Abstract
Chitosan is a natural polysaccharide which has been authorized for oenological practices for the treatment of musts and wines. This authorization is limited to chitosan of fungal origin while that of crustacean origin is prohibited. To guarantee its origin, a method based on the measurement of the stable isotope ratios (SIR) of carbon δ13C, nitrogen δ15N, oxygen δ18O and hydrogen δ2H of chitosan has been recently proposed without indicating the threshold authenticity limits of these parameters which, for the first time, were estimated in this paper. In addition, on part of the samples analysed through SIR, Fourier transform infrared spectrometry (FTIR) and thermogravimetric analysis (TGA) were performed as simple and rapid discrimination methods due to limited technological resources. Samples having δ13C values above −14.2‰ and below −125.1‰ can be considered as authentic fungal chitosan without needing to analyse other parameters. If the δ13C value falls between −25.1‰ and −24.9‰, it is necessary to proceed further with the evaluation of the parameter δ15N, which must be above +2.7‰. Samples having δ18O values lower than +25.3‰ can be considered as authentic fungal chitosan. The combination of maximum degradation temperatures (obtained using TGA) and peak areas of Amide I and NH2/Amide II (obtained using FTIR) also allows the discrimination between the two origins of the polysaccharide. Hierarchical cluster analysis (HCA) and principal component analysis (PCA) based on TGA, FTIR and SIR data successfully distributed the tested samples into informative clusters. Therefore, we present the technologies described as part of a robust analytical strategy for the correct identification of chitosan samples from crustaceans or fungi.
1. Introduction
Chitin is a linear polysaccharide polymer composed of D-glucosamine and N-acetyl-D-glucosamine monomers linked by β(1,4) covalent bonds [1]. Chitin is synthesized by a large number of living organisms, such as arthropods, insects, crustaceans, algae, plants and fungi [2]. On industrial scale, the main production comes from crustacean residues (exoskeletons), whereas fungi (cell walls) represent a less used alternative, despite being an abundant chitin source. Chitin is converted into chitosan by deacetylation involving the removal of the acetyl group through alkaline or enzymatic hydrolysis. Both chitin and chitosan are currently intensively used in pharmaceutical, cosmetic, biomedical, biotechnological, agricultural, food, and non-food industries (water treatment, paper and textile industries). These unique polymers are suitable for various applications in many fields due to their excellent biocompatibility, complete biodegradability, low toxicity, non-immunogenic properties and for having a wide range of interesting biological activities, including antimicrobial ones [3,4,5,6,7].
These characteristics make chitosan interesting for winemaking. Wine is a popular beverage consumed worldwide but its quality is influenced by several factors, including the control of Brettanomyces yeast population, which is responsible for altering the organoleptic characteristics of the wine. Chitosan has been approved for (1) wine treatment (to reduce heavy metal content, notably iron, lead, cadmium, and copper; to prevent iron haze and copper haze; to reduce possible contaminants, especially ochratoxin A; to reduce undesirable micro-organisms, notably Brettanomyces [8]), for (2) wine fining (to reduce turbidity by precipitating particles in suspension and to carry out a treatment to prevent protein haze by the partial precipitation of excess proteinaceous matter [9]) and for (3) must fining (to facilitate settling and clarification and to carry out a treatment to prevent protein haze [10]). However, only fungal chitosan (from Aspergillus niger and from Agaricus bisporus) is currently authorized for these oenological uses in order to avoid the risks of allergic reaction potentially caused by tropomyosin, the protein contained in shellfish products. In this context, the OIV specified three official methods for the identification of fungal chitosan in the chitosan monograph, i.e., content of residual glucans, viscosity of 1% chitosan solution and settled density [11]. The official methods for chitosan identification have specific limits, since the three mentioned parameters may be falsified to comply with the specifications.
Chitosan molecular mass distribution, solubility, degree of deacetylation, biological properties and quantity/density of residual saccharides bound to it represent additional techniques tested as tools for the characterisation of this product [12]. However, these procedures, as well as the official ones, are time-consuming and require many different equipment that are not available in all laboratories. For all these reasons, Perini et al. [13] developed a faster and easier method based on the analysis of the stable isotope ratios, expressed in delta, of different elements (13C/12C, 15N/14N, 18O/16O and 2H/1H). Among these parameters, δ13C and δ18O resulted as the most discriminant ones. Although this automated method showed promise as a discriminant tool between fungal and crustacean chitosan, the reduced number of samples analysed and their uncertain origin did not allow Perini et al. [13] to establish a limit value of δ13C and δ18O above or below which a sample could be assessed as ‘fungal’ or ‘not fungal’ chitosan.
In this study, for the first time, we analysed chitosan samples of certified origin obtained from crustacean and from A. niger and A. bisporus strains grown on different substrates with the aim to define threshold limits not only for the isotopic parameters δ13C and δ18O but also for δ15N. We also attempted to develop complementary tools to discriminate between fungal and crustacean chitosan, based on methods widely used in laboratories, namely thermogravimetric analysis (TGA) and Fourier transform infrared (FTIR) spectroscopy. Indeed, FTIR spectroscopy combined with chemometrics has previously been reported as an effective tool in quality control and food origin classification [14,15]. To evaluate the real discriminating ability of the different analytical approaches, we combined them using two different chemometric techniques, principal component analysis (PCA) and hierarchical cluster analysis (HCA).
2. Results and Discussion
2.1. Characterisation of Fungal and Crustacean Chitosan
2.1.1. Characterisation of Chitosan from Different Origins Using SIR Analysis and Definition of Threshold Limits
Based on the δ13C and δ15N found in chitosan samples from crustacean (CC) and from fungal (strain A. bisporus and A. niger grown on substrate from plants with C3 photosynthetic cycle (FC3) and C4 (FC4)), it was possible to separate not only these two types of products but also to characterise two subclasses (FC3 and FC4) of fungal-derived chitosan. As shown in Table 1, the δ13C of chitosan from animal exoskeleton (reported as CC-1 to CC-17) ranges from −24.6‰ to −17.2‰ while the δ15N ranges from −5.7‰ to 0.0‰. With the current dataset, it is not possible to detect statistically significant differences among samples from different exoskeletons (shrimp vs. crab vs. squid) (p > 0.05). All the δ13C and δ15N found in this study fell within the range of variability reported by Perini et al. for animal chitosan, thus confirming the method validity [13].

Table 1.
Description of the chitosan samples used for SIR, FTIR and TGA analysis, viscosity (cP, 1% in 1% acetic acid) and degree of deacetylation (%) declared and experimental values (mean, SD, min and max, low and high threshold value 95%) of δ13C (‰, vs. V-PDB), δ15N (‰, vs. AIR), δ2H and δ18O (‰ vs. V-SMOW) isotopic parameters; of the main TGA results (maximum temperature of degradation and weight loss) and of the main FTIR results (amide I and NH2/amide II peak areas) obtained from chitosan samples from crustacean (CC) and from fungal (strain A. bisporus and A. niger grown on substrate C3 (FC3) and C4 (FC4)).
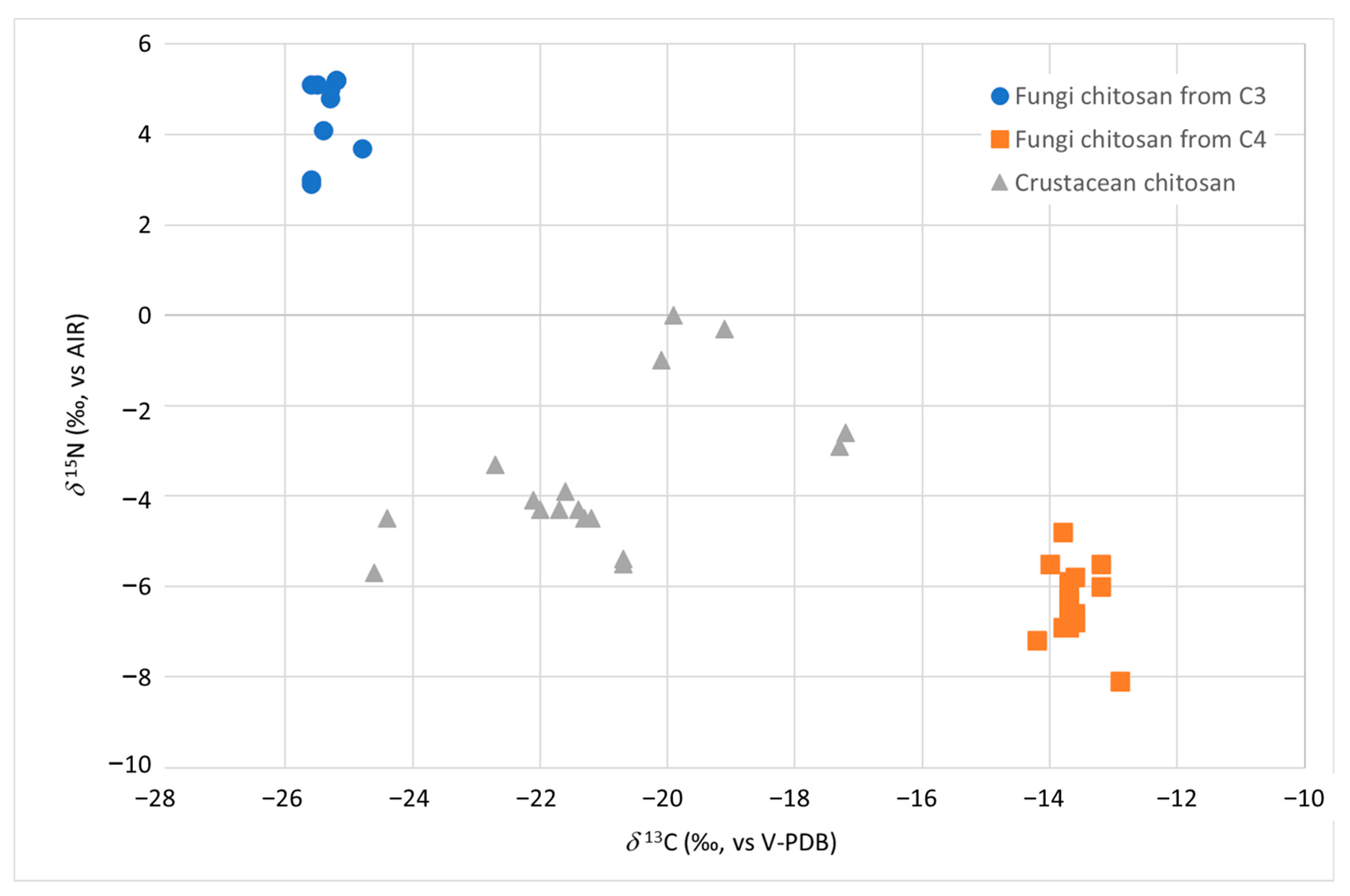
As for fungal chitosan samples (reported as FC3 and FC4 in Table 1), the isotopic values δ13C and δ15N allowed to identify not only their fungal origin but also to differentiate them according to the carbon and nitrogen source used by A. niger and A. bisporus during the growth of their mycelium (Figure 1).
