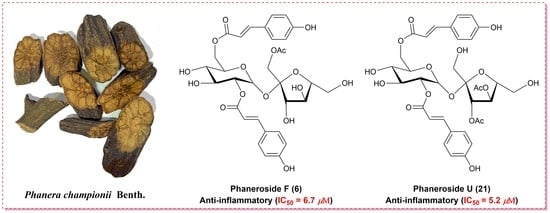
Phanerosides A–X, Phenylpropanoid Esters of Sucrose from the Rattans of Phanera championii Benth
Abstract
:1. Introduction
2. Results and Discussion
Structural Elucidation
- In Vitro Anti-inflammatory Effects of Compounds 1–24
- b.
- Antioxidant Effects of Compounds 1–24
3. Materials and Methods
General Experimental Procedures
- Plant Material
- b.
- Extraction and Isolation
- c.
- Physicochemical Properties and Spectroscopic Data of Compounds 1–24
- d.
- Acid Hydrolysis of Compound 1
- e.
- NO Production Measurements and Cell Viability Assays
- f.
- Antioxidant Activity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Zhao, W.; Huang, X.X.; Yu, L.H.; Liu, Q.B.; Li, L.Z.; Sun, Q.; Song, S.J. Tomensides A–D, new antiproliferative phenylpropanoid sucrose esters from Prunus tomentosa leaves. Bioorg. Med. Chem. Lett. 2014, 24, 2459–2462. [Google Scholar] [CrossRef] [PubMed]
- Vargas, J.A.M.; Ortega, J.O.; Metzker, G.; Larrahondo, J.E.; Boscolo, M. Natural sucrose esters: Perspectives on the chemical and physiological use of an under investigated chemical class of compounds. Phytochemistry 2020, 177, 112433. [Google Scholar] [CrossRef] [PubMed]
- Panda, P.; Appalashetti, M.; Judeh, Z.M.A. Phenylpropanoid sucrose esters: Plant-derived natural products as potential leads for new therapeutics. Curr. Med. Chem. 2011, 18, 3234–3251. [Google Scholar] [CrossRef]
- Kim, D.; Wang, C.Y.; Hu, R.; Lee, J.Y.; Luu, T.T.T.; Park, H.J.; Lee, S.K. Antitumor activity of vanicoside B isolated from Persicaria dissitiflora by targeting CDK8 in triple-negative breast cancer cells. J. Nat. Prod. 2019, 82, 3140–3149. [Google Scholar] [CrossRef] [PubMed]
- Panda, P.; Appalashetti, M.; Natarajan, M.; Chan-Park, M.B.; Venkatraman, S.S.; Judeh, Z.M. Synthesis and antitumor activity of lapathoside D and its analogs. A. Eur. J. Med. Chem. 2012, 53, 1–12. [Google Scholar] [CrossRef]
- Takasaki, M.; Kuroki, S.; Kozuka, M.; Konoshima, T. New phenylpropanoid esters of sucrose from Polygonum lapathifolium. J. Nat. Prod. 2001, 64, 1305–1308. [Google Scholar] [CrossRef]
- Chang, C.L.; Zhang, L.J.; Chen, R.Y.; Kuo, L.M.Y.; Huang, J.P.; Huang, H.C.; Lee, K.H.; Wu, Y.C.; Kuo, Y.H. Antioxidant and anti-inflammatory phenylpropanoid derivatives from Calamus quiquesetinervius. J. Nat. Prod. 2010, 73, 1482–1488. [Google Scholar] [CrossRef]
- Wang, N.; Yao, X.; Ishii, R.; Kitanaka, S. Bioactive sucrose esters from Bidens parviflora. Phytochemistry 2003, 62, 741–746. [Google Scholar] [CrossRef]
- Zhang, L.; Liao, C.C.; Huang, H.C.; Shen, Y.C.; Yang, L.M.; Kuo, Y.H. Antioxidant phenylpropanoid glycosides from Smilax bracteate. Phytochemistry 2008, 69, 1398–1404. [Google Scholar] [CrossRef]
- Qian-Cutrone, J.; Huang, S.; Trimble, J.; Li, H.; Lin, P.F.; Alam, M.; Klohr, S.E.; Kadow, K.F. Niruriside, a new HIV REV/RRE binding inhibitor from Phyllanthus niruri. J. Nat. Prod. 1996, 59, 196–199. [Google Scholar] [CrossRef]
- Daude, D.; Remaud-Simeon, M.; Andre, I. Sucrose analogs: An attractive (bio)source for glycodiversification. Nat. Prod. Rep. 2012, 29, 945–960. [Google Scholar] [CrossRef] [PubMed]
- Ong, L.L.; Wong, P.W.K.; Raj, S.D.; Khong, D.T.; Panda, P.; Santoso, M.; Judeh, Z.M.A. An orthogonal approach for the precise synthesis of phenylpropanoid sucrose esters. New J. Chem. 2022, 46, 9710–9717. [Google Scholar] [CrossRef]
