Three New Benzophenone Derivatives from Selaginella tamariscina
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structure Determination
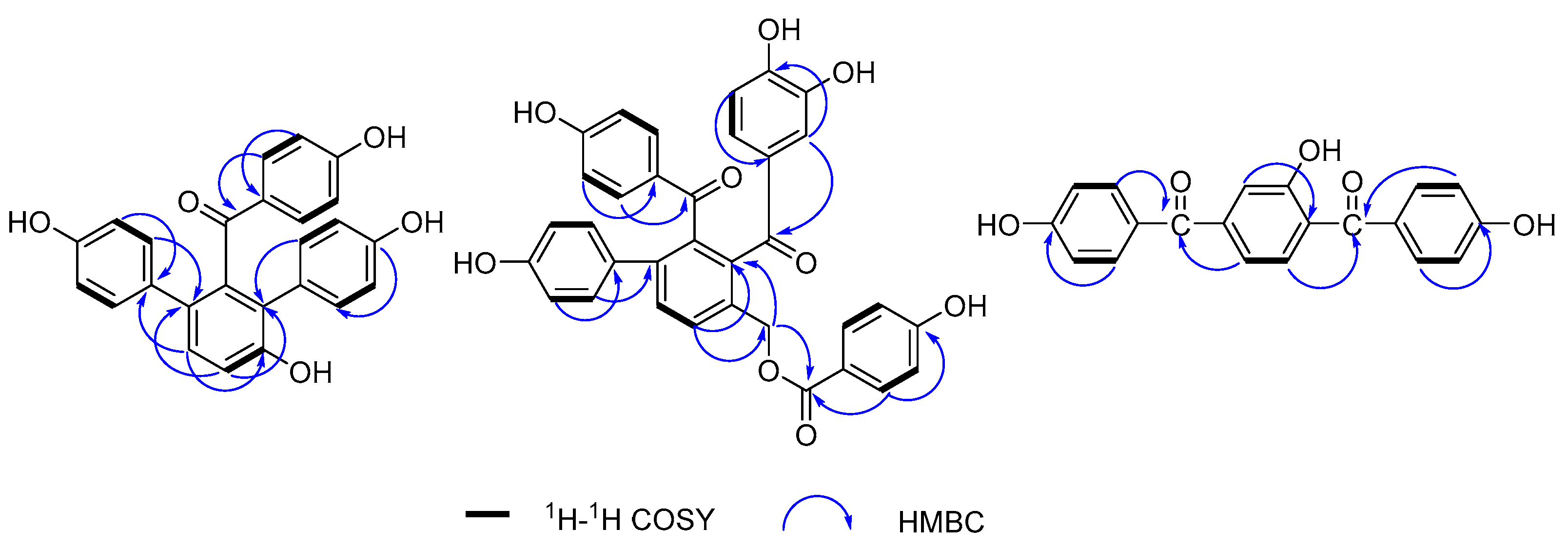
- Compound 1 was purified as a yellow amorphous powder. The molecular formula was elucidated as C25H18O5 based on its [M + H]+ quasi-molecular ion peak at 399.1243 (calcd for C25H19O5, 399.1232) in the HR–ESI–MS, which indicated 17 unsaturations. This molecular formula was consistent with the 1H and 13C NMR data (Table 1). The 1H NMR spectrum of 1 (Figure S1) exhibited signals for three para-substituted phenyls at δ 7.36 (2H, d, J = 8.7 Hz), 6.59 (2H, d, J = 8.7 Hz), δ 7.02 (2H, d, J = 8.5 Hz), 6.58 (2H, d, J = 8.5 Hz), and δ 6.96 (2H, d, J = 8.2 Hz), 6.60 (2H, d, J = 8.2 Hz), and one orthotetra-substituted phenyls at δ 7.20 (1H, d, J = 8.4 Hz), 7.05 (1H, d, J = 8.4 Hz) on the basis of 1H-1H COSY spectrum. The 13C NMR spectrum of 1 (Figure S2) showed 25 carbon resonances including the corresponding 24 aromatic carbon and one carbonyl carbon at δ 198.7 (C-7). According to these spectroscopic data compared with our reported selagibenzophenone C [20,21], compound 1 was inferred to be a benzophenone carrying two phenyl groups. The 1H-1H COSY spectrum confirmed that ring A was an orthotetra-substituted benzene ring. In the HMBC spectrum (Figure 2), the correlations were observed for H-10, 20, 24 to C-8, which indicated ring C was attached at C-8. The correlations for H-10, 14, 18 to C-12 indicated ring D was attached at C-12. The correlations for H-3, 5 to C-7 along with the weak correlation for H-11 to C-7 evidenced the benzophenone nucleus structure. Except for the above signals, the remaining hydroxyl should be located at C-9 because of the HMBC correlations for H-10, 11 to C-9. Therefore, the structure of compound 1 was elucidated and named selagibenzophenone D, and its 3D structure was shown in Figure 3. To our knowledge, compound 1 represents the second example of diarylbenzophenone from natural sources.