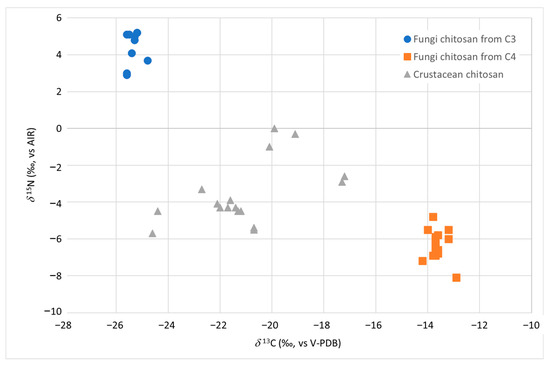
Figure 1.
δ13C vs. δ15N distribution of chitosan samples from fungi (strain A. bisporus and A. niger grown on C3 and C4 substrate in blue and orange, respectively) and from crustacean (in grey).
Fungal chitosan could be obtained by A. niger mycelium, a by-product of the citric acid synthesis through sugar fermentation carried by A. niger strains. Indeed, strains of A. niger are used in the production of citric acid since they can use low-cost raw materials (among which the most widespread are beet or cane molasses) and produce higher quantities of citric acid, resulting in a less expensive process [16].
This fermentation process involves the use of sugar (such as glucose and sucrose) both in the inoculum of the culture and in the fermentation media supplemented with a limited concentration of phosphate, nitrogen and trace elements. Therefore, the carbon used by the strain during its growth derives almost exclusively from beet or cane sugar and reflects the typical δ13C of a C3 (from −29 to −25‰) or C4 (between −14 and −12‰) photosynthetic cycle plant [17]. After the separation of the citric acid, the fungal mycelium, figuring as a waste product of citric acid synthesis, can be used as the starting raw material to produce chitin and, therefore, chitosan.
An alternative source used as raw material to produce fungal chitosan is the A. bisporus mushroom [18]. It is widely cultivated in Europe and represents about 32% of the world mushroom production [19], but its cultivation gives rise to several by-products, including waste and off-grade mushrooms with no suitable commercial use, whose amount ranges between 5% and 20% of production volume [20]. A strategy to reduce the environmental impact of the agro-food industries is the use of by-products of A. bisporus for the production of non-animal chitin and chitosan [21]. Additionally, in this case, the substrates used on an industrial level as a source of carbon for the growth of the fungal mycelium are cane or beer, as already discussed for A. niger.
The ranges of variability of the two isotopic ratios δ13C and δ15N were found to be significantly different (p < 0.01) between the samples of chitosan from fungi and crustaceans and also between the fungal chitosan from strains grown on substrates of C3 (indicated as FC3) and C4 photosynthetic cycle plants (referred to as FC4) (p < 0.01) (Table 1). It was therefore necessary to calculate and report separately in Table 1 the dispersion indexes (e.g., average) for the two types of samples, FC3 and FC4. If the carbon source used by the strain came from C3 sugar (samples FC3), the δ13C varied from −25.6‰ to −24.8‰, while if it came from C4 sugars (samples FC4), it varied from −14.2‰ to −12.9‰ (Table 1).
The δ13C alone resulted to be effective in discriminating between chitosan from crustaceans and from fungi given a C4 source for its growth. On the other hand, a slight overlap between the δ13C of the chitosan samples from crustacean and those from fungi given a C3 source was observed. In this case, the δ15N parameter allowed to clearly discriminate (p < 0.01) between the two groups, with the product from fungi having (average +4.5‰) higher values than in the crustacean one (average −3.4‰).
The cultivation of the A. bisporus mushroom requires the use of substrates, such as chicken and/or horse manure [22]. Like all animal fertilizers, these products have high δ15N, related to the trophic effect [23]. The same nitrogen sources with high δ15N could also be used in A. niger strains fermentation. These factors may explain the high δ15N values found in the FC3 samples. On the other hand, as already observed by Perini et al. [13], the δ15N alone was unable to discriminate between chitosan from crustaceans and chitosan obtained from fungi using C4 sugars as carbon source. Table 1 shows the threshold limit values (calculated as mean ±2 standard deviation) which can be proposed as authenticity limits. For δ13C values above −14.2‰ and below −25.1‰, the sample can be considered an authentic fungal chitosan without the need to analyse other parameters. If the value of δ13C falls between −25.1‰ and −24.9‰, it is necessary to proceed with the evaluation of the parameter δ15N, which must be above +2.7‰.
The isotopic values δ18O and δ2H measured in the samples under study are listed in Table 1. As already reported by Perini et al. [13], the two isotopic parameters are not correlated to each other (R2 = 0.3925), as they are differently influenced by the isotopic composition of diet and water. In particular, as reported by Nielson and Bowen, the δ2H of chitin showed a strong linear correlation with both food and water δ2H, with approximately 26% of the hydrogen signal reflecting food and approximately 38% reflecting water, while more than 69% of oxygen in chitin exchanged with environmental water and only 10% derived from food [24]. The δ18O parameter appeared to be the most discriminating one, having the lowest values in chitosan samples from fungi (average +23.5‰) and the highest values in samples from crustaceans (average +28.1‰). The use of carbon sources from C3 or C4 plants cannot be discriminated in the samples of fungal origin based on the δ18O (p > 0.05, Table 1). Therefore, the dispersion parameters (e.g., mean) were calculated considering the whole group of fungal chitosan samples (FC3 + FC4). The δ18O of chitosan is strictly related to the same value of the water used for the formulation of the fermentative medium. This value is supposed to normally range between −14‰ and −6‰ [25], while it is higher in the ocean water where marine invertebrates live (around 0‰ on average) [26]. This may explain the differences found between the samples of fungal origin (FC3 + FC4 together) and those of crustacean origin (CC). The use of the isotopic parameter δ18O could be therefore useful to identify the origin of chitosan. Calculating a threshold authenticity limit as mean ±2 standard deviation, a sample from fungi could be considered authentic for values lower than +25.3‰.
