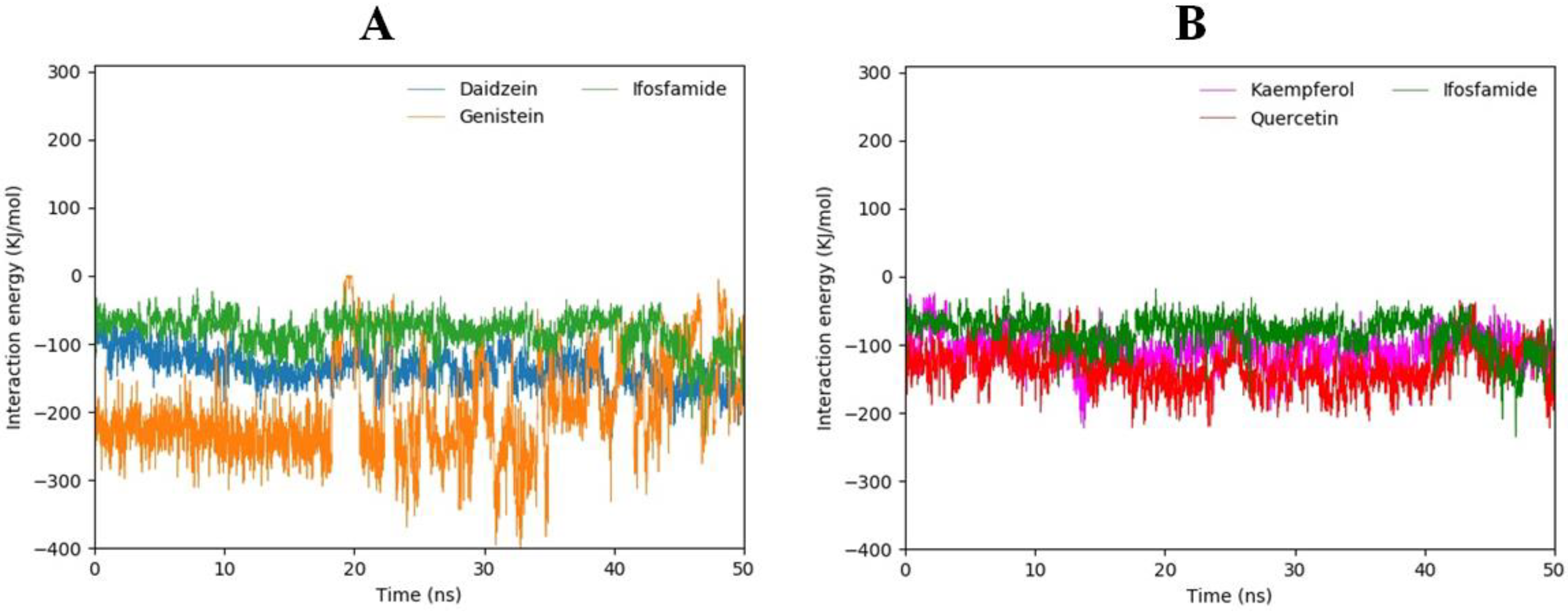
3.15. Interaction Energy Analysis
3.16. Binding Mode Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Li, W.; Dong, S.; Lin, Y.; Wu, H.; Chen, M.; Qin, C.; Li, K.; Zhang, J.; Tang, Z.-R.; Wang, H.; et al. A tool for predicting overall survival in patients with Ewing sarcoma: A multicenter retrospective study. BMC Cancer 2022, 22, 914. [Google Scholar] [CrossRef] [PubMed]
- Twardziok, M.; Kleinsimon, S.; Rolff, J.; Jäger, S.; Eggert, A.; Seifert, G.; Delebinski, C.I. Multiple active compounds from Viscum album L. synergistically converge to promote apoptosis in Ewing sarcoma. PLoS ONE 2016, 11, e0159749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Zhao, L.; Wang, Y.; Liu, X.; Wu, X.; Li, Y. Primary intracranial Ewing sarcoma/peripheral primitive neuroectodermal tumor mimicking meningioma: A case report and literature review. Front. Oncol. 2020, 10, 528073. [Google Scholar] [CrossRef] [PubMed]
- Mazzocca, A.; Paternostro, F.; Minelli, A.; Silletta, M.; Greco, C.; Valeri, S.; Ramella, S.; Tonini, G.; Vincenzi, B. How I Treat Localized Soft Tissue Sarcomas: Update on Diagnosis, Risk Stratification, and Treatment. Chemotherapy 2022, 67, 234–247. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Nguyen, P.T.; Shim, H.S.; Hyeon, S.J.; Im, H.; Choi, M.-H.; Chung, S.; Kowall, N.W.; Lee, S.B.; Ryu, H. EWSR1, a multifunctional protein, regulates cellular function and aging via genetic and epigenetic pathways. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 2019, 1865, 1938–1945. [Google Scholar] [CrossRef]
- Jiang, W.; Wu, T.; Shi, X.; Xu, J. Overexpression of EWSR1 (Ewing sarcoma breakpoint region 1/EWS RNA binding protein 1) predicts poor survival in patients with hepatocellular carcinoma. Bioengineered 2021, 12, 7941–7949. [Google Scholar] [CrossRef]
- Thway, K.; Noujaim, J.; Zaidi, S.; Miah, A.B.; Benson, C.; Messiou, C.; Jones, R.L.; Fisher, C. Desmoplastic small round cell tumor: Pathology, genetics, and potential therapeutic strategies. Int. J. Surg. Pathol. 2016, 24, 672–684. [Google Scholar] [CrossRef]
- Bale, T.A.; Oviedo, A.; Kozakewich, H.; Giannini, C.; Davineni, P.K.; Ligon, K.; Alexandrescu, S. Intracranial myxoid mesenchymal tumors with EWSR1–CREB family gene fusions: Myxoid variant of angiomatoid fibrous histiocytoma or novel entity? Brain Pathol. 2018, 28, 183–191. [Google Scholar] [CrossRef]