- Compound 2 was purified as yellow amorphous powder. The molecular formula was deduced as C34H24O9 from its [M + H]+ quasi-molecular ion peak at 577.1516 (calcd for C34H25O9, 577.1499) in the HR–ESI–MS spectrum. This molecular formula was consistent with the 1H and 13C NMR data (Table 1). The 1H NMR spectrum of 2 (Figure S7)showed two para-substituted phenyls at δ 7.35 (2H, d, J = 8.7 Hz), 6.56 (2H, J = 8.7 Hz), and δ 7.11 (2H, d, J = 8.6 Hz), 6.63 (2H, J = 8.7 Hz), and one orthotetra-substituted phenyls at δ 7.78 (2H, J = 7.9 Hz), 7.62 (2H, J = 7.9 Hz), which was confirmed by the 1H-1H COSY spectrum. These above 1H NMR spectral signals of 2 showed some similarity to those of 1 including the signals of rings A, B and C, implying the similar biphenylbenzophenone skeleton. The 1H NMR spectrum of 2 exhibited signals of two more benzene rings; one is p-hydroxyphenyl (ring E) at δ 7.60 (2H, d, J = 8.7 Hz) and 6.69 (2H, d, 8.7 Hz), and the other is a O-dihydroxyphenyl (ring D) at δ 6.84 (1H, dd, J = 8.6, 2.2 Hz), 6.66 (2H, m). One oxymethylene signal was also observed at 5.35 (2H, s), confirmed by DEPT experiment. The 13C NMR spectrum of 2 (Figure S8) showed 34 carbon resonances including the corresponding 30 aromatic carbon, three carbonyl carbon at δ 166.0 (C-28), 196.7 (C-7) and 202.2 (C-20), and one methylene carbon signal at 63.6 (C-27). In the HMBC spectrum (Figure 2), the correlations of H-21, 25 to C-20 and the weak correlations of H-10 to C-20 indicated that ring C and ring A were linked with C-20. The correlations of H-10 to C-13, 27 and H-27 to C-10, C-13 indicated that the methylene was attached to C-9. The correlations of H-29, 33 to C-28, C-31 and H-30, 32 to C-34 confirmed ring E was a p-hydroxybenzoyloxy, which was located at C-27 evidenced by the HMBC couplings of H-27 to C-28. Therefore, the structure of compound 2 was elucidated and named selagibenzophenone E, and its 3D structure was shown in Figure 4. Compound 2 possesses an unusual biphenyl-bisbenzophenone structure. It seems that compound 2 and compound 5 (selaginellin S) have a similar substitution pattern in ring A. Selaginellin S belongs to the selaginellin family with the parent nucleus structure of an alkynylphenol. Li et al. reviewed such compounds from the genus of Selaginella and summarized the proposed biosynthetic pathways [1]. These proposed that compound 2 originated from the similar precursor.
- Compound 3 was purified as yellow amorphous powder. The molecular formula of C20H14O5 was analyzed from its [M + H]+ quasi-molecular ion peak at 335.0918 (calcd for C20H15O5, 335.0919) in the HR–ESI–MS spectrum. The 1H NMR spectrum of 3 (Figure S14) exhibited signals for two para-substituted phenyls at δ 7.56 (2H, d, J = 8.5 Hz), 6.76 (2H, d, J = 8.5 Hz) and δ 7.59 (2H, d, J = 8.5 Hz), 6.78 (2H, d, J = 8.5 Hz), and one 1,2,4-trisubstituted phenyls at δ 7.53 (1H, d, J = 8.5 Hz), 7.01 (1H, dd, J = 8.5, 2.2 Hz), 6.92 (1H, d, J = 2.2 Hz), which was confirmed by the 1H-1H COSY spectrum. The 13C NMR spectrum of 3 (Figure S15) showed 20 carbon resonances including the corresponding 18 aromatic carbon and two carbonyl carbon at δ 195.3 (C-7) and 196.5 (C-14). In HMBC spectrum, the correlations of H-3, 5 to C-7 and H-12 to C-7 indicated that ring A and ring B were connected with C-7. The HMBC couplings of H-9, 11 to C-14 and H-15, 19 to C-14 indicated that ring B and ring C were linked with C-14. In addition, the HMBC couplings of H-9, 12 to C-8 defined the location of a hydroxyl at C-8. Thus, the structure of compound 3 was elucidated and given a successive name, selagibenzophenone F, and its 3D structure was shown in Figure 5.