2.1.2. Characterisation of Chitosan from Different Origins Using FTIR Analysis
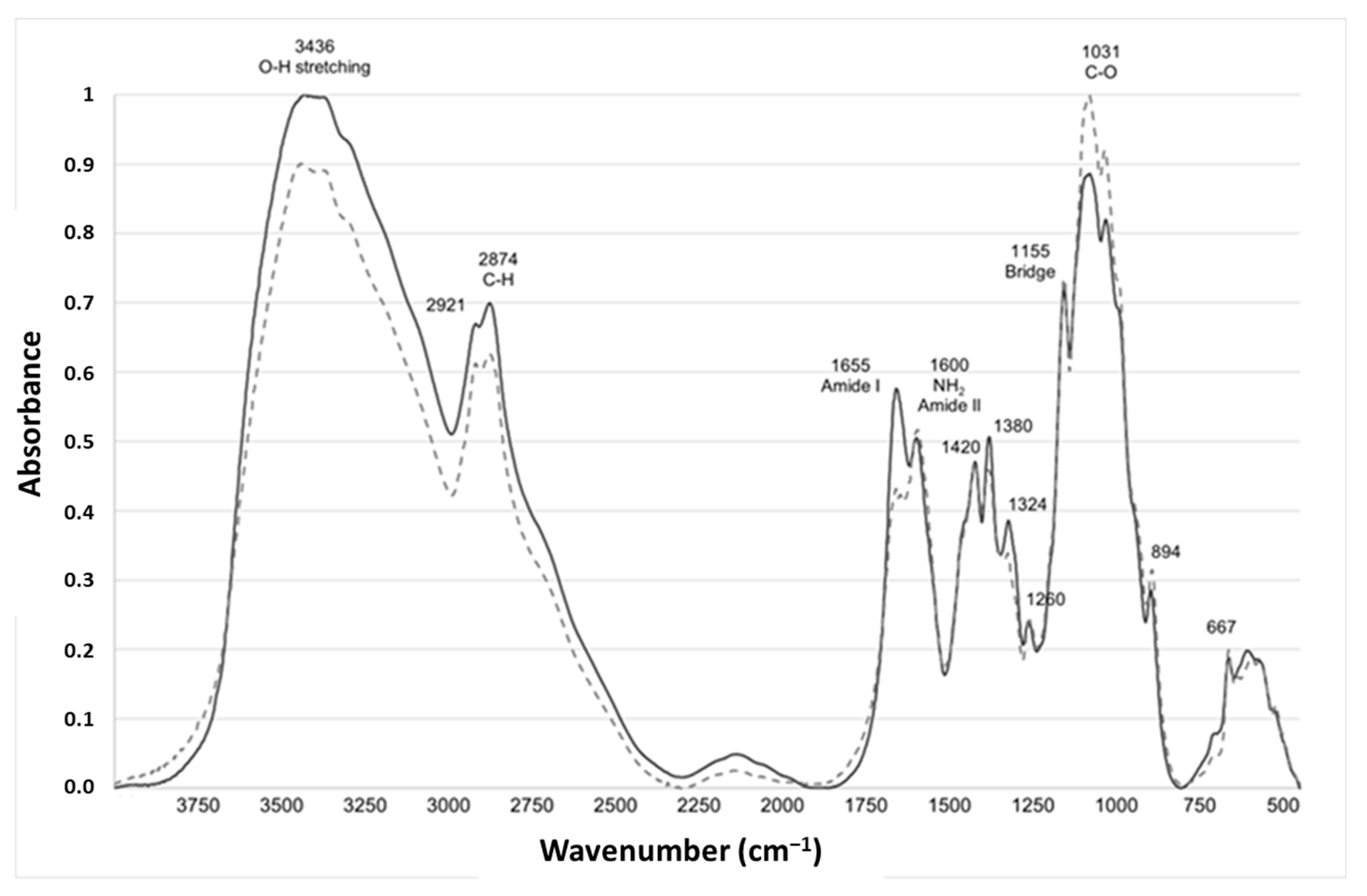
The FTIR is a simple technique performed using equipment that is widely available. FTIR has been used to study the composition and structure of chitin, to distinguish the β-form from the α-one [27,28]. A representative example of spectra from fungal and crustacean chitosan is given in Figure 2.
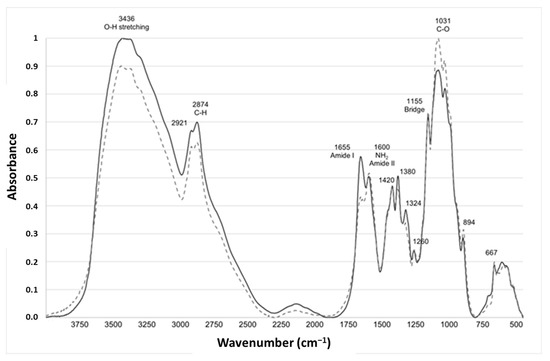
Figure 2.
Characteristic infrared absorption spectra of typical fungal (light grey dotted line, FC4-15) and crustacean (solid line dark grey, CC-5) chitosan.
Both chitosan spectra showed a series of narrow absorption bands, typical of crystalline polysaccharide samples. The C=O stretching region of the amide moiety of chitosan, evident between 1700 and 1500 cm−1, corresponds to the specific signature of α-chitin, the most common form, found in arthropods, including crustaceans, fungi and yeasts, whose amide I band is generally split into two signals at 1660 and 1630 cm−1 [29,30].
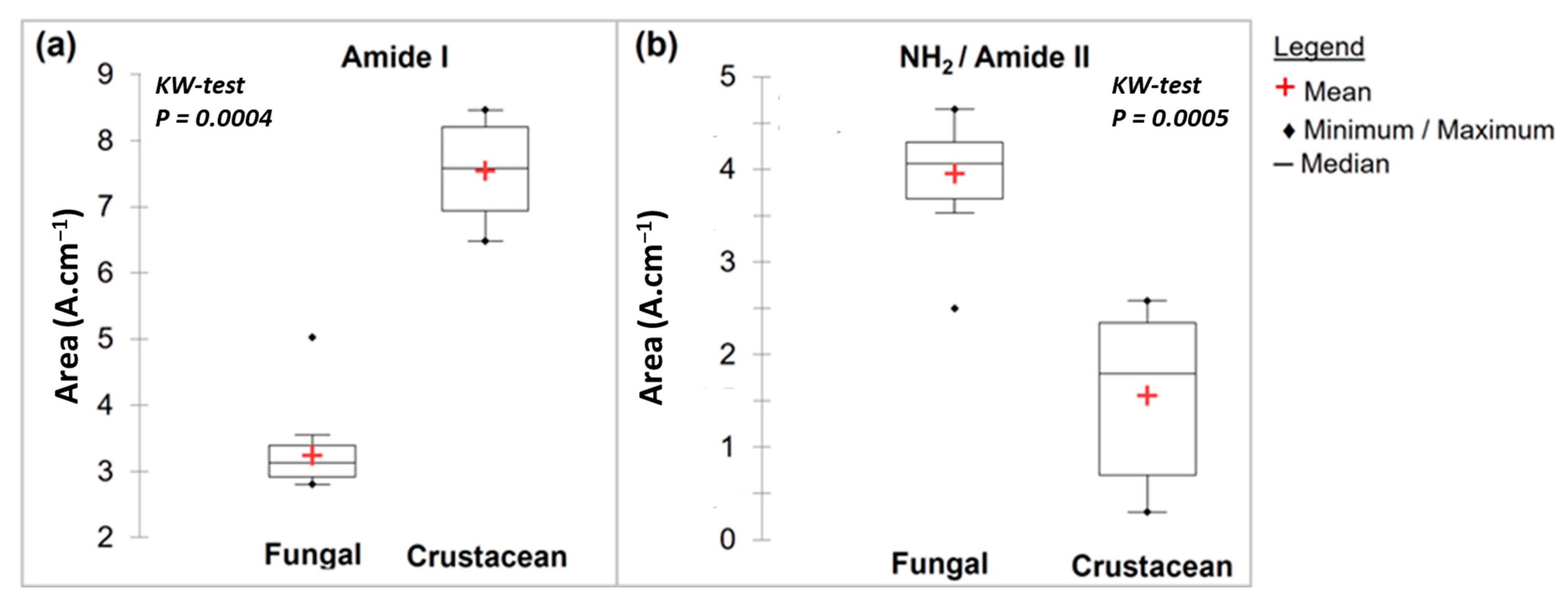
During the N-deacetylation of chitin into chitosan, this amide I band gradually decreases due to the removal of the acetate moiety. In this study, fungal chitosan spectra from A. niger are characterized by two typical absorption bands at 1655 cm−1 (C=O stretching of amide I band confirming the presence of residual N-acetyl groups) and at 1600 cm−1 (N-H bending of the primary amine NH2 resulting from the deacetylation of chitin and possible overlap with the N-H bending of the amide II) [31,32]. Hereinafter, these two bands at 1655 and 1600 cm−1 will be referred to as Amide I and NH2/Amide II, respectively. The IR spectra can be used to distinguish between fungal and crustacean chitosan. The respective areas of these two peaks were therefore calculated to obtain quantitative data to compare the chitosan according to their origin (see Table 1 and Figure 3). It appears that crustacean chitosan presents a larger peak area for amide I than fungal chitosan (on average 7.5 vs. 3.2 A·cm−1, respectively—p < 0.001) and a smaller peak area for NH2/Amide II (on average 1.6 vs. 4.0 A·cm−1, respectively—p < 0.001).