- Montanha, G.S.; Romeu, S.L.Z.; Marques, J.P.R.; Rohr, L.A.; de Almeida, E.; dos Reis, A.R.; Linhares, F.S.; Sabatini, S.; de Carvalho, H.W.P. Microprobe-XRF assessment of nutrient distribution in soybean, cowpea, and kidney bean seeds: A Fabaceae family case study. ACS Agric. Sci. Technol. 2022, 2, 1318–1324. [Google Scholar] [CrossRef]
- Maroyi, A. Medicinal uses of the Fabaceae family in Zimbabwe: A review. Plants 2023, 12, 1255. [Google Scholar] [CrossRef]
- Wang, M.; Huang, S.; Li, M.; Mckey, D.; Zhang, L. Staminodes influence pollen removal and deposition rates in nectar-rewarding self-incompatible Phanera yunnanensis (Caesalpinioideae). J. Trop. Ecol. 2019, 35, 34–42. [Google Scholar] [CrossRef]
- Xu, W.; Chu, K.; Li, H.; Zhang, Y.; Zheng, H.; Chen, R.; Chen, L. Ionic liquid-based microwave-assisted extraction of flavonoids from Bauhinia championii (Benth.) Benth. Molecules 2012, 17, 14323–14335. [Google Scholar] [CrossRef] [Green Version]
- Qin, X.Y.; Luo, J.Y.; Gao, Z.G. Yao Ethnic Medicinals in China; Ethnic Publish House: Beijing, China, 2002; p. 57. [Google Scholar]
- Xu, W.; Huang, M.; Zhang, Y.; Li, H.; Zheng, H.; Yu, L.; Chu, K. Extracts of Bauhinia championii (Benth.) Benth. inhibit NF-<kappa>B-signaling in a rat model of collagen-induced arthritis and primary synovial cells. J. Ethnopharmacol. 2016, 185, 140–146. [Google Scholar]
- Chen, C.C.; Chen, Y.P.; Hsu, H.Y.; Lee, K.H.; Tani, S.; McPhail, A.T. Bauhinin, a new nitrile glucoside from Bauhinia championii. J. Nat. Prod. 1985, 48, 933–937. [Google Scholar] [CrossRef]
- Hua, L.P.; Zhang, Y.Q.; Ye, M.; Xu, W.; Wang, X.Y.; Fu, Y.H.; Xu, W. Bioactive dibenzofurans from the rattans of Bauhinia championii (Benth.) Benth. Phytochem. Lett. 2018, 24, 154–157. [Google Scholar] [CrossRef]
- Hua, L.P.; Zhang, Y.Q.; Ye, M.; Xu, W.; Wang, X.Y.; Fu, Y.H.; Xu, W. A new polyoxygenated abietane diterpenoid from the rattans of Bauhinia championii (Benth.) Benth. Nat. Prod. Res. 2018, 32, 2577–2582. [Google Scholar] [CrossRef]
- Zhu, F.; Du, B.; Xu, B. Anti-inflammatory effects of phytochemicals from fruits, vegetables, and food legumes: A review. Crit. Rev. Food Sci. 2018, 58, 1260–1270. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, N.; Li, X.; Chen, G.; Wang, C.; Lin, B.; Hou, Y. Characteristic α-acid derivatives from Humulus lupulus with antineuroinflammatory activities. J. Nat. Prod. 2017, 80, 3081–3092. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.N.; Su, B.J.; Wang, Y.Q.; Liao, H.B.; Chen, Z.F.; Liang, D. Isolation, absolute configuration, and biological activities of chebulic acid and brevifolincarboxylic acid derivatives from Euphorbia hirta. J. Nat. Prod. 2020, 83, 985–995. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Su, B.J.; Hu, Y.J.; Liu, J.L.; Li, H.; Wang, Y.Q.; Liao, H.B.; Liang, D. Piperhancins A and B, two pairs of antineuroinflammatory cycloneolignane enantiomers from Piper hancei. J. Org. Chem. 2021, 86, 5284–5291. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.M.; Li, Y.H.; Hua, J.; Huang, F.P.; Wang, H.S.; Liang, D. Antiviral matrine-type alkaloids from the rhizomes of Sophora tonkinensis. J. Nat. Prod. 2015, 78, 1683−1688. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.G.; Ren, Y.J.; Liu, Y.L.; Wang, M.N.; He, C.; Chen, X.; Fan, X.L.; Zhang, Y.L.; Hao, Z.Y.; Li, H.W.; et al. Iridoid glycosides and lignans from the fruits of Gardenia jasminoides Eills. Phytochemistry 2021, 190, 112893. [Google Scholar] [CrossRef]