| 1 | 2 | 3 | ||||
|---|---|---|---|---|---|---|
| Position | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | δC |
| 1 | - | 162.4 | - | 162.4 | - | 162.2 |
| 2 | 6.59 (1H, d, 8.7) | 114.2 | 6.56 (1H, d, 8.7) | 114.4 | 6.76 (1H, d, 8.5) | 114.6 |
| 3 | 7.36 (1H, d, 8.7) | 132.0 | 7.35 (1H, d, 8.7) | 132.4 | 7.56 (1H, d, 8.5) | 132.2 |
| 4 | - | 131.8 | - | 128.8 | - | 128.8 |
| 5 | 7.36 (1H, d, 8.7) | 132.0 | 7.35 (1H, d, 8.7) | 132.4 | 7.56 (1H, d, 8.5) | 132.2 |
| 6 | 6.59 (1H, d, 8.7) | 114.2 | 6.56 (1H, d, 8.7) | 114.4 | 6.76 (1H, d, 8.5) | 114.6 |
| 7 | - | 198.7 | - | 196.7 | - | 195.3 |
| 8 | - | 126.6 | - | 137.9 | - | 160.0 |
| 9 | - | 153.2 | - | 137.4 | 6.92 (1H, d, 2.2) | 115.6 |
| 10 | 7.05 (1H, d, 8.4) | 115.7 | 7.62 (1H, d, 7.9) | 130.6 | - | 143.2 |
| 11 | 7.20 (1H, d, 8.4) | 129.4 | 7.78 (1H, d, 7.9) | 130.8 | 7.01 (1H, dd, 8.5, 2.2) | 115.6 |
| 12 | - | 131.4 | - | 141.0 | 7.53 (1H, d, 8.5) | 132.2 |
| 13 | - | 140.2 | - | 132.2 | - | 130.2 |
| 14 | 6.96 (1H, d, 8.2) | 131.5 | 7.11 (1H, d, 8.6) | 130.0 | - | 196.5 |
| 15 | 6.60 (1H, d, 8.2) | 114.4 | 6.63 (1H, d, 8.6) | 114.8 | 7.59 (1H, 8.5) | 132.0 |
| 16 | - | 156.0 | - | 157.0 | 6.78 (1H, 8.5) | 114.6 |
| 17 | 6.60 (1H, d, 8.2) | 114.4 | 6.63 (1H, d, 8.6) | 114.8 | - | 162.5 |
| 18 | 6.96 (1H, d, 8.2) | 131.5 | 7.11 (1H, d, 8.6) | 130.0 | 6.78 (1H, 8.5 | 114.6 |
| 19 | - | 126.8 | - | 137.5 | 7.59 (1H, 8.5) | 132.0 |
| 20 | 7.02 (1H, d, 8.5) | 130.0 | - | 202.2 | - | 129.2 |
| 21 | 6.58 (1H, d, 8.5) | 114.0 | 6.66 (1H, m) | 117.8 | ||
| 22 | - | 155.9 | - | 148.9 | ||
| 23 | 6.58 (1H, d, 8.5) | 114.0 | - | 155.2 | ||
| 24 | 7.02 (1H, d, 8.5) | 130.0 | 6.66 (1H, m) | 117.2 | ||
| 25 | - | 129.8 | 6.84 (1H, dd, 8.6, 2.2) | 124.9 | ||
| 26 | - | 120.2 | ||||
| 27 | 5.35 (2H, s) | 63.6 | ||||
| 28 | - | 166.0 | ||||
| 29 | 7.60 (1H, d, 8.7) | 131.4 | ||||
| 30 | 6.69 (1H, d, 8.7) | 114.6 | ||||
| 31 | - | 162.1 | ||||
| 32 | 6.69 (1H, d, 8.7) | 114.6 | ||||
| 33 | 7.60 (1H, d, 8.7) | 131.4 | ||||
| 34 | - | 120.0 | ||||



2.2. Cytotoxic Effects against Cancer Cells
2.3. NO Inhibitory Activities
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Plant Material
3.3. Extraction and Isolation
- Selagibenzophenone D (1): Yellow powder. UV (MeOH) λmax (nm; log ε): 276 (4.61). 1H NMR and 13C NMR (MeOH-d4) see Table 1; HR-ESI-MS calcd for C25H19O5 [M + H]+ 399.1243; found 399.1232.