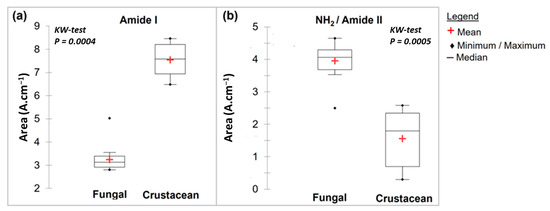
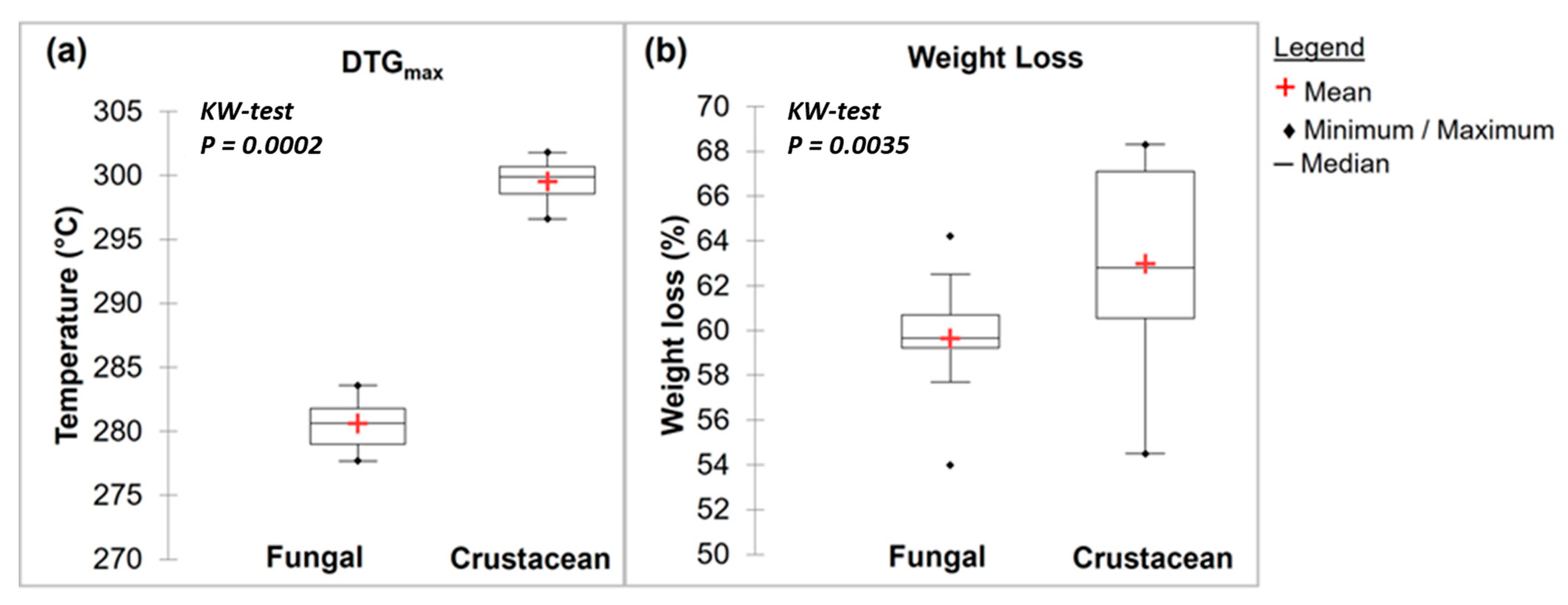
Figure 3.
Box plots of FTIR main results with a Kruskal–Wallis evaluation (p < 0.05) for fungal and crustacean samples. (a,b) correspond to Amide I and NH2/Amide II peak areas, respectively.
2.1.3. Characterisation of Chitosan from Different Origins Using TGA Analysis
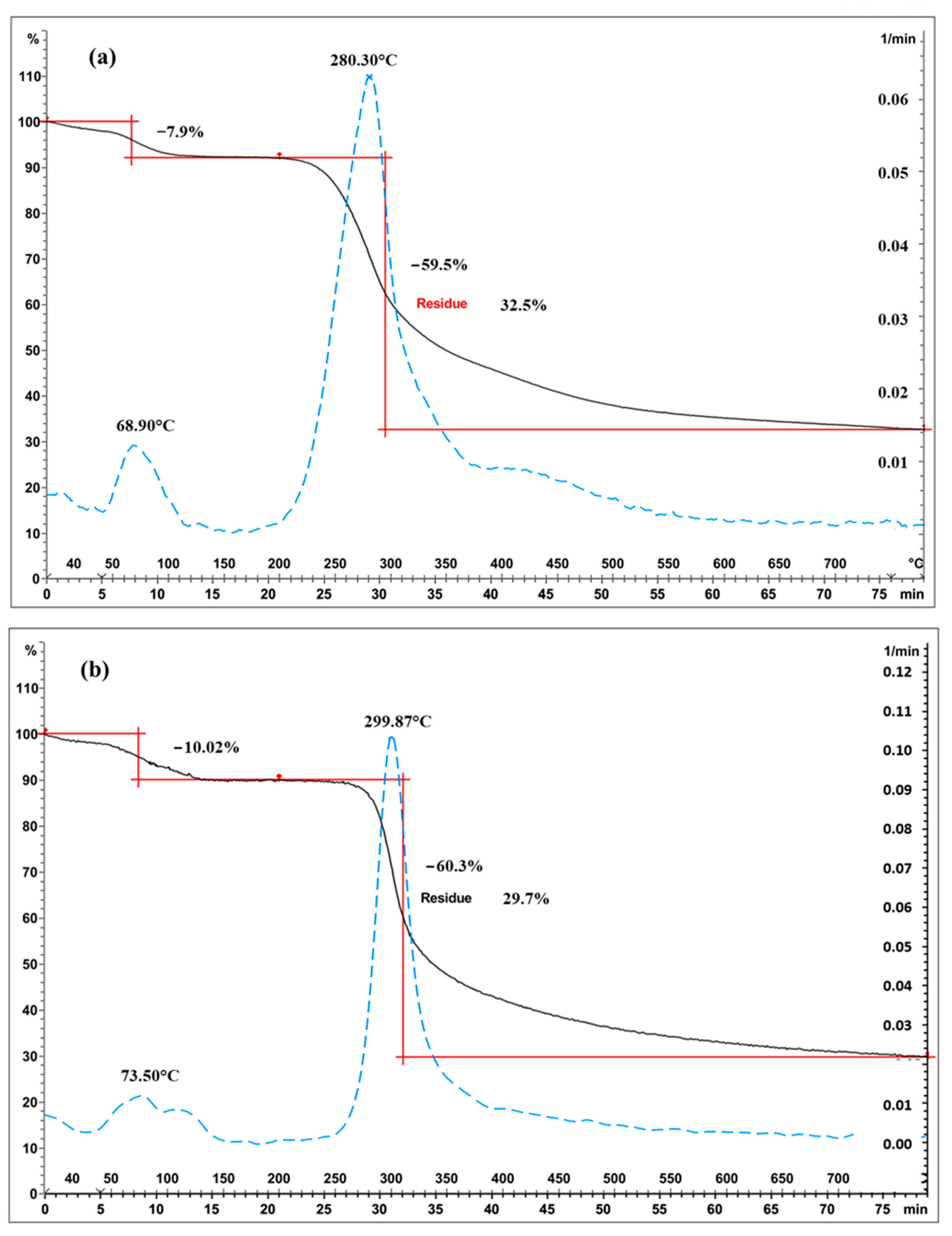
Representative TGA curves of chitosan are shown in Figure 4a for fungal and Figure 4b for crustacean chitosan. As found in the literature [33,34,35,36], both curves present two degradation stages: the first step occurs around 60 °C (weight loss WL about 5–10%) and is assigned to the evaporation of the residual water because of the strong affinity of polysaccharides for water. Then, the weight of partially deacetylated chitosan remained stable up to 250 °C followed by a rapid substantial loss of weight. The second step occurs around 300 °C (exothermic, WL about 60–70%) and is related to the pyrolytic decomposition of chitosan which is characteristic for the chitosan structure [37,38]. The experimental data obtained from the TGA characterisation of the chitosan samples are summarized in Table 1 and represented in Figure 5. The data show that crustacean chitosan exhibited higher DTGmax than fungal chitosan (average DTGmax value of 299.5 vs. 280.6 °C, respectively—p < 0.001). The differences in weight loss are less discriminatory than the maximum degradation temperatures (DTGmax). For this reason, only DTGmax values will be used for the following statistical analysis. It should be noted that few studies focus on the TGA analysis of fungal chitosan, but our results correspond to the DTGmax values reported by Kaya et al., asserting that the low DTGmax value of fungal chitin and chitosan stems from the glucan residues that could not be removed from the chitin structure [29].
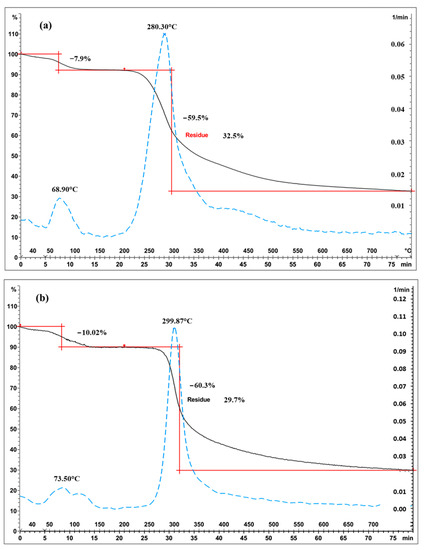
Figure 4.
Thermograms of chitosan ((a) fungal chitosan FC4-15, (b) crustacean chitosan CC-5). Black solid curves represent TGA, blue dotted lines represent the derivatives and red represents the weight loss.
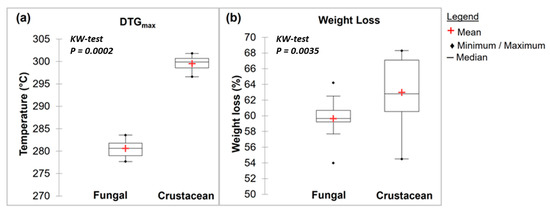
Figure 5.