| Position | 1 a | 2 b | 3 a | 4 a | 5 a |
|---|---|---|---|---|---|
| 1 | 3.51, d (12.0) 3.31, d (12.0) | 3.51, d (12.0) 3.31, overlapped | 3.52, d (12.0) 3.32, d (12.0) | 3.51, d (12.0) 3.30, d (12.0) | 3.51, d (11.6) 3.31, overlapped |
| 3 | 4.22, d (8.8) | 4.22, d (9.0) | 4.22, d (8.8) | 4.22, d (8.4) | 4.22, d (8.8) |
| 4 | 4.04, t (8.8) | 4.04, t (9.0) | 4.04, t (8.8) | 4.07, dd (9.2, 8.4) | 4.07, t (8.8) |
| 5 | 3.79, m | 3.78, m | 3.79, m | 3.79, m | 3.79, m |
| 6 | 3.88, dd (12.0, 7.2) 3.77, m | 3.88, dd (12.0, 7.2) 3.77, dd (12.0, 2.4) | 3.88, m 3.77, m | 3.88, m 3.77, m | 3.89, overlapped 3.78, m |
| 1′ | 5.60, d (3.6) | 5.60, d (3.6) | 5.60, d (4.0) | 5.60, d (3.6) | 5.61, d (3.6) |
| 2′ | 4.73, dd (10.0, 3.6) | 4.73, dd (9.6, 3.6) | 4.73, dd (10.0, 4.0) | 4.73, dd (9.6, 3.6) | 4.73, dd (9.6, 3.6) |
| 3′ | 4.01, dd (10.0, 9.2) | 4.01, t (9.6) | 4.01, t (10.0) | 4.01, t (9.6) | 4.01, t (9.6) |
| 4′ | 3.45, t (9.2) | 3.46, t (9.6) | 3.46, t (10.0) | 3.45, t (9.6) | 3.45, t (9.6) |
| 5′ | 4.19, ddd (9.2, 6.0, 2.0) | 4.20, m | 4.18, dd (10.0, 6.0, 2.0) | 4.20, m | 4.20, m |
| 6′ | 4.54, dd (12.0, 2.0) 4.31, dd (12.0, 6.0) | 4.56, br d (12.0) 4.34, dd (12.0, 6.6) | 4.54, dd (12.0, 2.0) 4.31, dd (12.0, 6.0) | 4.54, d (12.0) 4.31, dd (12.0, 6.4) | 4.54, br d (12.0) 4.31, dd (12.0, 6.4) |
| 2″ | 7.49, d (8.8) | 7.49, d (8.4) | 7.21, d (2.0) | 7.48, d (8.4) | 7.21, br s |
| 3″ | 6.82, d (8.8) | 6.81, d (8.4) | 6.81, d (8.4) | ||
| 5″ | 6.82, d (8.8) | 6.81, d (8.4) | 6.82, d (8.0) | 6.81, d (8.4) | 6.82, d (8.4) |
| 6″ | 7.49, d (8.8) | 7.49, d (8.4) | 7.11, dd (8.0, 2.0) | 7.48, d (8.4) | 7.11, br d (8.4) |
| 7″ | 7.72, d (16.0) | 7.72, d (16.2) | 7.72, d (16.0) | 7.72, d (16.0) | 7.72, d (16.0) |
| 8″ | 6.39, d (16.0) | 6.39, d (16.2) | 6.42, d (16.0) | 6.39, d (16.0) | 6.42, d (16.0) |
| 2‴ | 7.51, d (8.8) | 7.65, m | 7.50, d (8.4) | 7.25, br s | 7.25, br s |
| 3‴ | 6.81, d (8.8) | 7.41, overlapped | 6.81, d (8.4) | ||
| 4‴ | 7.41, overlapped | ||||
| 5‴ | 6.81, d (8.8) | 7.41, overlapped | 6.81, d (8.4) | 6.81, d (8.4) | 6.81, d (8.4) |
| 6‴ | 7.51, d (8.8) | 7.65, m | 7.50, d (8.4) | 7.10, br d (8.4) | 7.11, br d (8.4) |
| 7‴ | 7.67, d (16.0) | 7.75, d (16.2) | 7.67, d (16.0) | 7.65, d (16.0) | 7.66, d (16.0) |
| 8‴ | 6.43, d (16.0) | 6.64, d (16.2) | 6.43, d (16.0) | 6.46, d (16.0) | 6.46, d (16.0) |
| 3″-OMe | 3.90, s | 3.90, s | |||
| 3‴-OMe | 3.89, s | 3.90, s |
| Position | 1 a | 2 b | 3 a | 4 a | 5 a |
|---|---|---|---|---|---|
| 1 | 62.9 | 62.9 | 62.9 | 62.9 | 63.0 |
| 2 | 105.7 | 105.7 | 105.7 | 105.6 | 105.7 |
| 3 | 77.0 | 77.0 | 77.0 | 77.0 | 77.0 |
| 4 | 75.6 | 75.6 | 75.6 | 75.7 | 75.7 |
| 5 | 84.0 | 84.0 | 84.0 | 84.0 | 84.0 |
| 6 | 64.2 | 64.2 | 64.2 | 64.2 | 64.2 |
| 1′ | 90.6 | 90.6 | 90.6 | 90.5 | 90.5 |
| 2′ | 74.4 | 74.4 | 74.4 | 74.4 | 74.4 |
| 3′ | 71.9 | 71.9 | 71.9 | 72.0 | 72.0 |
| 4′ | 72.0 | 72.0 | 72.0 | 72.0 | 72.0 |
| 5′ | 71.9 | 71.9 | 71.9 | 71.9 | 71.9 |
| 6′ | 65.0 | 65.2 | 65.0 | 65.0 | 65.1 |
| 1″ | 127.1 | 127.0 | 127.6 | 127.0 | 127.7 |
| 2″ | 131.3 | 131.3 | 111.8 | 131.3 | 111.8 |
| 3″ | 116.9 | 116.9 | 149.4 | 116.9 | 149.4 |
| 4″ | 161.6 | 161.7 | 151.0 | 161.6 | 151.0 |
| 5″ | 116.9 | 116.9 | 116.5 | 116.9 | 116.5 |
| 6″ | 131.3 | 131.3 | 124.3 | 131.3 | 124.3 |
| 7″ | 147.5 | 147.5 | 147.8 | 147.5 | 147.8 |