- Baranov, E.; Black, M.A.; Fletcher, C.D.; Charville, G.W.; Hornick, J.L. Nuclear expression of DDIT3 distinguishes high-grade myxoid liposarcoma from other round cell sarcomas. Mod. Pathol. 2021, 34, 1367–1372. [Google Scholar] [CrossRef]
- Wang, J.; Thway, K. Clear cell sarcoma-like tumor of the gastrointestinal tract: An evolving entity. Arch. Pathol. Lab. Med. 2015, 139, 407–412. [Google Scholar] [CrossRef]
- Paronetto, M.P. Ewing sarcoma protein: A key player in human cancer. Int. J. Cell Biol. 2013, 2013, 642853. [Google Scholar] [CrossRef] [Green Version]
- Mackintosh, C.; Madoz-Gúrpide, J.J. Mining sarcomas by proteomics approaches: Ewing sarcoma on the spotlight. Recent Pat. Biotechnol. 2013, 7, 98–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassan, M.; Yasir, M.; Shahzadi, S.; Kloczkowski, A.J. Exploration of Potential Ewing Sarcoma Drugs from FDA-Approved Pharmaceuticals through Computational Drug Repositioning, Pharmacogenomics, Molecular Docking, and MD Simulation Studies. ACS Omega 2022, 7, 19243–19260. [Google Scholar] [CrossRef] [PubMed]
- Karimi, A.; Majlesi, M.; Rafieian-Kopaei, M. Herbal versus synthetic drugs; beliefs and facts. J. Nephropharmacol. 2015, 4, 27. [Google Scholar]
- Havsteen, B.H. The biochemistry and medical significance of the flavonoids. Pharmacol. Ther. 2002, 96, 67–202. [Google Scholar] [PubMed]
- Nageen, B.; Sarfraz, I.; Rasul, A.; Hussain, G.; Rukhsar, F.; Irshad, S.; Riaz, A.; Selamoglu, Z.; Ali, M. Eupatilin: A natural pharmacologically active flavone compound with its wide range applications. J. Asian Nat. Prod. Res. 2020, 22, 1–16. [Google Scholar] [CrossRef]
- Ginwala, R.; Bhavsar, R.; Chigbu, D.G.I.; Jain, P.; Khan, Z.K. Potential role of flavonoids in treating chronic inflammatory diseases with a special focus on the anti-inflammatory activity of apigenin. Antioxidants 2019, 8, 35. [Google Scholar] [CrossRef] [Green Version]
- Kopustinskiene, D.M.; Jakstas, V.; Savickas, A.; Bernatoniene, J. Flavonoids as anticancer agents. Nutrients 2020, 12, 457. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Studio, D.J.A. Discovery Studio. 2008. Available online: https://www.researchgate.net/profile/Tanweer-Alam-2/post/hi_can_somebody_plz_tell_me_how_to_import_a_database_into_Discovery_Studio_for_a_3D_database_search/attachment/59d63bb879197b8077998bbd/AS%3A412232203685889%401475295224962/download/ds-overview-20.pdf (accessed on 15 October 2022).