Box plots of TGA main results with a Kruskal–Wallis evaluation (p < 0.05) for fungal and crustacean samples (a,b) correspond to DTGmax and weight loss, respectively.
2.2. Discrimination and Clustering Based on the Origin of Chitosan
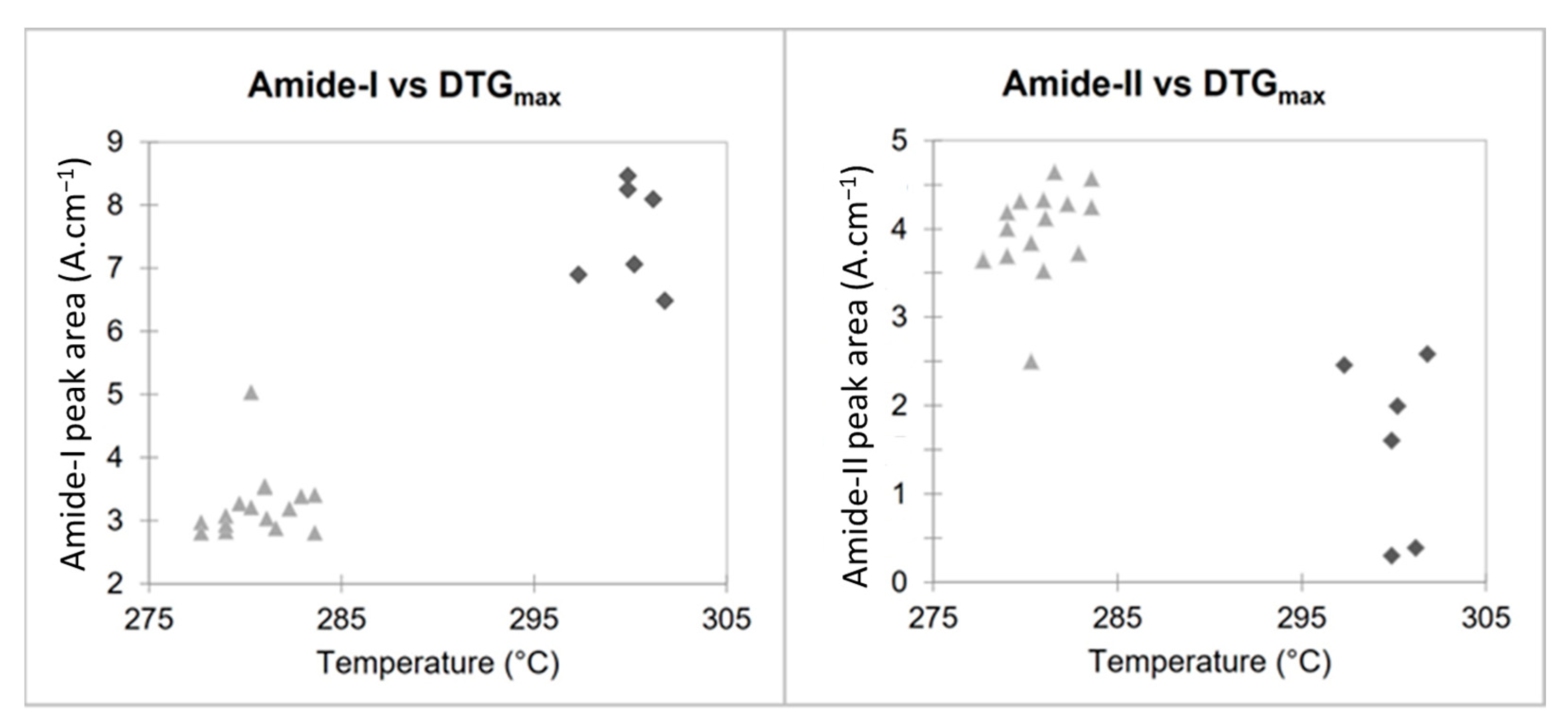
Analysis of FTIR and TGA data (Table 1) shows that the origin of the chitosan impacts the intrinsic characteristics of the biopolymer, with significant differences for each parameter studied (KW-test < 0.05). However, the maximum degradation temperature measured using TGA, as well as amide I and NH2/amide II peak areas using FTIR seem to be the most promising parameters for determining the origin of chitosan (p < 0.001).
These two parameters, plotted in the scatter plots in Figure 6, show how it is possible to classify the two types of chitosan in a simple and fast way and without the need for specific software. Figure 6 highlights the relationship between the maximum degradation temperatures (obtained using TGA) and areas of the amide I and NH2/Amide II peaks (obtained using FTIR). Fungal and crustacean chitosan samples are well clustered into two separate groups.
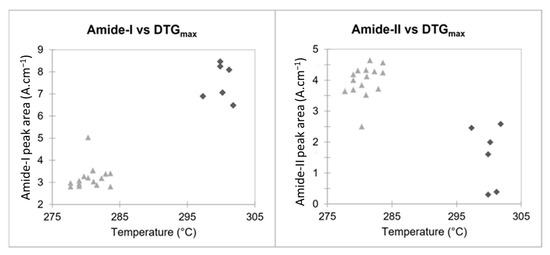
Figure 6.
Scatter plots of FTIR and TGA results for fungal (light grey triangle) and crustacean (dark grey losange).
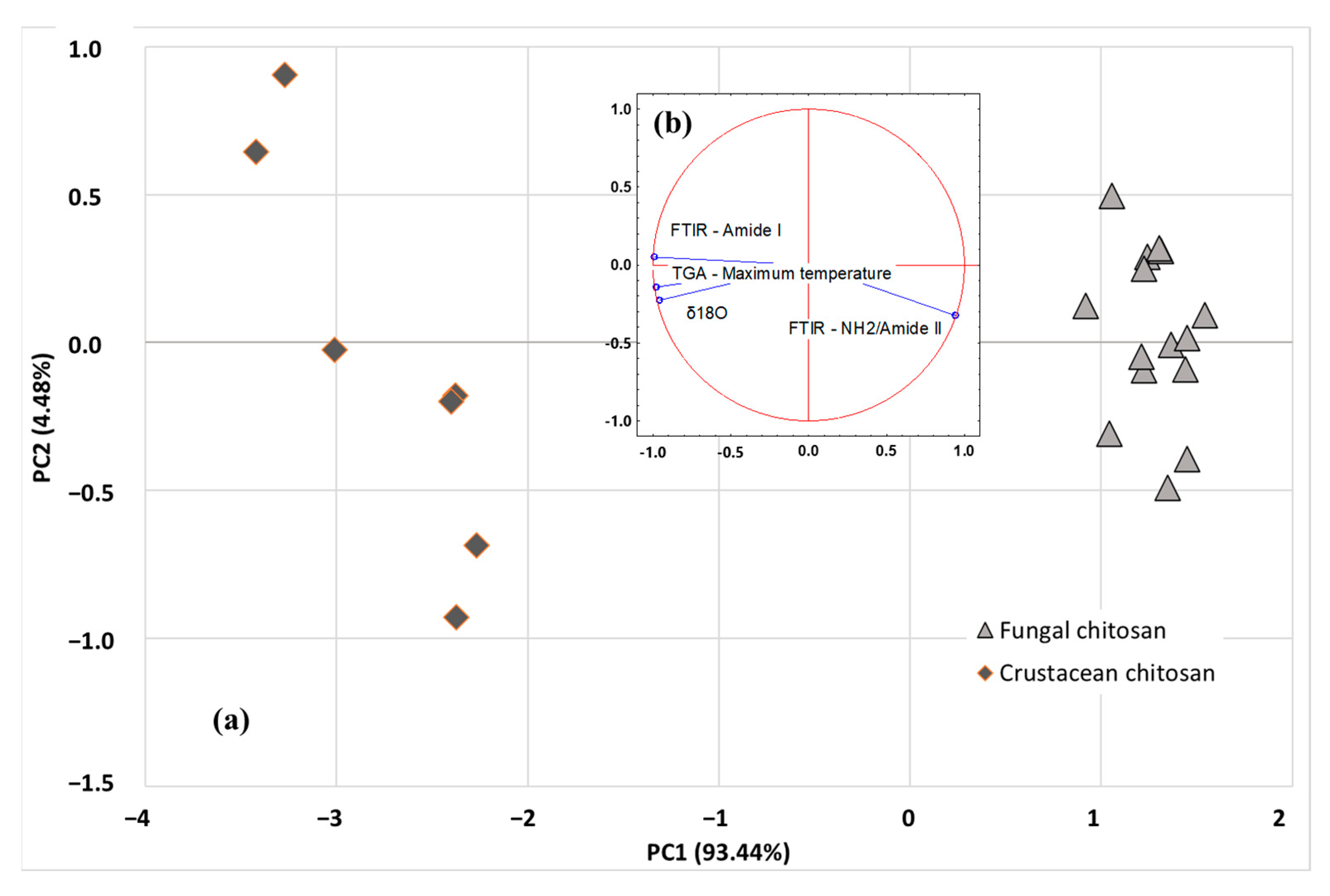
More advanced multivariate statistical analysis of the different investigated parameters (SIR, TGA, and FTIR) were obtained using PCA or HCA exploratory methods. Among the isotopic ratios, δ18O was selected as SIR parameter as it is independent of the carbon source used by the fungi strain, while maximum degradation temperature and ratio of amide I and NH2/amide II peak areas were selected as TGA and FTIR parameters, respectively. Unsupervised methods, also named clustering or displays methods, are used to study the data structure and to evaluate whether clustering exists in a dataset. The PCA scatter plot involving the first 2 PCs (i.e., PC1 and PC2) is shown in Figure 7a. The analysis discriminated the origin of chitosan using the first two components: PC1 (93.44%) and PC2 (4.48%), explaining 97.92% of total variance. PCA results revealed that the acquired data points were clearly grouped into two classes (fungal chitosan, represented by light grey triangles or crustacean chitosan, represented by dark grey losange), based on the SIR, TGA and FTIR parameters of the samples. As indicated in Figure 7b, PC1 axis is linked to FTIR parameter amide I, TGA parameter (DTGmax) and δ18O, whereas PC2 axis is more linked to FTIR parameter NH2/Amide II peaks. Once the representative PCs were found, based on sample differentiation/grouping and variance explained, loading analysis is started to find the underlying relationships in the original data structure. The positive factor loadings indicate that the factor will be higher in the positive axis of that PC. For example, for NH2/Amide II, a factor loading of 0.942 was obtained with PC1 and, for Amide I, a factor loading of 0.050 was obtained with PC2. It means that the samples located in the right-hand side (i.e., fungi origin) of the graph have higher mean NH2/Amide II and Amide I area values than the samples located in the left-hand side (i.e., crustacean origin). Similarly, the negative factor loadings indicate that the factor will be higher in the positive axis of that PC. For example, for DTGmax, a factor loading of −0.976 was obtained with PC1, meaning that the samples located in the right-hand side (i.e., crustacean origin) of the graph have lower mean DTGmax values than the samples located in the left-hand side (i.e., crustacean origin). The study of the regression vectors (see Figure 7b) shows vectors of correlation coefficients between the original variables with each PC-score.