| 8″ | 114.7 | 114.7 | 114.9 | 114.7 | 115.0 |
| 9″ | 168.8 | 168.8 | 168.8 | 168.8 | 168.8 |
| 1‴ | 127.0 | 135.8 | 127.1 | 127.7 | 127.6 |
| 2‴ | 131.3 | 129.4 | 131.3 | 111.6 | 111.6 |
| 3‴ | 116.8 | 130.0 | 116.8 | 149.4 | 149.4 |
| 4‴ | 161.4 | 131.6 | 161.4 | 150.7 | 150.8 |
| 5‴ | 116.8 | 130.0 | 116.8 | 116.4 | 116.4 |
| 6‴ | 131.3 | 129.4 | 131.3 | 124.3 | 124.3 |
| 7‴ | 146.9 | 146.7 | 146.9 | 147.1 | 147.1 |
| 8‴ | 114.9 | 118.7 | 115.0 | 115.2 | 115.2 |
| 9‴ | 169.2 | 168.5 | 169.2 | 169.1 | 169.1 |
| 3″-OMe | 56.4 | 56.5 | |||
| 3‴-OMe | 56.5 | 56.5 |
| Position | 6 | 7 | 8 | 9 | 10 | 11 |
|---|---|---|---|---|---|---|
| 1 | 4.11, d (11.6) 4.06, d (11.6) | 3.54, d (12.0) 3.35, d (12.0) | 3.57, d (12.0) 3.40, d (12.0) | 4.11, d (12.0) 4.06, d (12.0) | 3.54, d (12.0) 3.34, d (12.0) | 3.57, br d (12.0) 3.40, br d (12.0) |
| 3 | 4.08, d (8.8) | 5.43, d (8.4) | 4.47, d (8.4) | 4.08, d (6.8) | 5.44, d (8.4) | 4.47, d (8.4) |
| 4 | 4.04, t (8.8) | 4.35, t (8.4) | 5.30, t (8.4) | 4.07, t (6.8) | 4.39, t (8.4) | 5.28, dd (8.4, 7.6) |
| 5 | 3.79, m | 3.90, m | 3.91, m | 3.79, m | 3.91, m | 3.91, overlapped |
| 6 | 3.88, dd (11.6, 7.2) 3.75, m | 3.84, dd (12.0, 6.4) 3.78, dd (12.0, 3.2) | 3.92, m 3.77, dd (11.2, 2.4) | 3.89, overlapped 3.76, m | 3.85, dd (11.6, 6.8) 3.78, dd (11.6, 2.8) | 3.93, m 3.80, dd (11.2, 2.8) |
| 1-OAc | 2.01, s | 2.01, s | ||||
| 3-OAc | 2.18, s | 2.18, s | ||||
| 4-OAc | 2.05, s | 2.04, s | ||||
| 1′ | 5.65, d (3.6) | 5.63, d (3.6) | 5.65, d (3.6) | 5.66, d (3.6) | 5.63, d (3.6) | 5.65, d (3.6) |
| 2′ | 4.79, dd (10.4, 3.6) | 4.72, dd (10.0, 3.6) | 4.76, dd (10.0, 3.6) | 4.80, dd (10.0, 3.6) | 4.73, dd (10.4, 3.6) | 4.76, dd (10.0, 3.6) |
| 3′ | 4.00, dd (10.4, 9.2) | 3.88, t (10.0) | 4.01, dd (10.0, 9.6) | 4.00, dd (10.0, 8.8) | 3.88, overlapped | 4.01, t (10.0) |
| 4′ | 3.46, dd (10.0, 9.2) | 3.47, t (10.0) | 3.43, t (9.6) | 3.45, dd (10.0, 8.8) | 3.45, dd (10.0, 8.8) | 3.44, t (10.0) |
| 5′ | 4.20, ddd (10.0, 6.0, 2.0) | 4.16, ddd (10.0, 6.0, 2.0) | 4.23, m | 4.21, ddd (10.0, 6.8, 2.0) | 4.19, ddd (10.0, 6.8, 2.0) | 4.24, ddd (10.0, 6.4, 1.6) |
| 6′ | 4.54, dd (12.0, 2.0) 4.31, dd (12.0, 6.0) | 4.56, dd (12.0, 2.0) 4.32, dd (12.0, 6.0) | 4.57, dd (12.0, 1.6) 4.31, dd (12.0, 6.4) | 4.54, dd (12.0, 2.0) 4.30, dd (12.0, 6.8) | 4.56, dd (12.0, 2.0) 4.31, dd (12.0, 6.8) | 4.57, dd (12.0, 1.6) 4.32, dd (12.0, 6.4) |
| 2″ | 7.49, d (8.8) | 7.48, d (8.8) | 7.49, d (8.4) | 7.49, d (8.8) | 7.48, d (8.8) | 7.49, d (8.8) |
| 3″ | 6.80, d (8.8) | 6.82, d (8.8) | 6.81, d (8.4) | 6.81, d (8.8) | 6.81, d (8.8) | 6.81, d (8.8) |
| 5″ | 6.80, d (8.8) | 6.82, d (8.8) | 6.81, d (8.4) | 6.81, d (8.8) | 6.81, d (8.8) | 6.81, d (8.8) |
| 6″ | 7.49, d (8.8) | 7.48, d (8.8) | 7.49, d (8.4) | 7.49, d (8.8) | 7.48, d (8.8) | 7.49, d (8.8) |
| 7″ | 7.71, d (16.0) | 7.70, d (16.0) | 7.70, d (16.0) | 7.71, d (16.0) | 7.69, d (16.0) | 7.70, d (16.0) |
| 8″ | 6.39, d (16.0) | 6.37, d (16.0) | 6.40, d (16.0) | 6.41, d (16.0) | 6.37, d (16.0) | 6.41, d (16.0) |
| 2‴ | 7.50, d (8.8) | 7.50, d (8.8) | 7.51, d (8.4) | 7.26, d (2.0) | 7.25, d (2.0) | 7.25, d (2.0) |
| 3‴ | 6.80, d (8.8) | 6.80, d (8.8) | 6.80, d (8.4) | |||
| 5‴ | 6.80, d (8.8) | 6.80, d (8.8) | 6.80, d (8.4) | 6.81, d (8.0) | 6.81, d (8.0) | 6.81, d (8.4) |
| 6‴ | 7.50, d (8.8) | 7.50, d (8.8) | 7.51, d (8.4) | 7.11, dd (8.0, 2.0) | 7.09, dd (8.0, 2.0) | 7.11, dd (8.4, 2.0) |