- Stank, A.; Kokh, D.B.; Fuller, J.C.; Wade, R.C. Protein binding pocket dynamics. Acc. Chem. Res. 2016, 49, 809–815. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.T.H.; Orhan, I.; Şenol, F.; Kartal, M.; Şener, B.; Dvorská, M.; Šmejkal, K.; Šlapetová, T. Cholinesterase inhibitory activities of some flavonoid derivatives and chosen xanthone and their molecular docking studies. Chem.-Biol. Interact. 2009, 181, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Zanoaga, O.; Braicu, C.; Jurj, A.; Rusu, A.; Buiga, R.; Berindan-Neagoe, I. Progress in research on the role of flavonoids in lung cancer. Int. J. Mol. Sci. 2019, 20, 4291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Zhang, Z.; Dai, E.N.; Tian, J.X.; Xin, J.Z.; Xu, L. Modeling osteosarcoma progression by measuring the connectivity dynamics using an inference of multiple differential modules algorithm. Mol. Med. Rep. 2017, 16, 1047–1054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Wei, Q.; An, R.; Zhang, H.-M.; Shen, J.-Y.; Qin, X.-Y.; Han, X.-L.; Li, J.; Li, X.-W.; Gao, X.-M.; et al. Anti-osteoporosis effect of Semen Cuscutae in ovariectomized mice through inhibition of bone resorption by osteoclasts. J. Ethnopharmacol. 2022, 285, 114834. [Google Scholar] [CrossRef]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef] [Green Version]
- Pasha, A.; Vignoli, M.; Subbiani, A.; Nocentini, A.; Selleri, S.; Gratteri, P.; Dabraio, A.; Casini, T.; Filippi, L.; Fotzi, I.; et al. β3-Adrenoreceptor activity limits apigenin efficacy in Ewing sarcoma cells: A dual approach to prevent cell survival. Int. J. Mol. Sci. 2019, 20, 2149. [Google Scholar] [CrossRef] [Green Version]
- Shanmugavadivu, A.; Balagangadharan, K.; Selvamurugan, N. Angiogenic and osteogenic effects of flavonoids in bone regeneration. Biotechnol. Bioeng. 2022, 119, 2313–2330. [Google Scholar] [CrossRef]
- Kristo, A.S.; Klimis-Zacas, D.; Sikalidis, A. Protective role of dietary berries in cancer. Antioxidants 2016, 5, 37. [Google Scholar] [CrossRef] [Green Version]
- Yuan, S.; Chan, H.S.; Hu, Z. Using PyMOL as a platform for computational drug design. WIREs Comput. Mol. Sci. 2017, 7, e1298. [Google Scholar] [CrossRef]
- Ozaki, T. Diagnosis and treatment of Ewing sarcoma of the bone: A review article. J. Orthop. Sci. Off. J. Jpn. Orthop. Assoc. 2015, 20, 250–263. [Google Scholar] [CrossRef] [Green Version]
- Hassan, M.; Abbasi, M.A.; Siddiqui, S.Z.; Hussain, G.; Shah, S.A.A.; Shahid, M.; Seo, S.-Y. Exploration of synthetic multifunctional amides as new therapeutic agents for Alzheimer’s disease through enzyme inhibition, chemoinformatic properties, molecular docking and dynamic simulation insights. J. Theor. Biol. 2018, 458, 169–183. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Kohli, D.; Chaturvedi, S.; Sharma, S. Molecular Modelling Studies of Some Substitued 2-Butylbenzimidazoles Angiotensin ii Receptor a Ntagonists as Antihypertensive Agents. Dig. J. Nanomater. Biostruct. 2009, 4, 843–856. [Google Scholar]
- Park, J.-Y.; Lee, Y.; Lee, H.J.; Kwon, Y.-S.; Chun, W. In silico screening of GABA aminotransferase inhibitors from the constituents of Valeriana officinalis by molecular docking and molecular dynamics simulation study. J. Mol. Model. 2020, 26, 228. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.; van der Spoel, D.; van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Gao, Y.; Dong, Y.; Cheng, P.; Chen, A.; Huang, H. Flavonoids active against osteosarcoma: A review of the molecular mechanisms involved. Curr. Pharm. Des. 2017, 23, 1993–2001. [Google Scholar] [CrossRef] [PubMed]