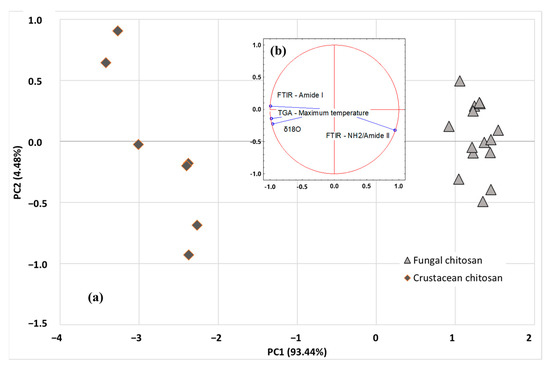
Figure 7.
PCA scatter plot (a) and related biplots (b) of SIR (δ18O), TGA (DTGmax) and FTIR (Amide I, NH2/Amide II) data. Actives observations are represented by light grey triangles (fungal chitosan) or dark grey losange (crustacean chitosan).
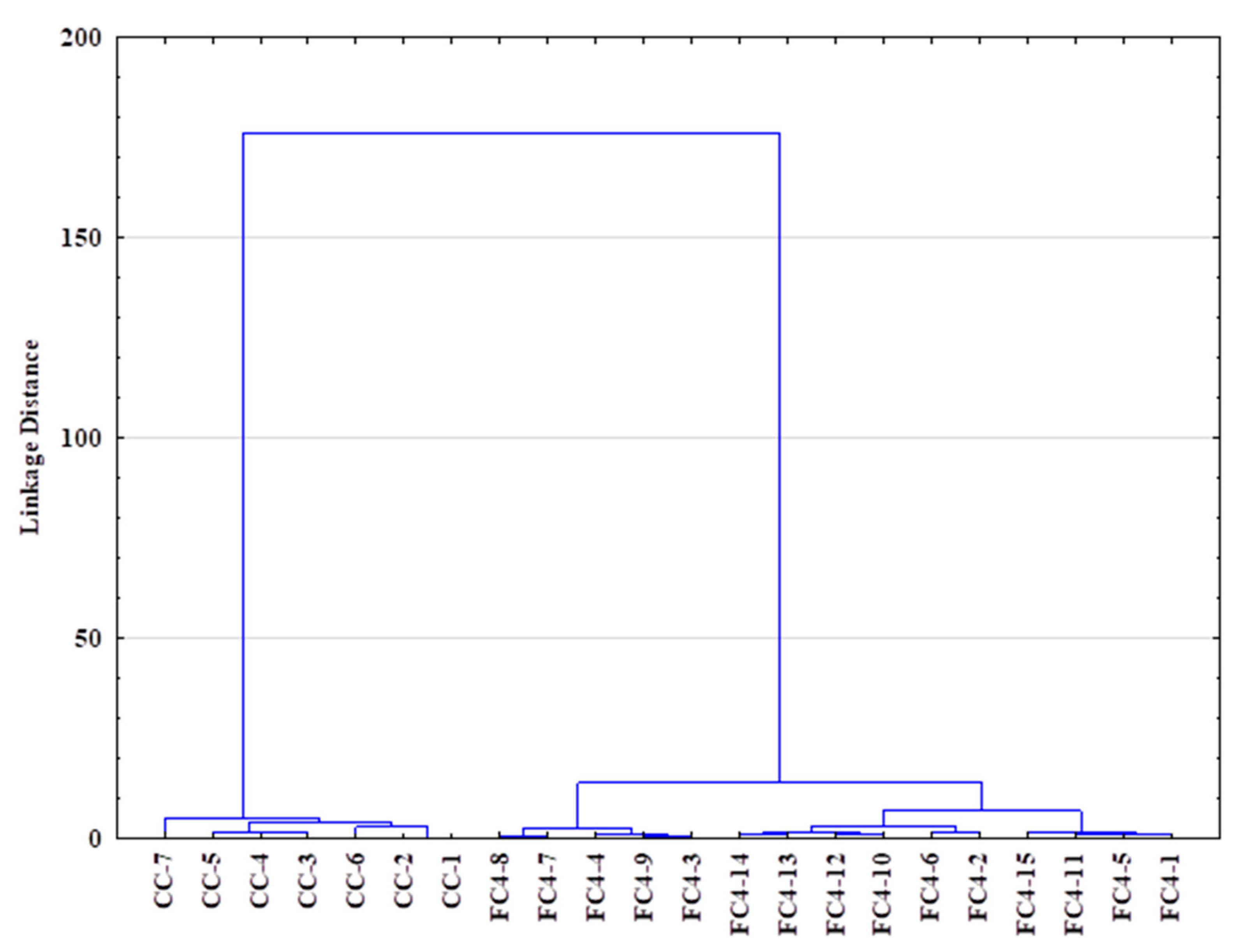
In addition, a hierarchical clustering analysis (HCA) was performed to explore the organization of samples in groups and among groups depicting relationships in tree form (Figure 8). The agglomerative approach was used on the SIR, TGA and FTIR data: the complete linkage method was used for cluster building, and the distance between clusters was computed using the Euclidean method (Ward method, Euclidean distances). Figure 8 shows the similarity dendrogram obtained, highlighting two main clusters: (I) crustacean chitosan and (II) fungal chitosan, thus confirming the PCA results while providing more investigatory outcomes. The obtained HCA dendrogram reveals that fungal samples could be readily distinguished from the crustacean samples.
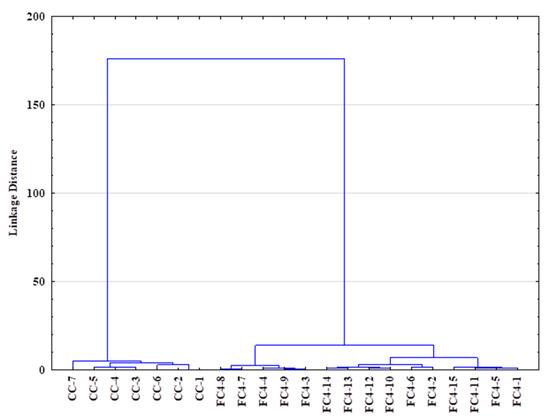
Figure 8.
Dendrogram obtained via hierarchical clustering analysis (HCA—Euclidean distances) using SIR, TGA and FTIR data of crustacean (CC) and fungal chitosan sample (FC).
3. Materials and Methods
3.1. Sampling
To build the isotopic database of chitosan, 50 samples of chitosan from different fungal sources and crustacean exoskeleton were obtained from different producers (Table 1). The samples provided by the producers were accompanied by a certificate of analysis highlighting the origin of chitosan (from fungi or crustaceans) and their main characteristics, as well as a description of the production methods, including information about the strain used to produce the fungi (A. bisporus and A. niger) and the type of substrate used for the growth of the fungal strain.
Since the detailed industrial process is confidential, the exact recipes and the quantities of the individual ingredients have not been provided.
For TGA and FTIR analysis, 15 samples of fungal origin and 7 of animal origin were selected (Table 1).
In this study, chitosan obtained from fungus is identified as “FC” and more specifically as “FC3” if the substrate used for the growth of the fungi strain was from C3 photosynthetic cycle plants or “FC4” if it was from C4 photosynthetic cycle plants.