| 7‴ | 7.66, d (16.0) | 7.66, d (16.0) | 7.65, d (16.0) | 7.65, d (15.6) | 7.65, d (16.0) | 7.65, d (16.0) |
| 8‴ | 6.43, d (16.0) | 6.43, d (16.0) | 6.49, d (16.0) | 6.47, d (15.6) | 6.47, d (16.0) | 6.52, d (16.0) |
| 3″-OMe | 3.90, s | |||||
| 3‴-OMe | 3.90, s | 3.89, s |
| Position | 12 a | 13 a | 14 a | 15 a | 16 b | 17 a | 18 b | 19 b |
|---|---|---|---|---|---|---|---|---|
| 1 | 4.14, d (12.4) 4.03, d (12.4) | 3.55, d (11.6) 3.36, d (11.6) | 3.59, d (12.0) 3.42, d (12.0) | 4.14, d (11.6) 4.02, d (11.6) | 3.54, d (12.0) 3.34, d (12.0) | 3.59, d (12.0) 3.42, d (12.0) | 3.55, d (12.0) 3.39, d (12.0) | 3.58, d (12.0) 3.42, d (12.0) |
| 3 | 4.09, d (8.4) | 5.43, d (8.4) | 4.47, d (8.0) | 4.09, d (8.4) | 5.44, d (8.4) | 4.48, d (8.8) | 4.47, d (8.4) | 4.46, d (8.4) |
| 4 | 4.04, t (8.4) | 4.35, t (8.4) | 5.31, t (8.0) | 4.08, t (8.4) | 4.39, t (8.4) | 5.29, dd (8.8, 7.6) | 5.26, t (8.4) | 5.29, t (8.4) |
| 5 | 3.79, m | 3.90, overlapped | 3.91, overlapped | 3.80, m | 3.91, overlapped | 3.91, overlapped | 3.90, overlapped | 3.90, overlapped |
| 6 | 3.88, m 3.75, dd (11.6, 2.8) | 3.84, dd (12.0, 6.4) 3.78, dd (12.0, 3.2) | 3.92, overlapped 3.78, m | 3.89, overlapped 3.76, dd (11.6, 2.8) | 3.85, dd (12.0, 6.6) 3.79, dd (12.0, 2.4) | 3.92, overlapped 3.81, dd (11.2, 2.4) | 3.92, overlapped 3.78, dd (10.8, 2.4) | 3.90, overlapped 3.78, m |
| 1-OAc | 2.02, s | 2.02, s | ||||||
| 3-OAc | 2.18, s | 2.18, s | ||||||
| 4-OAc | 2.04, s | 2.03, s | 2.08, s | 2.05, s | ||||
| 1′ | 5.64, d (3.6) | 5.63, d (4.0) | 5.67, d (3.6) | 5.65, d (4.0) | 5.64, d (3.6) | 5.66, d (4.0) | 5.62, d (3.6) | 5.66, d (3.6) |
| 2′ | 4.82, dd (10.4, 3.6) | 4.72, dd (10.4, 4.0) | 4.76, dd (10.0, 3.6) | 4.82, dd (10.0, 4.0) | 4.72, dd (10.2, 3.6) | 4.76, dd (10.0, 4.0) | 4.75, dd (10.2, 3.6) | 4.73, dd (10.2, 3.6) |
| 3′ | 4.00, dd (10.4, 9.2) | 3.88, overlapped | 4.01, t (10.0) | 4.00, dd (10.0, 8.8) | 3.87, t (10.2) | 4.01, dd (10.0, 8.8) | 3.98, dd (10.2, 9.6) | 3.99, dd (10.2, 9.0) |
| 4′ | 3.46, dd (10.0, 9.2) | 3.47, t (10.0) | 3.43, t (10.0) | 3.46, dd (10.0, 8.8) | 3.45, t (10.2) | 3.44, dd (10.0, 8.8) | 3.43, t (9.6) | 3.43, dd (10.2, 9.0) |
| 5′ | 4.20, ddd (10.0, 6.4, 2.0) | 4.16, ddd (10.0, 6.0, 2.0) | 4.23, dd (10.0, 6.4) | 4.22, ddd (10.0, 6.4, 2.0) | 4.19, m | 4.23, ddd (10.0, 6.4, 1.6) | 4.23, ddd (9.6, 6.6, 1.8) | 4.19, ddd (10.2, 5.4, 1.8) |
| 6′ | 4.54, dd (12.0, 2.0) 4.31, dd (12.0, 6.4) | 4.56, dd (12.0, 2.0) 4.32, dd (12.0, 6.0) | 4.57, br d (12.0) 4.31, dd (12.0, 6.4) | 4.54, dd (12.0, 2.0) 4.30, dd (12.0, 6.4) | 4.56, br d (12.0) 4.31, dd (12.0, 6.6) | 4.57, dd (12.0, 1.6) 4.32, dd (12.0, 6.4) | 4.56, dd (12.0, 1.8) 4.31, dd (12.0, 6.6) | 4.53, dd (12.0, 1.8) 4.30, dd (12.0, 5.4) |
| 2″ | 7.26, d (2.0) | 7.21, d (2.0) | 7.23, d (2.0) | 7.26, d (2.4) | 7.21, br s | 7.23, d (2.0) | 7.93, d (2.4) | 7.23, d (1.8) |
| 5″ | 6.81, d (8.4) | 6.82, d (8.0) | 6.82, d (8.0) | 6.81, d (8.0) | 6.82, d (7.8) | 6.81, d (8.0) | 6.77, d (8.4) | 6.81, d (8.4) |
| 6″ | 7.09, dd (8.4, 2.0) | 7.10, dd (8.0, 2.0) | 7.09, dd (8.0, 2.0) | 7.09, dd (8.0, 2.4) | 7.10, overlapped | 7.09, dd (8.0, 2.0) | 7.19, dd (8.4, 2.4) | 7.09, dd (8.4, 1.8) |
| 7″ | 7.70, d (15.6) | 7.69, d (15.6) | 7.69, d (16.0) | 7.70, d (16.0) | 7.69, d (16.2) | 7.69, d (16.0) | 6.92, d (12.6) | 7.68, d (15.6) |
| 8″ | 6.45, d (15.6) | 6.39, d (15.6) | 6.44, d (16.0) | 6.45, d (16.0) | 6.40, d (16.2) | 6.44, d (16.0) | 5.88, d (12.6) | 6.44, d (15.6) |