- Becquemont, L. Pharmacogenomics of adverse drug reactions: Practical applications and perspectives. Pharmacogenomics 2009, 10, 961–969. [Google Scholar] [CrossRef]
- Morrissey, C.; True, L.D.; Roudier, M.P.; Coleman, I.M.; Hawley, S.; Nelson, P.S.; Coleman, R.; Wang, Y.-C.; Corey, E.; Lange, P.H.; et al. Differential expression of angiogenesis associated genes in prostate cancer bone, liver and lymph node metastases. Clin. Exp. Metastasis 2008, 25, 377–388. [Google Scholar] [CrossRef]
- Amary, F.; Markert, E.; Berisha, F.; Ye, H.; Gerrand, C.; Cool, P.; Tirabosco, R.; Lindsay, D.; Pillay, N.; O’Donnell, P.; et al. FOS expression in osteoid osteoma and osteoblastoma: A valuable ancillary diagnostic tool. Am. J. Surg. Pathol. 2019, 43, 1661–1667. [Google Scholar] [CrossRef]
- Selleri, L.; Hermanson, G.G.; Eubanks, J.H.; Lewis, K.A.; Evans, G.A. Molecular localization of the t (11; 22)(q24; q12) translocation of Ewing sarcoma by chromosomal in situ suppression hybridization. Proc. Natl. Acad. Sci. USA 1991, 88, 887–891. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Zhang, Z.; Zheng, L.; You, C.; Chi, H.; Zhang, T.; Xu, G. Calcium-sensing receptor activating ERK1/2 and PI3K-Akt pathways to induce the proliferation of osteosarcoma cells. Clin. Exp. Pharmacol. Physiol. 2020, 47, 517–519. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-C.; Hsiao, Y.-M.; Wu, M.-F.; Ou, C.-C.; Lin, Y.-W.; Lue, K.-H.; Ko, J.-L. Interruption of lung cancer cell migration and proliferation by fungal immunomodulatory protein FIP-fve from Flammulina velutipes. J. Agric. Food Chem. 2013, 61, 12044–12052. [Google Scholar] [CrossRef] [PubMed]
- Liao, P.; Wang, W.; Li, Y.; Wang, R.; Jin, J.; Pang, W.; Chen, Y.; Shen, M.; Wang, X.; Jiang, D.; et al. Correction: Palmitoylated SCP1 is targeted to the plasma membrane and negatively regulates angiogenesis. Elife 2017, 6, e32342. [Google Scholar] [CrossRef] [PubMed]
- Saxon, J.A.; Yu, H.; Polosukhin, V.V.; Stathopoulos, G.T.; Gleaves, L.A.; McLoed, A.G.; Massion, P.P.; Yull, F.E.; Zhao, Z.; Blackwell, T.S. p52 expression enhances lung cancer progression. Sci. Rep. 2018, 8, 6078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Almeida Magalhães, T.; Borges, K.S.; de Sousa, G.R.; Brandalise, S.R.; Seidinger, A.L.; Scrideli, C.A.; Oba-Shinjo, S.M.; Yunes, J.A.; Tone, L.G. The TP53 p. R337H mutation is uncommon in a Brazilian cohort of pediatric patients diagnosed with ependymoma. Neurol. Sci. 2020, 41, 691–694. [Google Scholar] [CrossRef]
- Edwards, S.K.; Han, Y.; Liu, Y.; Kreider, B.Z.; Liu, Y.; Grewal, S.; Desai, A.; Baron, J.; Moore, C.R.; Luo, C.; et al. Signaling mechanisms of bortezomib in TRAF3-deficient mouse B lymphoma and human multiple myeloma cells. Leuk. Res. 2016, 41, 85–95. [Google Scholar] [CrossRef] [Green Version]
- Ke, H.-L.; Ke, R.-H.; Li, S.-T.; Li, B.; Lu, H.-T.; Wang, X.-Q. Expression of RACGAP1 in high grade meningiomas: A potential role in cancer progression. J. Neuro-Oncol. 2013, 113, 327–332. [Google Scholar] [CrossRef]