As per the certificate of analysis provided by the suppliers, FC3-1 to FC3-9 are fungal chitosan samples extracted from A. bisporus. FC3-10 and FC3-11 and FC4-1 to FC4-22 are fungal chitosan samples extracted from A. niger.
Chitosan derived from crustacean is identified as “CC”. CC-1 to CC-17 are crustacean chitosan samples derived from crab, shrimp or squid. A more detailed description of the chitosan products, as well as values of the main molecular characteristics including viscosity in solution and degree ad deacetylation, are reported in Table 1, based on information available from the suppliers.
3.2. Stable Isotope Analysis
The stable isotope ratios of H, C, N and O were measured in pure (95%) bulk lyophilized and ground chitosan, previously washed with a water/alcohol solution (90:10 v/v). This approach is considerably fast and automated (<10 min for each analysis).
The 13C/12C and 15N/14N ratios were measured in one run (around 0.5 mg) using an isotope ratio mass spectrometer (IRMS) (Isoprime Ltd., Cheadle Hulme, UK) following total combustion in an elemental analyser (VARIO CUBE, Elementar Analysensysteme GmbH, Langenselbold, Germany). The 2H/1H and 18O/16O ratios were measured in one go (around 0.5 mg) using an IRMS (Finnigan DELTA XP, Thermo Scientific, Waltham, MA, USA) coupled with a pyrolyser (Finnigan TC/EA, high temperature conversion elemental analyser, Thermo Scientific).
Based on the IUPAC protocol [39], the different stable isotope ratios were expressed in the delta scale (δ‰) against the international V-PDB (Vienna PeeDee Belemnite) standard according to Equation (1):
where, ref is the international measurement standard, sample is the analysed sample, and iE/jE is the ratio of heavier to lighter isotopes. The delta values were multiplied by 1000 and expressed in “per mil” (‰) units.
The sample analysis was carried out in duplicate. For δ13C and δ15N, the isotopic values were calculated against working in-house standards (protein), which were themselves calibrated against international reference materials: fuel oil NBS-22 (δ13C = −30.03‰), IAEA-CH-6 (IAEA-International Atomic Energy Agency, Vienna, Austria) (δ13C = −10.45‰) for 13C/12C, L-glutamic acid USGS 40 (U.S. Geological Survey, Reston, VA, USA) (δ13C = −26.39‰ and δ15N = −4.52‰) for 13C/12C and 15N/14N and potassium nitrate IAEA-NO3 (δ15N = +4.70‰) for δ15N.
Both natural chitin and its deacetylation product chitosan contain some strongly adsorbed water that can isotopically exchange its hydrogen with ambient H2O and add isotopic noise to the δ2H of organic hydrogen during the measure. In this study, an equilibration of exchangeable hydrogen in chitin with the H2O of the known δ2H was carried out [40].
The δ2H and δ18O values were calculated against CBS (Caribou Hoof Standard δ2H = −157 ± 2‰ and δ18O = +3.8 ± 0.1‰) and KHS (Kudu Horn Standard, δ2H = −35.3 ± 1‰ and δ18O = +20.3 ± 0.2‰) through the creation of a linear equation and by adopting a comparative equilibration procedure [41]. We used these two keratinous standards because of the absence of any international organic reference material with a similar matrix to our samples (chitosan).
The uncertainty of measurements, calculated as standard reproducibility (that we obtain from the analysis of the same sample over time) multiplied for the coverage factor 2, was <0.3‰ for δ13C and δ15N analysis, <1‰ for δ18O, and <3‰ for δ2H.
3.3. Thermogravimetric Analysis
Thermogravimetric analysis (TGA) was performed using a TGA 2 (Mettler-Toledo Garvens GmbH, Giesen, Germany) coupled with the STARe Excellence V16.10 software. A quantity of 3–5 mg of each sample was placed in an open crucible and the temperature was raised from 40 °C to 600 °C at a heating rate of 10 °C per minute.
3.4. Fourier Transform Infrared Analysis
The Fourier Transform Infrared (FTIR) analyses were performed using a Spectrum One spectrophotometer (Perkin Elmer, Waltham, MA, USA) operating in transmission mode. FTIR analysis was carried out using the sample dispersed in KBr (1:20 weight ratio). The wave-number range was 4000–400 cm−1 and the resolution were 4 cm−1, with 12 scans performed on each sample. The system was coupled with the Spectrum V. 3.02.01 software (Perkin Elmer Inc., Wellesley, MA, USA) for further analysis, such as obtaining the area of the characteristic peaks (i.e., 1655 cm−1 and 1600 cm−1 corresponding to the Amide I and NH2/Amide II bands, respectively).
3.5. Statistical Analysis
The isotopic data were evaluated using R software, vers. 4.0.4 (R Foundation for Statistical Computing, Vienna, Austria). The FTIR and TGA data were evaluated statistically using XLSTAT (XLSTAT version 2019.2.2, Addinsoft, Paris, France). Data normality was tested (Shapiro–Wilk, p < 0.05) and statistical origin differences were checked using the Kruskal–Wallis test (KW-test, p < 0.05).
Principal component analysis (PCA) of samples was carried out to further investigate the differences between the fungal and animal origin of chitosan based on the SIR, FTIR and TGA parameters. Finally, hierarchical clustering analysis (HCA) was carried out based on the SIR, TGA and FTIR data to reveal relationships between samples from the same origin and thus confirm the chitosan origin using XLSTAT.
4. Conclusions
In this study, it was demonstrated that δ13C and δ15N of the chitosan from fungi are closely related to the substrate used for the growth of the strains (e.g., A. niger and A. bisporus). Sugars from C3 photosynthetic cycle plants resulted in isotopic ratios different from those from C4 cycle plant, as well as the use of manure instead of urea as a nitrogen source. In both cases, the chitosan from fungi had isotope ratios different from those derived from crustaceans. For the first time, the threshold limits of the isotopic parameters δ13C, δ15N and δ18O of fungal chitosan samples were here defined.
The data of maximum degradation temperatures (obtained using TGA) combined with those of the peak areas of amide I and NH2/Amide II (obtained using FTIR) seemed to allow the differentiation of the two types of chitosan (fungal vs. animal) into well-defined clusters.
Chemometric analyses (PCA and HCA) based on SIR, TGA and FTIR successfully distributed the tested samples into informative clusters, allowing for the differentiation of the samples according to their origin. The results have highlighted a strong similarity between HCA and PCA findings. Chemometric techniques based on SIR, TGA and FTIR provided an efficient, robust methodology for the assessment of chitosan origin. The set of methods provided a multi- approach strategy that appears to be more reliable than the official methods reported in the current monograph of chitosan to prove its authenticity. We therefore present the described technologies as part of an analytical strategy for the correct identification of chitosan samples sourced from crustaceans or fungi. The proposed order of analysis would flow from FTIR analysis, using equipment which is most widely available in labs and has a high throughput, to TGA, which is less widely available and more time consuming, and to SIR, which requires more technological expertise and specialized equipment. Once a sample can clearly be identified with one or more of the technologies, further resources can be spared. Additionally, in any case, upon completion of the full strategy the identification will be without significant doubt for even the most challenging samples.
Author Contributions
Conceptualization, R.C.A.O., M.P. and E.C.; Methodology, R.C.A.O. and M.P.; Software, E.C.; Validation, E.C.; Formal Analysis, S.P., S.R. and B.Y.; Investigation, E.C., M.P. and R.C.A.O.; Resources, R.L.; Data Curation, M.P. and E.C.; Writing—Original Draft Preparation, E.C., R.C.A.O. and M.P.; Writing—Review and Editing, R.W. and R.L.; Supervision, R.L. and R.W.; Project Administration, R.L. and R.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially funded by “Service Public de Wallonie” (SPW-DGO6, Belgium) under Grant No. 8141.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw data presented in this study are available upon request.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of chitosan are not available.
References
- Yang, Q.; Fukamizo, T. Targeting Chitin-Containing Organisms; Springer: Berlin/Heidelberg, Germany, 2019; ISBN 9789811373183. [Google Scholar]
- Santos, V.P.; Marques, N.S.; Maia, P.C.; Lima, M.A.B.D.; Franco, L.D.O.; Campos-Takaki, G.M.D. Seafood Waste as Attractive Source of Chitin and Chitosan Production and Their Applications. Int. J. Mol. Sci. 2020, 21, 4290. [Google Scholar] [CrossRef] [PubMed]
- Abo Elsoud, M.M.; El Kady, E.M. Current Trends in Fungal Biosynthesis of Chitin and Chitosan. Bull. Natl. Res. Cent. 2019, 43, 59. [Google Scholar] [CrossRef]
- Mujtaba, M.; Khawar, K.M.; Camara, M.C.; Carvalho, L.B.; Fraceto, L.F.; Morsi, R.E.; Elsabee, M.Z.; Kaya, M.; Labidi, J.; Ullah, H.; et al. Chitosan-Based Delivery Systems for Plants: A Brief Overview of Recent Advances and Future Directions. Int. J. Biol. Macromol. 2020, 154, 683–697. [Google Scholar] [CrossRef] [PubMed]
- Rabea, E.I.; Badawy, M.E.-T.; Stevens, C.V.; Smagghe, G.; Steurbaut, W. Chitosan as Antimicrobial Agent: Applications and Mode of Action. Biomacromolecules 2003, 4, 1457–1465. [Google Scholar] [CrossRef]
- Rinaudo, M. Chitin and Chitosan: Properties and Applications. Prog. Polym. Sci. 2006, 31, 603–632. [Google Scholar] [CrossRef]
- Rocha, M.A.M.; Coimbra, M.A.; Nunes, C. Applications of Chitosan and Their Derivatives in Beverages: A Critical Review. Curr. Opin. Food Sci. 2017, 15, 61–69. [Google Scholar] [CrossRef]
- International Code of Oenological Practices OIV/OENO, 338A/2009; Chitosan Monography. Organisation Internationale de la vigne et du vin: Dijon, France, 2009.