| 2‴ | 7.51, d (8.4) | 7.50, d (8.8) | 7.51, d (8.0) | 7.26, d (2.4) | 7.26, br s | 7.24, d (2.0) | 7.25, d (2.4) | 7.87, d (1.8) |
| 3‴ | 6.81, d (8.4) | 6.80, d (8.8) | 6.80, d (8.0) | |||||
| 5‴ | 6.81, d (8.4) | 6.80, d (8.8) | 6.80, d (8.0) | 6.81, d (8.0) | 6.80, d (7.8) | 6.81, d (8.0) | 6.81, d (8.4) | 6.77, d (8.4) |
| 6‴ | 7.51, d (8.4) | 7.50, d (8.8) | 7.51, d (8.0) | 7.11, dd (8.0, 2.4) | 7.10, overlapped | 7.11, dd (8.0, 2.0) | 7.11, dd (8.4, 2.4) | 7.15, dd (8.4, 1.8) |
| 7‴ | 7.66, d (15.6) | 7.66, d (15.6) | 7.65, d (16.0) | 7.65, d (16.0) | 7.65, d (16.2) | 7.65, d (16.0) | 7.65, d (16.2) | 6.88, d (13.2) |
| 8‴ | 6.43, d (15.6) | 6.43, d (15.6) | 6.49, d (16.0) | 6.47, d (16.0) | 6.47, d (16.2) | 6.52, d (16.0) | 6.53, d (16.2) | 5.90, d (13.2) |
| 3″-OMe | 3.92, s | 3.90, s | 3.92, s | 3.92, s | 3.90, s | 3.92, s | 3.88, s | 3.92, s |
| 3‴-OMe | 3.90, s | 3.89, s | 3.90, s | 3.90, s | 3.89, s |
| Position | 6 a | 7 a | 8 a | 9 a | 10 a | 11 a | 12 a | 13 a | 14 a | 15 a | 16 b | 17 a | 18 b | 19 b |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 64.8 | 64.4 | 62.5 | 64.8 | 64.5 | 62.5 | 64.8 | 64.5 | 62.5 | 64.8 | 64.5 | 62.5 | 62.5 | 62.3 |
| 2 | 103.9 | 105.0 | 106.0 | 103.8 | 104.9 | 106.0 | 103.9 | 105.0 | 106.1 | 103.8 | 104.9 | 106.1 | 105.9 | 106.2 |
| 3 | 78.5 | 78.6 | 75.2 | 78.5 | 78.6 | 75.2 | 78.4 | 78.7 | 75.2 | 78.4 | 78.6 | 75.2 | 75.2 | 75.2 |
| 4 | 75.3 | 73.8 | 78.0 | 75.4 | 73.8 | 78.1 | 75.4 | 73.8 | 78.0 | 75.4 | 73.8 | 78.2 | 78.2 | 77.9 |
| 5 | 84.1 | 84.2 | 82.2 | 84.1 | 84.2 | 82.3 | 84.2 | 84.2 | 82.3 | 84.2 | 84.2 | 82.3 | 82.3 | 82.3 |
| 6 | 64.1 | 63.9 | 64.8 | 64.2 | 64.0 | 64.8 | 64.2 | 63.9 | 64.9 | 64.3 | 64.0 | 64.9 | 64.8 | 64.8 |
| 1-OAc | 172.0 20.6 | 172.0 20.6 | 172.0 20.6 | 172.0 20.6 | ||||||||||
| 3-OAc | 172.2 20.8 | 172.2 20.8 | 172.2 20.8 | 172.2 20.8 | ||||||||||
| 4-OAc | 172.4 20.8 | 172.4 20.8 | 172.3 20.8 | 172.4 20.8 | 172.4 20.9 | 172.3 20.8 | ||||||||
| 1′ | 90.9 | 90.5 | 91.1 | 90.9 | 90.4 | 91.1 | 91.0 | 90.6 | 91.2 | 90.9 | 90.4 | 91.2 | 91.0 | 91.3 |
| 2′ | 74.1 | 74.3 | 74.2 | 74.1 | 74.3 | 74.3 | 74.1 | 74.3 | 74.2 | 74.1 | 74.3 | 74.2 | 73.9 | 74.2 |
| 3′ | 72.1 | 72.3 | 72.0 | 72.2 | 72.3 | 72.0 | 72.2 | 72.3 | 72.0 | 72.2 | 72.3 | 72.1 | 71.9 | 72.0 |
| 4′ | 72.0 | 71.8 | 72.0 | 72.0 | 71.9 | 72.1 | 72.1 | 72.0 | 72.1 | 72.0 | 71.9 | 72.0 | 72.1 | 71.9 |
| 5′ | 72.0 | 72.0 | 72.0 | 72.0 | 72.0 | 72.0 | 72.0 | 71.8 | 72.0 | 72.0 | 72.0 | 72.0 | 72.0 | 71.8 |
| 6′ | 65.0 | 64.9 | 65.1 | 65.1 | 65.1 | 65.1 | 65.0 | 64.9 | 65.1 | 65.1 | 65.1 | 65.1 | 65.1 | 64.5 |
| 1″ | 127.1 | 127.1 | 127.0 | 127.2 | 127.0 | 127.1 | 127.7 | 127.5 | 127.7 | 127.8 | 127.5 | 127.6 | 127.9 | 127.5 |
| 2″ | 131.3 | 131.3 | 131.3 | 131.4 | 131.3 | 131.4 | 111.7 | 111.8 | 111.7 | 111.6 | 111.8 | 111.8 | 115.3 | 111.7 |
| 3″ | 116.9 | 116.8 | 116.9 | 116.8 | 116.9 | 116.8 | 149.4 | 149.5 | 149.4 | 149.4 | 149.5 | 149.4 | 148.3 | 149.5 |
| 4″ | 161.7 | 161.6 | 161.6 | 161.4 | 161.6 | 161.4 | 150.8 | 151.5 | 150.8 | 150.8 | 151.1 | 150.9 | 149.9 | 150.9 |
| 5″ | 116.9 | 116.8 | 116.9 | 116.8 | 116.9 | 116.8 | 116.4 | 116.6 | 116.4 | 116.4 | 116.5 | 116.4 | 115.6 | 116.5 |
| 6″ | 131.3 | 131.3 | 131.3 | 131.4 | 131.3 | 131.4 | 124.5 | 124.3 | 124.4 | 124.5 | 124.3 | 124.4 | 127.4 | 124.5 |