- Selagibenzophenone E (2): Yellow powder. UV (MeOH) λmax (nm; log ε): 264 (4.26). 1H NMR and 13C NMR (MeOH-d4) see Table 1; HR-ESI-MS calcd for C34H25O9 [M + H]+ 577.1516; found 577.1499.
- Selagibenzophenone F (3): Yellow powder. UV (MeOH) λmax (nm; log ε): 290 (4.22). 1H NMR and 13C NMR (MeOH-d4) see Table 1; HR-ESI-MS calcd for C20H15O5 [M + H]+ 335.0918; found 335.0919.
- Selaginellin H (4): Light yellow amorphous powder. UV (MeOH) λmax (nm): 226, 264, 334. 1H NMR (MeOH-d4, 400 MHz): δH 7.86 (1H, d, J = 8.0 Hz, H-16), 7.58 (1H, d, J = 8.0 Hz, H-17), 6.83 (4H, d, J = 8.5 Hz, H-3, 5, 8, 12), 6.64 (4H, d, J = 8.5 Hz, H-2, 6, 9, 11), 6.51 (2H, d, J = 8.5 Hz, H-20, 24), 6.47 (2H, d, J = 8.5 Hz, H-21, 23), 5.20 (2H, s, H-26). 13C NMR (MeOH-d4, 100 MHz): δC 170.6 (C-27), 157.4 (C-1, 10), 156.6 (C-22), 151.1 (C-19), 141.5 (C-15), 138.1 (C-18), 137.5 (C-17), 130.5 (C-4, 20, 24), 130.2 (C-3, 5), 129.9 (C-8, 12), 129.8 (C-13), 129.5 (C-16), 129.4 (C-25), 122.0 (C-14), 114.1 (C-2, 6, 9, 11, 21, 23), 93.7 (C-7), 59.3 (C-26).
- Selaginellin S (5): Yellow powder. UV (MeOH) λmax (nm): 280. 1H NMR (MeOH-d4, 400 MHz): δH 7.70 (1H, d, J = 8.0 Hz, H-10), 7.64 (2H, d, J = 8.5 Hz, H-3, 5), 7.42 (1H, d, J = 8.0 Hz, H-11), 7.11 (2H, d, J = 8.0 Hz, H-14, 18), 6.98 (2H, dd, J = 7.5, 2.5 Hz, H-22, 26), 6.78 (2H, d, J = 8.5 Hz, H-2, 6), 6.67 (2H, dd, J = 7.5, 2.5 Hz, H-23, 25), 6.66 (1H, d, J = 8.0 Hz, H-15, 17), 4.89 (2H, s, H-28). 13C NMR (MeOH-d4, 100 MHz): δC 197.9 (C-7), 162.9 (C-1), 158.0 (C-24), 156.7 (C-16), 141.3 (C-9), 141.0 (C-13), 138.9 (C-12), 132.4 (C-22, 26), 132.2 (C-3, 5), 130.8 (C-19), 129.8 (C-14, 18), 129.3 (C-11), 129.1 (C-4), 126.8 (C-10), 119.0 (C-8), 115.0 (C-2, 6), 114.9 (C-23, 25), 114.6 (C-15, 17), 113.2 (C-27), 99.2 (C-21), 82.5 (C-20), 61.7 (C-28).