- Zhou, H.; Yi, W.; Li, A.; Wang, B.; Ding, Q.; Xue, L.; Zeng, X.; Feng, Y.; Li, Q.; Wang, T.; et al. Specific Small-Molecule NIR-II Fluorescence Imaging of Osteosarcoma and Lung Metastasis. Adv. Healthc. Mater. 2020, 9, 1901224. [Google Scholar] [CrossRef]
- Ahn, J.; Truesdell, P.; Meens, J.; Kadish, C.; Yang, X.; Boag, A.H.; Craig, A.W. Fer protein-tyrosine kinase promotes lung adenocarcinoma cell invasion and tumor metastasis. Mol. Cancer Res. MCR 2013, 11, 952–963. [Google Scholar] [CrossRef] [Green Version]
- Shekhar, R.; Priyanka, P.; Kumar, P.; Ghosh, T.; Khan, M.M.; Nagarajan, P.; Saxena, S. The microRNAs miR-449a and miR-424 suppress osteosarcoma by targeting cyclin A2 expression. J. Biol. Chem. 2019, 294, 4381–4400. [Google Scholar] [CrossRef] [Green Version]
- Cen, J.; Shen, J.; Wang, X.; Kang, H.; Wang, L.; Sun, L.; Li, Y.; Yu, L. Association between lymphoma prognosis and aberrant methylation of ID4 and ZO-1 in bone marrow and paraffin-embedded lymphoma tissues of treatment-naive patients. Oncol. Rep. 2013, 30, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Liu, Z.; Lu, C.; Wang, Q. Clinical and laboratory features of adult biphenotypic acute leukemia. Asia-Pac. J. Clin. Oncol. 2013, 9, 146–154. [Google Scholar] [CrossRef]
- Chen, L.; Shern, J.F.; Wei, J.S.; Yohe, M.E.; Song, Y.K.; Hurd, L.; Liao, H.; Catchpoole, D.; Skapek, S.X.; Barr, F.G.; et al. Clonality and evolutionary history of rhabdomyosarcoma. PLoS Genet. 2015, 11, e1005075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fürstenau, C.R.; de Souza, I.C.C.; de Oliveira, M.R. Tanshinone I Induces Mitochondrial Protection by a Mechanism Involving the Nrf2/GSH Axis in the Human Neuroblastoma SH-SY5Y Cells Exposed to Methylglyoxal. Neurotox. Res. 2019, 36, 491–502. [Google Scholar] [CrossRef]
- Wan, T.; Shao, J.; Hu, B.; Liu, G.; Luo, P.; Zhou, Y. Prognostic role of HSF1 overexpression in solid tumors: A pooled analysis of 3,159 patients. OncoTargets Ther. 2018, 11, 383–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, E.; Zhang, C.; Polavaram, N.; Liu, F.; Wu, G.; Schroeder, M.A.; Lau, J.S.; Mukhopadhyay, D.; Jiang, S.W.; O’Neill, B.P.; et al. The role of factor inhibiting HIF (FIH-1) in inhibiting HIF-1 transcriptional activity in glioblastoma multiforme. PLoS ONE 2014, 9, e86102. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Jelkmann, W.; Depping, R. Prolyl-4-hydroxylase 2 enhances hypoxia-induced glioblastoma cell death by regulating the gene expression of hypoxia-inducible factor-α. Cell Death Dis. 2014, 5, e1322. [Google Scholar] [CrossRef] [Green Version]
- Obermann, E.C.; Eward, K.L.; Dogan, A.; Paul, E.A.; Loddo, M.; Munson, P.; Williams, G.H.; Stoeber, K. DNA replication licensing in peripheral B-cell lymphoma. J. Pathol. 2005, 205, 318–328. [Google Scholar] [CrossRef] [PubMed]


| No | Flavonoids | CDOCKER Energy (kcal/mol) | CDOCKER Interaction Energy (kcal/mol) |
|---|---|---|---|
| 1 | Daidzein | −48.34 | −52.15 |
| 2 | Kaempferol | −44.60 | −56.40 |
| 3 | Genistein | −39.86 | −45.43 |
| 4 | Quercetin | −30.14 | −39.42 |
| 5 | Pelargonidin | −29.67 | −47.85 |
| 6 | Pulchellidin | −28.66 | −44.59 |
| 7 | Baicalein | −23.63 | −24.15 |
| 8 | Butin | −22.99 | −28.45 |
| 9 | Sterubin | −19.87 | −25.75 |
| 10 | Wogonin | −16.57 | −24.84 |
| 11 | Biochanin_A | −16.55 | −28.38 |
