In Silico Identification of Natural Product-Based Inhibitors Targeting IL-1β/IL-1R Protein–Protein Interface
Abstract
:1. Introduction
2. Results
2.1. Docking-Based Virtual Screening
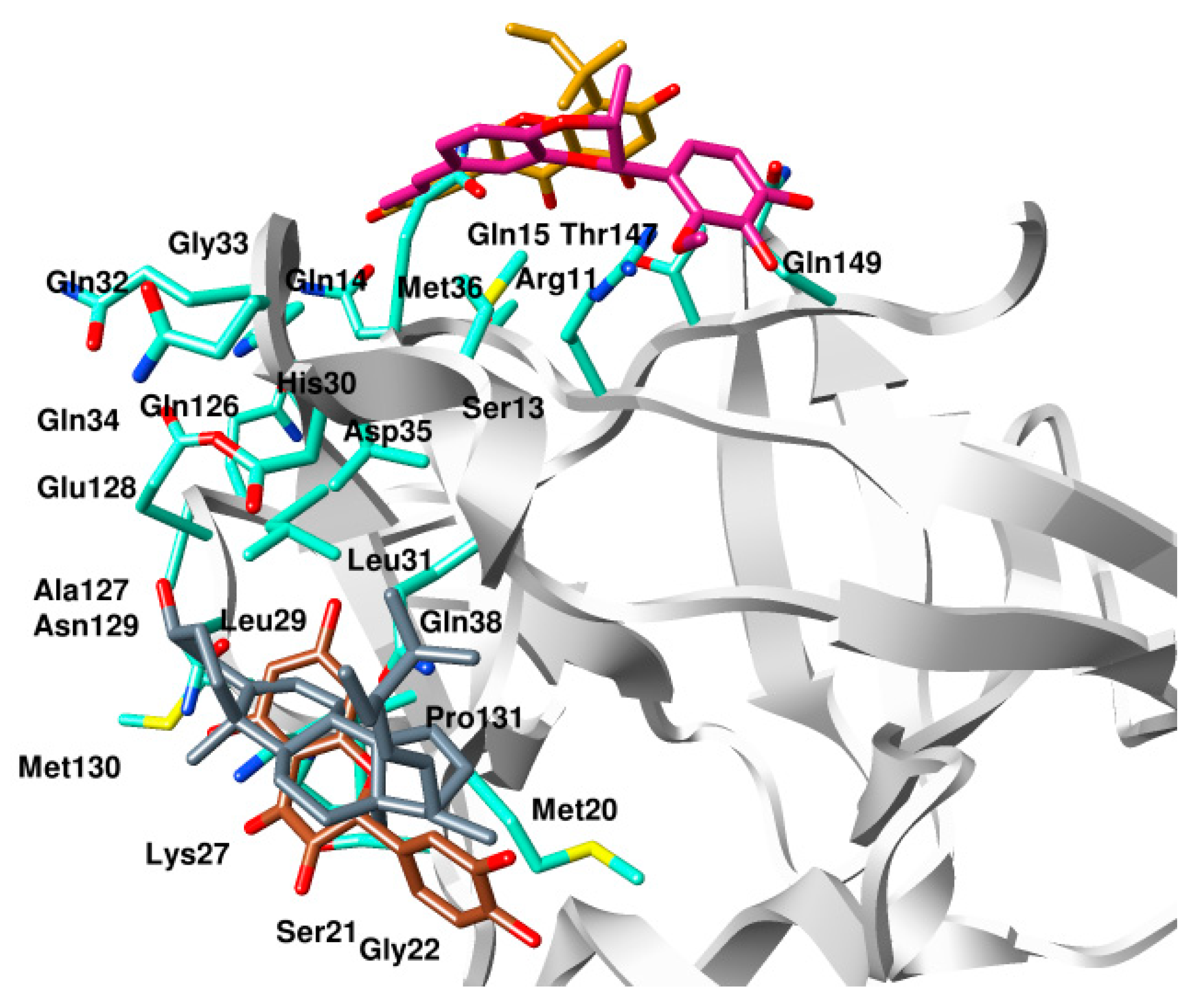
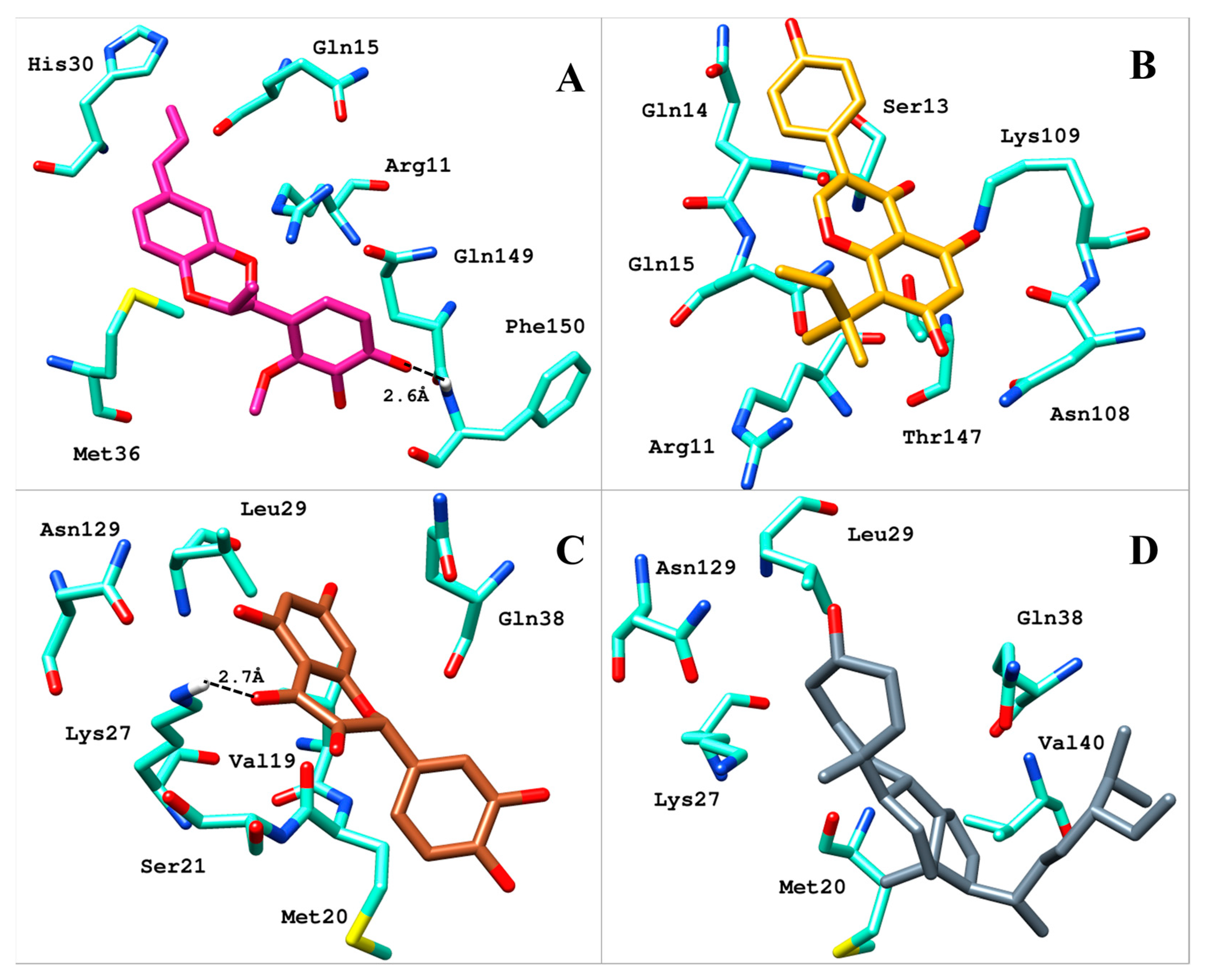
2.2. Analysis of Binding Mode
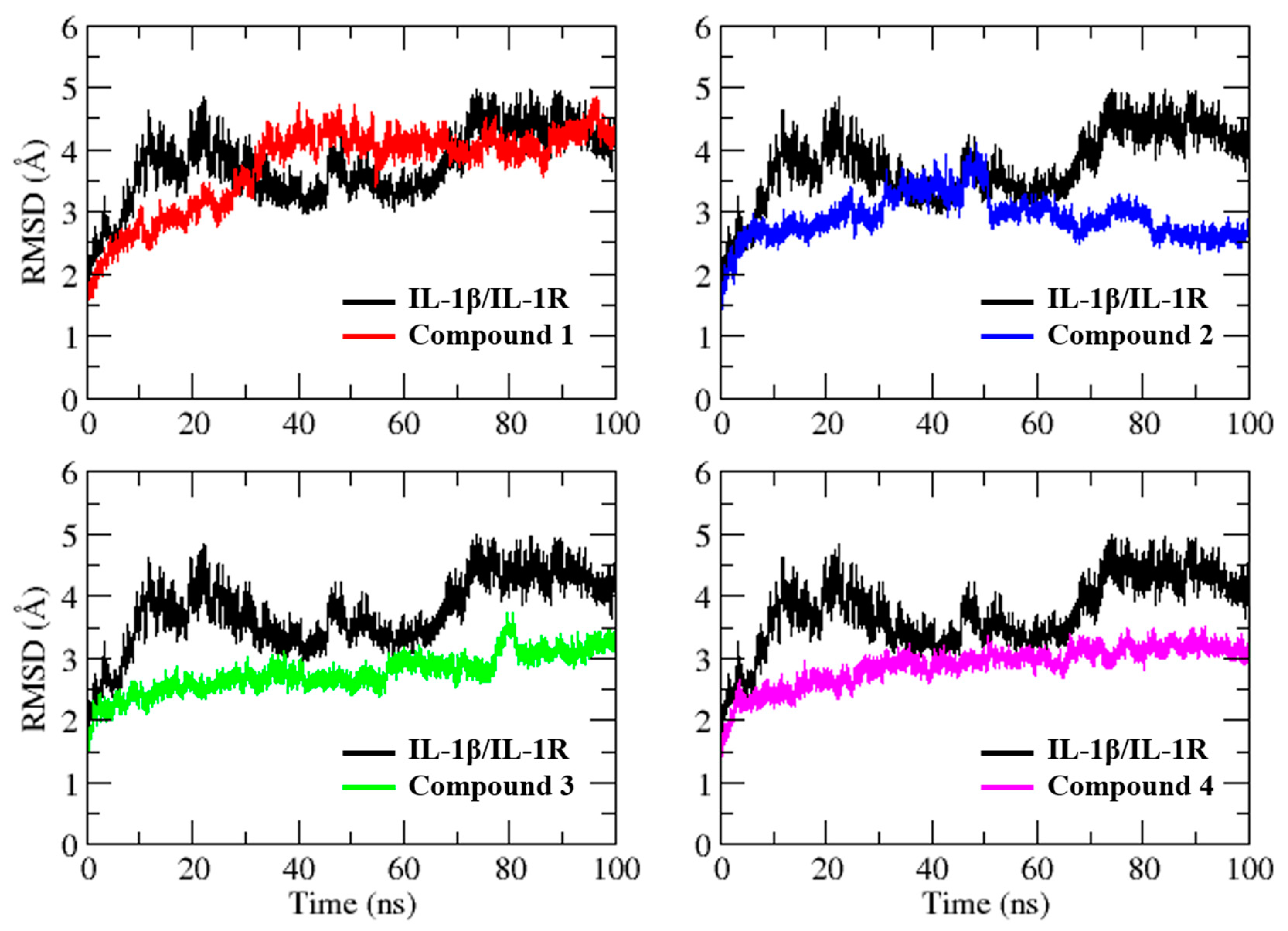
2.3. Molecular Dynamics Simulation
3. Discussion
4. Materials and Methods
4.1. Target Preparation
4.2. Database Preparation
4.3. Docking-Based Virtual Screening
4.4. Molecular Dynamics Simulation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Hunter, P. The Inflammation Theory of Disease. The Growing Realization That Chronic Inflammation Is Crucial in Many Diseases Opens New Avenues for Treatment. EMBO Rep. 2012, 13, 968–970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Yang, G.; Thompson-Lanza, J.A.; Glassman, A.; Hayes, K.; Patterson, A.; Marquez, R.T.; Auersperg, N.; Yu, Y.; Hahn, W.C.; et al. A Genetically Defined Model for Human Ovarian Cancer. Cancer Res. 2004, 64, 1655–1663. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, H.; Ogata, H.; Nishigaki, R.; Broide, D.H.; Karin, M. Tobacco Smoke Promotes Lung Tumorigenesis by Triggering IKKβ and JNK1 Dependent Inflammation. Cancer Cell 2010, 17, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teufel, L.U.; Arts, R.J.W.; Netea, M.G.; Dinarello, C.A.; Joosten, L.A.B. IL-1 Family Cytokines as Drivers and Inhibitors of Trained Immunity. Cytokine 2022, 150, 155773. [Google Scholar] [CrossRef] [PubMed]
- Gabay, C.; Lamacchia, C.; Palmer, G. IL-1 Pathways in Inflammation and Human Diseases. Nat. Rev. Rheumatol. 2010, 6, 232–241. [Google Scholar] [CrossRef]
- Dinarello, C.A.; Simon, A.; van der Meer, J.W.M. Treating Inflammation by Blocking Interleukin-1 in a Broad Spectrum of Diseases. Nat. Rev. Drug Discov. 2012, 11, 633–652. [Google Scholar] [CrossRef] [Green Version]
- Dinarello, C.A. Biologic Basis for Interleukin-1 in Disease. Blood 1996, 87, 2095–2147. [Google Scholar] [CrossRef] [Green Version]
- Thornberry, N.A.; Bull, H.G.; Calaycay, J.R.; Chapman, K.T.; Howard, A.D.; Kostura, M.J.; Miller, D.K.; Molineaux, S.M.; Weidner, J.R.; Aunins, J. A Novel Heterodimeric Cysteine Protease Is Required for Interleukin-1 Beta Processing in Monocytes. Nature 1992, 356, 768–774. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Yamamoto, K.; Saido, T.; Kawasaki, H.; Oppenheim, J.J.; Matsushima, K. Identification of Calcium-Activated Neutral Protease as a Processing Enzyme of Human Interleukin 1 Alpha. Proc. Natl. Acad. Sci. USA 1990, 87, 5548–5552. [Google Scholar] [CrossRef] [Green Version]