- Unciflavone D (6): Light yellow amorphous powder. UV (MeOH) λmax (nm): 226, 264, 324. 1H NMR (MeOH-d4, 400 MHz): δH 8.00 (1H, dd, J = 8.5, 2.5 Hz, H-4″), 7.98 (1H, d, J = 2.5 Hz, H-6″), 7.54 (2H, d, J = 8.5 Hz, H-2′, 6′), 7.02 (1H, d, J = 8.5 Hz, H-3″), 6.77 (2H, d, J = 8.5 Hz, H-3′, 5′), 6.62 (2H, s, H-3), 6.37 (1H, s, H-6). 13C NMR (MeOH-d4, 100 MHz): δC 182.9 (C-4), 165.6 (C-7″), 164.7 (C-2), 162.7 (C-7), 161.2 (C-2″), 160.8 (C-5), 156.1 (C-1″), 159.1 (C-9), 134.8 (C-6″), 130.8 (C-4″), 128.0 (C-2′, 6′), 121.9 (C-5″), 121.8 (C-1′), 118.8 (C-4′), 115.4 (C-3′, 5′), 114.8 (C-3″), 104.9 (C-8), 103.9 (C-10), 101.8 (C-3), 98.8 (C-6).
3.4. Cytotoxicity Assay
3.5. Bioassay for NO Inhibitory Activities
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Li, W.; Tang, G.H.; Yin, S. Selaginellins from the genus Selaginella: Isolation, structure, biological activity, and synthesis. Nat. Prod. Rep. 2021, 38, 822–842. [Google Scholar] [CrossRef] [PubMed]
- Okigawa, M.; Hwa, C.W.; Kawano, N.; Rahman, W. Biflavones in Selaginella species. Phytochemistry 1971, 10, 3286–3287. [Google Scholar] [CrossRef]
- Ha, L.M.; Thao, D.T.; Huong, H.T.; Minh, C.V.; Dat, N.T. Toxicity and anticancer effects of an extract from Selaginella tamariscina on a mice model. Nat. Prod. Res. 2011, 26, 1130–1134. [Google Scholar] [CrossRef]
- Yin, D.; Li, J.; Lei, X.; Liu, Y.; Yang, Z.; Chen, K. Antiviral activity of total flavonoid extracts from Selaginella moellendorffii Hieron against coxsackie virus B3 in vitro and in vivo. Evid.-Based Complement. Altern. Med. 2014, 2014, 950817. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Chen, K.-L.; Zhang, G.-L.; Deng, G.-R.; Li, J. Pharmacological Basis for Use of Selaginella moellendorffii in Gouty Arthritis: Antihyperuricemic, Anti-Inflammatory, and Xanthine Oxidase Inhibition. Evid.-Based Complement. Altern. Med. 2017, 2017, 2103254. [Google Scholar] [CrossRef]
- Zeng, W.; Yao, C.P.; Xu, P.S.; Zhang, G.G.; Liu, Z.Q.; Xu, K.P.; Zou, Z.X.; Tan, G.S. A new neolignan from Selaginella moellendorffii Hieron. Nat. Prod. Res. 2017, 31, 2223–2227. [Google Scholar] [CrossRef]
- Nguyen, P.-H.; Zhao, B.-T.; Ali, Y.; Choi, J.-S.; Rhyu, D.-Y.; Min, B.-S.; Woo, M.-H. Insulin-mimetic selaginellins from Selaginella tamariscina with protein tyrosine phosphatase 1B (PTP1B) inhibitory activity. J. Nat. Prod. 2015, 78, 34–42. [Google Scholar] [CrossRef]
- Chinese Pharmacopoeia Commission. Chinese Pharmacopoeia; China Medical Science Press: Beijing, China, 2020; Volume 1, p. 226. [Google Scholar]
- Jung, Y.-J.; Lee, E.H.; Lee, C.G.; Rhee, K.-J.; Jung, W.-S.; Choi, Y.; Pan, C.-H.; Kang, K. AKR1B10-inhibitory Selaginella tamariscina extract and amentoflavone decrease the growth of A549 human lung cancer cells in vitro and in vivo. J. Ethnopharmacol. 2017, 202, 78–84. [Google Scholar] [CrossRef]
- Zhang, J.; Li, A.; Sun, H.; Xiong, X.; Qin, S.; Wang, P.; Dai, L.; Zhang, Z.; Li, X.; Liu, Z. Amentoflavone triggers cell cycle G2/M arrest by interfering with microtubule dynamics and inducing DNA damage in SKOV3 cells. Oncol. Lett. 2020, 20, 168. [Google Scholar] [CrossRef]
- Liu, H.; Yue, Q.; He, S. Amentoflavone suppresses tumor growth in ovarian cancer by modulating Skp2. Life Sci. 2017, 189, 96–105. [Google Scholar] [CrossRef]