- International Code of Oenological Practices OIV/OENO, 337A/2009; Wines—Fining Using Chitosan. Organisation Internationale de la vigne et du vin: Dijon, France, 2009.
- International Code of Oenological Practices OIV/OENO 336A/2009; Musts—Fining Using Chitosan. Organisation Internationale de la vigne et du vin: Dijon, France, 2009.
- International Organisation of Vine and Wine. Monograph on Chitosan (No. Resolution OIV/OENO 368/2009). 2017. Available online: http://www.oiv.int/public/medias/5119/code-2017-En.pdf (accessed on 13 February 2023).
- Pellis, A.; Guebitz, G.M.; Nyanhongo, G.S. Chitosan: Sources, Processing and Modification Techniques. Gels 2022, 8, 393. [Google Scholar] [CrossRef]
- Perini, M.; Nardin, T.; Venturelli, M.; Pianezze, S.; Larcher, R. Stable Isotope Ratio Analysis as a Fast and Simple Method for Identifying the Origin of Chitosan. Food Hydrocol. 2020, 101, 105516. [Google Scholar] [CrossRef]
- Christou, C.; Agapiou, A.; Kokkinofta, R. Use of FTIR Spectroscopy and Chemometrics for the Classification of Carobs Origin. J. Advert. Res. 2018, 10, 1–8. [Google Scholar] [CrossRef]
- Gad, H.A.; Bouzabata, A. Application of Chemometrics in Quality Control of Turmeric (Curcuma longa) Based on Ultra-Violet, Fourier Transform-Infrared and 1H NMR Spectroscopy. Food Chem. 2017, 237, 857–864. [Google Scholar] [CrossRef]
- Guc, S.; Erkmen, O. Citric Acid Production from Nontreated Beet Molasses by a Novel Aspergillus niger Strain: Effects of pH, Sugar and Ingredients. J. Food Microbiol. Saf. Hyg. 2017, 2, 3. [Google Scholar] [CrossRef]
- O’Leary, M.H. Carbon Isotopes in Photosynthesis. Bioscience 1988, 38, 328–336. [Google Scholar] [CrossRef]
- Hassainia, A.; Satha, H.; Sami, B. Two Routes to Produce Chitosan from Agaricus Bisporus. J. Renew. Mater. 2020, 8, 101–111. [Google Scholar] [CrossRef]
- Chakravarty Bipasha Trends in Mushroom Cultivation and Breeding. Aust. J. Agric. Res. 2011, 2, 102–109.
- Zivanovic, S. Identification of Opportunities for Production of Ingredients Based on Further Processed Fresh Mushrooms, off-Grade Mushrooms, Bi-Products, and Waste; Mushroom Council: Knoxville, TN, USA; University of Tennessee: Knoxville, TN, USA, 2006. [Google Scholar]
- Wu, T.; Zivanovic, S.; Draughon, F.A.; Sams, C.E. Chitin and Chitosan--Value-Added Products from Mushroom Waste. J. Agric. Food Chem. 2004, 52, 7905–7910. [Google Scholar] [CrossRef]
- Zicari, G.; Rivetti, D.; Soardo, V.; Cerrato, E. The Cultivation of the Mushroom Agaricus bisporus (Champignon) and Some Environmental and Health Aspects. Ig. Sanita Pubbl. 2012, 68, 435–446. [Google Scholar]
- Inácio, C.T.; Chalk, P.M.; Magalhães, A.M.T. Principles and Limitations of Stable Isotopes in Differentiating Organic and Conventional Foodstuffs: 1. Plant Products. Crit. Rev. Food Sci. Nutr. 2015, 55, 1206–1218. [Google Scholar] [CrossRef]
- Nielson, K.E.; Bowen, G.J. Hydrogen and Oxygen in Brine Shrimp Chitin Reflect Environmental Water and Dietary Isotopic Composition. Geochim. Cosmochim. Acta 2010, 74, 1812–1822. [Google Scholar] [CrossRef]
- Sodemann, H. Tropospheric Transport of Water Vapour: Lagrangian and Eulerian Perspectives; Logos Verlag: Berlin, Germany, 2006; ISBN 9783832513849. [Google Scholar]
- Reverdin, G.; Waelbroeck, C.; Pierre, C.; Akhoudas, C.; Aloisi, G.; Benetti, M.; Bourlès, B.; Danielsen, M.; Demange, J.; Diverrès, D.; et al. The CISE-LOCEAN Seawater Isotopic Database (1998–2021). Earth Syst. Sci. Data 2022, 14, 2721–2735. [Google Scholar] [CrossRef]
- Cardenas, G.; Patricia Miranda, S. FTIR and TGA studies of chitosan composite films. J. Chil. Chem. 2004, 49, 291–295. [Google Scholar] [CrossRef]
- Duarte, M.L.; Ferreira, M.C.; Marvão, M.R.; Rocha, J. An Optimised Method to Determine the Degree of Acetylation of Chitin and Chitosan by FTIR Spectroscopy. Int. J. Biol. Macromol. 2002, 31, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kaya, M.; Akata, I.; Baran, T.; Menteş, A. Physicochemical Properties of Chitin and Chitosan Produced from Medicinal Fungus (Fomitopsis pinicola). Food Biophys. 2015, 10, 162–168. [Google Scholar] [CrossRef]
- Pearson, F.G.; Marchessault, R.H.; Liang, C.Y. Infrared Spectra of Crystalline Polysaccharides. V. Chitin. J. Polym. Sci. 1960, 43, 101–116. [Google Scholar] [CrossRef]
- Khodaei, S.; Ghaedmohammadi, S.; Mohammadi, M.; Rigi, G.; Ghahremanifard, P.; Zadmard, R.; Ahmadian, G. Covalent Immobilization of Protein A on Chitosan and Aldehyde Double-Branched Chitosan as Biocompatible Carriers for Immunoglobulin G (Igg) Purification. J. Chromatogr. Sci. 2018, 56, 933–940. [Google Scholar] [CrossRef] [PubMed]
- Pawlak, A.; Mucha, M. Thermogravimetric and FTIR Studies of Chitosan Blends. Thermochim. Acta 2003, 396, 153–166. [Google Scholar] [CrossRef]
- Corazzari, I.; Nisticò, R.; Turci, F.; Faga, M.G.; Franzoso, F.; Tabasso, S.; Magnacca, G. Advanced Physico-Chemical Characterization of Chitosan by Means of TGA Coupled on-Line with FTIR and GCMS: Thermal Degradation and Water Adsorption Capacity. Polym. Degrad. Stab. 2015, 112, 1–9. [Google Scholar] [CrossRef]
- Diab, M.A.; El-Sonbati, A.Z.; Bader, D.M.D. Thermal Stability and Degradation of Chitosan Modified by Benzophenone. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2011, 79, 1057–1062. [Google Scholar] [CrossRef]
- Abdou, E.S.; Nagy, K.S.A.; Elsabee, M.Z. Extraction and Characterization of Chitin and Chitosan from Local Sources. Bioresour. Technol. 2008, 99, 1359–1367. [Google Scholar] [CrossRef]
- Szymańska, E.; Winnicka, K. Stability of Chitosan—A Challenge for Pharmaceutical and Biomedical Applications. Mar. Drugs 2015, 13, 1819–1846. [Google Scholar] [CrossRef]
- de Britto, D.; de Britto, D.; Campana-Filho, S.P. Kinetics of the Thermal Degradation of Chitosan. Thermochim. Acta 2007, 465, 73–82. [Google Scholar] [CrossRef]
- Romano, P.; Fabritius, H.; Raabe, D. The Exoskeleton of the Lobster Homarus Americanus as an Example of a Smart Anisotropic Biological Material. Acta Biomater. 2007, 3, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Coplen, T.B. Guidelines and Recommended Terms for Expression of Stable-Isotope-Ratio and Gas-Ratio Measurement Results. Rapid Commun. Mass Spectrom. 2011, 25, 2538–2560. [Google Scholar] [CrossRef] [PubMed]
- Wassenaar, L.I.; Hobson, K.A.; Sisti, L. An Online Temperature-Controlled Vacuum-Equilibration Preparation System for the Measurement of δ2H Values of Non-Exchangeable-H and of δ18O Values in Organic Materials by Isotope-Ratio Mass Spectrometry. Rapid Commun. Mass Spectrom. 2015, 29, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Wassenaar, L.I.; Hobson, K.A. Comparative Equilibration and Online Technique for Determination of Non-Exchangeable Hydrogen of Keratins for Use in Animal Migration Studies. Isot. Environ. Health Stud. 2003, 39, 211–217. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).