| 12 | Ifosfamide | −16.23 | −23.50 |
| 13 | Hirsutidin | −8.08 | −42.58 |
| 14 | Natsudaidain | −4.84 | −49.36 |
| 15 | Nobiletin | −0.23 | −35.30 |
| 16 | Naringin | 0.92 | −46.53 |
| 17 | Hesperidin | 5.97 | −42.60 |
| Drug | Gene | Interaction Score | Disease | Reference |
|---|---|---|---|---|
| Daidzein | IBSP | 3.25 | Malignant Neoplasm of bone | [39] |
| FOS | 0.65 | Osteosarcoma of bone | [40] | |
| LIF | 0.59 | Ewing Sarcoma | [41] | |
| PIK3CG | 0.07 | Osteosarcoma of bone | [42] | |
| RACGPA1 | 0.05 | Carcinoma of lung | [43] | |
| Kaempferol | CTDSP1 | 0.19 | Neoplasm | [44] |
| NFKB2 | 0.18 | Carcinogenesis | [45] | |
| RELA | 0.14 | Childhood Ependymoma | [46] | |
| NFKB1 | 0.08 | Childhood Lymphoma | [47] | |
| RACGAP1 | 0.05 | Childhood Grade III Meningioma | [48] | |
| Genistein | PAEP | 2.47 | Osteosarcoma of bone | [49] |
| EPHA8 | 1.24 | Adenocarcinoma of lung | [50] | |
| PTGES3 | 0.82 | Osteosarcoma of bone | [24] | |
| CCNA2 | 0.62 | Osteosarcoma of bone | [51] | |
| TJP1 | 0.62 | Childhood Lymphoma | [52] | |
| CEL | 0.62 | Childhood Acute Lymphoblastic Leukemia | [53] | |
| Quercetin | PKN1 | 0.78 | Childhood Rhabdomyosarcoma | [54] |
| GABPA | 0.52 | Childhood Neuroblastoma | [55] | |
| HSF1 | 0.52 | Osteosarcoma of bone | [56] | |
| Baicalein | HIF1AN | 1.65 | Childhood Glioblastoma | [57] |
| EGLN1 | 0.62 | Childhood Glioblastoma | [58] | |
| Wogonin | GMNN | 0.13 | Childhood Burkitt Lymphoma | [59] |
| Sr No | Compound | Interaction energy | Total Energy | |
|---|---|---|---|---|
| Coul-SR | LJ-SR | |||
| 1 | Daidzein | −36.6801 | −99.5852 | −136.2653 |
| 2 | Kaempferol | −26.2828 | −79.1068 | −105.3896 |
| 3 | Genistein | −141.839 | −57.418 | −199.257 |
| 4 | Pazopanib | −47.6918 | −139.997 | −187.6888 |
| 5 | Quercetin | −47.2754 | −87.048 | −134.3234 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yasir, M.; Park, J.; Han, E.-T.; Park, W.S.; Han, J.-H.; Kwon, Y.-S.; Lee, H.-J.; Hassan, M.; Kloczkowski, A.; Chun, W. Exploration of Flavonoids as Lead Compounds against Ewing Sarcoma through Molecular Docking, Pharmacogenomics Analysis, and Molecular Dynamics Simulations. Molecules 2023, 28, 414. https://doi.org/10.3390/molecules28010414
Yasir M, Park J, Han E-T, Park WS, Han J-H, Kwon Y-S, Lee H-J, Hassan M, Kloczkowski A, Chun W. Exploration of Flavonoids as Lead Compounds against Ewing Sarcoma through Molecular Docking, Pharmacogenomics Analysis, and Molecular Dynamics Simulations. Molecules. 2023; 28(1):414. https://doi.org/10.3390/molecules28010414
Chicago/Turabian StyleYasir, Muhammad, Jinyoung Park, Eun-Taek Han, Won Sun Park, Jin-Hee Han, Yong-Soo Kwon, Hee-Jae Lee, Mubashir Hassan, Andrzej Kloczkowski, and Wanjoo Chun. 2023. "Exploration of Flavonoids as Lead Compounds against Ewing Sarcoma through Molecular Docking, Pharmacogenomics Analysis, and Molecular Dynamics Simulations" Molecules 28, no. 1: 414. https://doi.org/10.3390/molecules28010414
APA StyleYasir, M., Park, J., Han, E.-T., Park, W. S., Han, J.-H., Kwon, Y.-S., Lee, H.-J., Hassan, M., Kloczkowski, A., & Chun, W. (2023). Exploration of Flavonoids as Lead Compounds against Ewing Sarcoma through Molecular Docking, Pharmacogenomics Analysis, and Molecular Dynamics Simulations. Molecules, 28(1), 414. https://doi.org/10.3390/molecules28010414