- Huang, W.; Liu, C.; Liu, F.; Liu, Z.; Lai, G.; Yi, J. Hinokiflavone induces apoptosis and inhibits migration of breast cancer cells via EMT signalling pathway. Cell Biochem. Funct. 2020, 38, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Kedi, P.B.E.; Meva, F.E.A.; Kotsedi, L.; Nguemfo, E.L.; Zangueu, C.B.; Ntoumba, A.A.; Mohamed, H.E.A.; Dongmo, A.B.; Maaza, M. Eco-friendly synthesis, characterization, in vitro and in vivo anti-inflammatory activity of silver nanoparticle-mediated Selaginella myosurus aqueous extract. Int. J. Nanomed. 2018, 13, 8537–8548. [Google Scholar] [CrossRef] [PubMed]
- Won, A.-N.; Kim, S.A.; Ahn, J.Y.; Han, J.-H.; Kim, C.-H.; Lee, J.-H.; Kim, D.-I. HO-1 Induction by Selaginella tamariscina Extract Inhibits Inflammatory Response in Lipopolysaccharide-Stimulated RAW 264.7 Macrophages. Evid.-Based Complement. Altern. Med. 2018, 2018, 7816923. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C. The traditional and modern uses of Selaginella tamariscina (P.Beauv.) Spring, in medicine and cosmetic: Applications and bioactive ingredients. J. Ethnopharmacol. 2021, 280, 114444. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.; Kang, K.B.; Kim, J.; Sung, S.H. Molecular Networking Reveals the Chemical Diversity of Selaginellin Derivatives, Natural Phosphodiesterase-4 Inhibitors from Selaginella tamariscina. J. Nat. Prod. 2019, 82, 1820–1830. [Google Scholar] [CrossRef] [PubMed]
- Long, H.-P.; Liu, J.; Xu, P.-S.; Xu, K.-P.; Li, J.; Tan, G.-S. Hypoglycemic flavonoids from Selaginella tamariscina (P.Beauv.) Spring. Phytochemistry 2022, 195, 113073. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Yan, X.-J.; Nie, C.-D.; Wang, Q.-X.; Li, W.-L. Two new flavonol glycosides from Selaginella tamariscina. J. Asian Nat. Prod. Res. 2022, 24, 496–502. [Google Scholar] [CrossRef]
- Shim, S.Y.; Lee, S.G.; Lee, M. Biflavonoids Isolated from Selaginella tamariscina and Their Anti-Inflammatory Activities via ERK 1/2 Signaling. Molecules 2018, 23, 926. [Google Scholar] [CrossRef]
- Woo, S.; Chae, H.-S.; Kim, J.; Chin, Y.-W. Selaginellin Derivatives from Selaginella tamariscina and Their Upregulating Effects on Low-Density Lipoprotein Receptor Expression. J. Nat. Prod. 2021, 84, 857–864. [Google Scholar] [CrossRef]
- Zhu, Q.-F.; Shao, L.-D.; Wu, X.-D.; Liu, J.-X.; Zhao, Q.-S. Isolation, Structural Assignment of Isoselagintamarlin A from Selaginella tamariscina and Its Biomimetic Synthesis. Nat. Prod. Bioprospect. 2019, 9, 69–74. [Google Scholar] [CrossRef]
- Dat, L.D.; Zhao, B.T.; Hung, N.D.; Lee, J.H.; Min, B.S.; Woo, M.H. Lignan derivatives from Selaginella tamariscina and their nitric oxide inhibitory effects in LPS-stimulated RAW 264.7 cells. Bioorg. Med. Chem. Lett. 2016, 27, 524–529. [Google Scholar] [CrossRef] [PubMed]