- Berda-Haddad, Y.; Robert, S.; Salers, P.; Zekraoui, L.; Farnarier, C.; Dinarello, C.A.; Dignat-George, F.; Kaplanski, G. Sterile Inflammation of Endothelial Cell-Derived Apoptotic Bodies Is Mediated by Interleukin-1α. Proc. Natl. Acad. Sci. USA 2011, 108, 20684–20689. [Google Scholar] [CrossRef] [Green Version]
- Dinarello, C.A.; Ikejima, T.; Warner, S.J.; Orencole, S.F.; Lonnemann, G.; Cannon, J.G.; Libby, P. Interleukin 1 Induces Interleukin 1. I. Induction of Circulating Interleukin 1 in Rabbits In Vivo and in Human Mononuclear Cells In Vitro. J. Immunol. 1987, 139, 1902–1910. [Google Scholar] [CrossRef]
- Martinon, F.; Mayor, A.; Tschopp, J. The Inflammasomes: Guardians of the Body. Annu. Rev. Immunol. 2009, 27, 229–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agostini, L.; Martinon, F.; Burns, K.; McDermott, M.F.; Hawkins, P.N.; Tschopp, J. NALP3 Forms an IL-1beta-Processing Inflammasome with Increased Activity in Muckle-Wells Autoinflammatory Disorder. Immunity 2004, 20, 319–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrei, C.; Dazzi, C.; Lotti, L.; Torrisi, M.R.; Chimini, G.; Rubartelli, A. The Secretory Route of the Leaderless Protein Interleukin 1beta Involves Exocytosis of Endolysosome-Related Vesicles. Mol. Biol. Cell 1999, 10, 1463–1475. [Google Scholar] [CrossRef]