| 7″ | 147.5 | 147.5 | 147.4 | 147.5 | 147.5 | 147.3 | 147.7 | 147.8 | 147.5 | 147.7 | 147.8 | 147.6 | 147.0 | 147.5 |
| 8″ | 114.8 | 114.6 | 114.9 | 114.9 | 114.6 | 114.9 | 115.2 | 114.9 | 115.2 | 115.2 | 114.9 | 115.2 | 115.8 | 115.1 |
| 9″ | 168.8 | 168.7 | 168.8 | 168.8 | 168.7 | 168.8 | 168.7 | 168.7 | 168.8 | 168.7 | 168.7 | 168.8 | 167.5 | 168.8 |
| 1‴ | 127.1 | 127.0 | 127.2 | 127.7 | 127.6 | 127.8 | 127.2 | 127.1 | 127.3 | 127.7 | 127.6 | 127.8 | 127.7 | 128.0 |
| 2‴ | 131.4 | 131.3 | 131.4 | 111.6 | 111.5 | 111.9 | 131.3 | 131.3 | 131.3 | 111.7 | 111.5 | 111.7 | 111.8 | 115.1 |
| 3‴ | 116.9 | 116.9 | 116.8 | 149.4 | 149.4 | 149.4 | 116.8 | 116.9 | 116.8 | 149.4 | 149.5 | 149.4 | 149.4 | 148.4 |
| 4‴ | 161.6 | 161.4 | 161.4 | 150.7 | 150.8 | 150.6 | 161.4 | 161.5 | 161.3 | 150.7 | 150.9 | 150.7 | 150.8 | 149.7 |
| 5‴ | 116.9 | 116.9 | 116.8 | 116.4 | 116.4 | 116.4 | 116.8 | 116.9 | 116.8 | 116.4 | 116.4 | 116.5 | 116.4 | 115.7 |
| 6‴ | 131.4 | 131.3 | 131.4 | 124.4 | 124.4 | 124.4 | 131.3 | 131.3 | 131.3 | 124.4 | 124.5 | 124.4 | 124.4 | 127.0 |
| 7‴ | 146.9 | 146.9 | 146.8 | 147.1 | 147.1 | 147.1 | 146.9 | 146.9 | 146.8 | 147.1 | 147.2 | 147.1 | 147.1 | 146.0 |
| 8‴ | 114.9 | 114.9 | 115.2 | 115.3 | 115.2 | 115.4 | 115.0 | 114.9 | 115.3 | 115.3 | 115.2 | 115.4 | 115.4 | 116.1 |
| 9‴ | 169.2 | 169.2 | 169.3 | 169.1 | 169.1 | 169.3 | 169.2 | 169.2 | 169.3 | 169.1 | 169.1 | 169.3 | 169.3 | 168.1 |
| 3″-OMe | 56.5 | 56.5 | 56.4 | 56.5 | 56.5 | 56.5 | 56.5 | 56.4 | 56.4 | |||||
| 3‴-OMe | 56.5 | 56.5 | 56.5 | 56.4 | 56.5 | 56.5 | 56.4 |
| Position | 20 a | 21 a | 22 b | 23 a | 24 a |
|---|---|---|---|---|---|
| 1 | 4.17, d (12.0) 4.00, d (12.0) | 3.57, d (12.0) 3.45, d (12.0) | 3.58, d (12.0) 3.47, d (12.0) | 4.16, d (11.6) 3.99, d (11.6) | 3.59, d (12.0) 3.48, d (12.0) |
| 3 | 5.29, d (8.0) | 5.65, d (7.6) | 5.65, d (7.8) | 5.30, d (8.4) | 5.65, d (7.6) |
| 4 | 4.34, t (8.0) | 5.48, t (7.6) | 5.50, t (7.8) | 4.38, t (8.4) | 5.49, t (7.6) |
| 5 | 3.89, m | 4.07, m | 4.08, m | 3.90, overlapped | 4.08, td (7.6, 4.4) |
| 6 | 3.84, dd (11.6, 6.8) 3.77, dd, (11.6, 2.8) | 3.87, dd (12.0, 6.8) 3.77, dd (12.0, 4.4) | 3.87, dd (12.0, 7.2) 3.78, dd (12.0, 4.2) | 3.85, dd (12.0, 6.8) 3.77, dd (12.0, 2.8) | 3.87, overlapped 3.80, dd (12.0, 4.4) |
| 1-OAc | 2.02, s | 2.02, s | |||
| 3-OAc | 2.17, s | 2.11, s | 1.98, s | 2.18, s | 2.11, s |
| 4-OAc | 1.99, s | 2.10, s | 1.97, s | ||
| 1′ | 5.66, d (3.6) | 5.62, d (3.6) | 5.62, d (3.6) | 5.66, d (3.6) | 5.62, d (3.6) |
| 2′ | 4.78, dd (10.0, 3.6) | 4.76, dd (10.0, 3.6) | 4.77, dd (10.2, 3.6) | 4.78, dd (10.0, 3.6) | 4.77, dd (10.0, 3.6) |
| 3′ | 3.88, dd (10.0, 9.2) | 3.91, dd (10.0, 9.2) | 3.91, m | 3.88, m | 3.91, overlapped |
| 4′ | 3.47, t (9.2) | 3.47, t (9.2) | 3.46, t (10.2) | 3.46, dd (10.0, 9.2) | 3.47, m |
| 5′ | 4.16, m | 4.19, ddd (10.0, 6.0, 2.0) | 4.18, ddd (10.2, 6.0, 1.8) | 4.18, m | 4.19, ddd (10.0, 6.4, 2.0) |
| 6′ | 4.57, dd (12.0, 2.0) 4.32, dd (12.0, 6.0) | 4.60, dd (12.0, 2.0) 4.33, dd (12.0, 6.0) | 4.59, dd (12.0, 1.8) 4.32, dd (12.0, 6.0) | 4.57, dd (12.0, 2.0) 4.30, dd (12.0, 6.8) | 4.60, dd (12.0, 2.0) 4.33, dd (12.0, 6.4) |
| 2″ | 7.49, d (8.8) | 7.49, d (8.8) | 7.25, br s | 7.49, d (8.8) | 7.24, d (2.0) |
| 3″ | 6.81, d (8.8) | 6.81, d (8.8) | 6.81, d (8.8) | ||
| 5″ | 6.81, d (8.8) | 6.81, d (8.8) | 6.81, d (8.4) | 6.81, d (8.8) | 6.82, d (8.0) |