- Sales, L.; Pezuk, J.A.; Borges, K.S.; Brassesco, M.S.; Scrideli, C.A.; Tone, L.G.; dos Santos, M.H.; Ionta, M.; de Oliveira, J.C. Anticancer activity of 7-epiclusianone, a benzophenone from Garcinia brasiliensis, in glioblastoma. BMC Complement. Altern. Med. 2015, 15, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Santa-Cecília, F.V.; Freitas, L.A.; Vilela, F.C.; Veloso, C.D.C.; da Rocha, C.Q.; Moreira, M.E.; Dias, D.F.; Giusti-Paiva, A.; dos Santos, M.H. Antinociceptive and anti-inflammatory properties of 7-epiclusianone, a prenylated benzophenone from Garcinia brasiliensis. Eur. J. Pharmacol. 2011, 670, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tang, G.-H.; Weng, H.-Z.; Zhang, J.-S.; Xu, Y.-K.; Yin, S. A new selaginellin derivative and a new triarylbenzophenone analog from the whole plant of Selaginella pulvinata. J. Asian Nat. Prod. Res. 2017, 20, 1123–1128. [Google Scholar] [CrossRef]
- Liu, R.; Zou, H.; Zou, Z.-X.; Cheng, F.; Yu, X.; Xu, P.-S.; Li, X.-M.; Li, D.; Xu, K.-P.; Tan, G.-S. Two new anthraquinone derivatives and one new triarylbenzophenone analog from Selaginella tamariscina. Nat. Prod. Res. 2020, 34, 2709–2714. [Google Scholar] [CrossRef]
- Chen, W.; Peng, Y.; Huang, W.; Zhou, L.; Quan, X.; Zhao, Q.; Zhang, D.; Sheng, X.; Luo, Y.; Zou, H. A New Diarylbenzophenone from Selaginella tamariscina. Rec. Nat. Prod. 2020, 14, 421–426. [Google Scholar] [CrossRef]
- Chu, P.; Wang, S.; Zhu, X.; Yang, Y.; Li, H.; Tesfaldet, T.; Shopit, A.; Yang, Y.; Ma, X.; Peng, J.; et al. Selaginellin B induces apoptosis and autophagy in pancreatic cancer cells via the JAK2/STAT3 signaling pathway. Am. J. Transl. Res. 2020, 12, 7127–7143. [Google Scholar]
- Liu, X.; Luo, H.-B.; Huang, Y.-Y.; Bao, J.-M.; Tang, G.-H.; Chen, Y.-Y.; Wang, J.; Yin, S. Selaginpulvilins A-D, new phosphodiesterase-4 inhibitors with an unprecedented skeleton from Selaginella pulvinata. Org. Lett. 2014, 16, 282–285. [Google Scholar] [CrossRef]
- Cao, Y.; Chen, J.J.; Tan, N.H.; Wu, Y.P.; Yang, J.; Wang, Q. Structure determination of selaginellins G and H from Selaginella pulvinata by NMR spectroscopy. Magn. Reson. Chem. 2010, 48, 656–659. [Google Scholar] [CrossRef]
- Cao, Y.; Wu, Y.-P.; Duan, J.-A. A new selaginellin derivative from Selaginella pulvinata. Yao Xue Xue Bao 2015, 50, 199–202. [Google Scholar]
- Zou, H.; Xu, K.-P.; Li, F.-S.; Zou, Z.-X.; Liu, R.; Li, J.; Tan, L.-H.; Tan, G.-S. Unciflavones A–F, six novel flavonoids from Selaginella uncinata (Desv.) Spring. Fitoterapia 2014, 99, 328–333. [Google Scholar] [CrossRef] [PubMed]


| Compounds | IC50 (μM) | |
|---|---|---|
| HepG2 | SMCC-7721 | |
| 1 | >80 | >80 |
| 2 | 32.575 | 15.816 |
| 3 | >80 | >80 |
| 4 | 40.928 | >80 |
| 5 | 61.521 | >80 |
| 6 | >80 | >80 |
| sorafenib | 4.796 | 2.089 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Long, J.; Mao, Q.; Peng, Y.; Liu, L.; Hong, Y.; Xiang, H.; Ma, M.; Zou, H.; Kuang, J. Three New Benzophenone Derivatives from Selaginella tamariscina. Molecules 2023, 28, 4582. https://doi.org/10.3390/molecules28124582
Long J, Mao Q, Peng Y, Liu L, Hong Y, Xiang H, Ma M, Zou H, Kuang J. Three New Benzophenone Derivatives from Selaginella tamariscina. Molecules. 2023; 28(12):4582. https://doi.org/10.3390/molecules28124582
Chicago/Turabian StyleLong, Jiayin, Qingqing Mao, Yujie Peng, Lei Liu, Yin Hong, Honglin Xiang, Ming Ma, Hui Zou, and Junwei Kuang. 2023. "Three New Benzophenone Derivatives from Selaginella tamariscina" Molecules 28, no. 12: 4582. https://doi.org/10.3390/molecules28124582