| 6″ | 7.49, d (8.8) | 7.49, d (8.8) | 7.10, dd (8.4, 1.8) | 7.49, d (8.8) | 7.10, dd (8.0, 2.0) |
| 7″ | 7.68, d (15.6) | 7.69, d (16.0) | 7.69, d (16.2) | 7.68, d (16.0) | 7.69, d (16.0) |
| 8″ | 6.38, d (15.6) | 6.41, d (16.0) | 6.45, d (16.2) | 6.38, d (16.0) | 6.45, d (16.0) |
| 2‴ | 7.50, d (8.8) | 7.50, d (8.8) | 7.49, d (8.4) | 7.26, d (2.0) | 7.24, d (2.0) |
| 3‴ | 6.81, d (8.8) | 6.81, d (8.8) | 6.80, d (8.4) | ||
| 5‴ | 6.81, d (8.8) | 6.81, d (8.8) | 6.80, d (8.4) | 6.81, d (8.0) | 6.82, d (8.0) |
| 6‴ | 7.50, d (8.8) | 7.50, d (8.8) | 7.49, d (8.4) | 7.09, dd (8.0, 2.0) | 7.10, dd (8.0, 2.0) |
| 7‴ | 7.66, d (16.0) | 7.65, d (16.0) | 7.65, d (16.2) | 7.65, d (16.0) | 7.65, d (16.0) |
| 8‴ | 6.43, d (16.0) | 6.45, d (16.0) | 6.44, d (16.2) | 6.47, d (16.0) | 6.49, d (16.0) |
| 3″-OMe | 3.91, s | 3.90, s | |||
| 3‴-OMe | 3.89, s | 3.91, s |
| Position | 20 a | 21 a | 22 b | 23 a | 24 a |
|---|---|---|---|---|---|
| 1 | 66.1 | 63.6 | 63.4 | 66.2 | 63.5 |
| 2 | 103.3 | 106.1 | 106.2 | 103.2 | 106.2 |
| 3 | 79.5 | 76.6 | 76.6 | 79.5 | 76.7 |
| 4 | 73.5 | 76.5 | 76.5 | 73.5 | 76.6 |
| 5 | 84.2 | 82.7 | 82.7 | 84.3 | 82.7 |
| 6 | 63.8 | 64.1 | 64.2 | 63.9 | 64.3 |
| 1-OAc | 172.0 20.6 | 171.9 20.6 | |||
| 3-OAc | 172.1 20.7 | 171.8 20.7 | 172.0 20.7 | 172.1 20.7 | 171.7 20.7 |
| 4-OAc | 172.0 20.7 | 171.8 20.7 | 172.0 20.7 | ||
| 1′ | 91.0 | 91.3 | 91.4 | 90.9 | 91.4 |
| 2′ | 74.0 | 74.1 | 74.1 | 74.0 | 74.1 |
| 3′ | 72.3 | 72.2 | 72.2 | 72.4 | 72.2 |
| 4′ | 71.8 | 71.8 | 71.9 | 71.9 | 71.9 |
| 5′ | 72.2 | 72.2 | 72.2 | 72.2 | 72.2 |
| 6′ | 64.9 | 64.9 | 64.9 | 65.1 | 64.9 |
| 1″ | 127.1 | 127.2 | 127.4 | 127.1 | 127.6 |
| 2″ | 131.3 | 131.3 | 111.7 | 131.4 | 111.7 |
| 3″ | 116.8 | 116.8 | 149.5 | 116.8 | 149.4 |
| 4″ | 161.5 | 161.6 | 151.2 | 161.5 | 150.7 |
| 5″ | 116.8 | 116.8 | 116.5 | 116.8 | 116.4 |
| 6″ | 131.3 | 131.3 | 124.6 | 131.4 | 124.3 |
| 7″ | 147.5 | 147.5 | 147.8 | 147.5 | 147.8 |
| 8″ | 114.7 | 114.8 | 115.0 | 114.8 | 115.1 |
| 9″ | 168.7 | 168.7 | 168.7 | 168.7 | 168.7 |
| 1‴ | 127.1 | 127.0 | 127.1 | 127.7 | 127.8 |
| 2‴ | 131.4 | 131.4 | 131.3 | 111.5 | 111.8 |
| 3‴ | 116.8 | 116.9 | 116.9 | 149.4 | 149.4 |
| 4‴ | 161.4 | 161.4 | 161.6 | 150.7 | 150.8 |
| 5‴ | 116.8 | 116.9 | 116.9 | 116.4 | 116.4 |
| 6‴ | 131.4 | 131.4 | 131.3 | 124.5 | 124.5 |
| 7‴ | 146.9 | 146.8 | 146.9 | 147.1 | 147.1 |
| 8‴ | 114.9 | 115.1 | 115.0 | 115.3 | 115.4 |
| 9‴ | 169.2 | 169.2 | 169.2 | 169.1 | 169.2 |
| 3″-OMe | 56.4 | 56.5 | |||
| 3‴-OMe | 56.5 | 56.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Y.-J.; Lan, Q.; Su, B.-J.; Liang, D. Phanerosides A–X, Phenylpropanoid Esters of Sucrose from the Rattans of Phanera championii Benth. Molecules 2023, 28, 4767. https://doi.org/10.3390/molecules28124767
Hu Y-J, Lan Q, Su B-J, Liang D. Phanerosides A–X, Phenylpropanoid Esters of Sucrose from the Rattans of Phanera championii Benth. Molecules. 2023; 28(12):4767. https://doi.org/10.3390/molecules28124767
Chicago/Turabian StyleHu, Ya-Jie, Qian Lan, Bao-Jun Su, and Dong Liang. 2023. "Phanerosides A–X, Phenylpropanoid Esters of Sucrose from the Rattans of Phanera championii Benth" Molecules 28, no. 12: 4767. https://doi.org/10.3390/molecules28124767
APA StyleHu, Y.-J., Lan, Q., Su, B.-J., & Liang, D. (2023). Phanerosides A–X, Phenylpropanoid Esters of Sucrose from the Rattans of Phanera championii Benth. Molecules, 28(12), 4767. https://doi.org/10.3390/molecules28124767