- Gardella, S.; Andrei, C.; Costigliolo, S.; Olcese, L.; Zocchi, M.R.; Rubartelli, A. Secretion of Bioactive Interleukin-1beta by Dendritic Cells Is Modulated by Interaction with Antigen Specific T Cells. Blood 2000, 95, 3809–3815. [Google Scholar] [CrossRef]
- Perregaux, D.G.; McNiff, P.; Laliberte, R.; Conklyn, M.; Gabel, C.A. ATP Acts as an Agonist to Promote Stimulus-Induced Secretion of IL-1 Beta and IL-18 in Human Blood. J. Immunol. 2000, 165, 4615–4623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Netea, M.G.; Simon, A.; van de Veerdonk, F.; Kullberg, B.-J.; Van der Meer, J.W.M.; Joosten, L.A.B. IL-1β Processing in Host Defense: Beyond the Inflammasomes. PLoS Pathog. 2010, 6, e1000661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamhane, M.; Cabrera-Ghayouri, S.; Abelian, G.; Viswanath, V. Review of Biomarkers in Ocular Matrices: Challenges and Opportunities. Pharm. Res. 2019, 36, 40. [Google Scholar] [CrossRef] [Green Version]
- Na, K.-S.; Mok, J.-W.; Kim, J.Y.; Rho, C.R.; Joo, C.-K. Correlations between Tear Cytokines, Chemokines, and Soluble Receptors and Clinical Severity of Dry Eye Disease. Investig. Ophthalmol. Vis. Sci. 2012, 53, 5443–5450. [Google Scholar] [CrossRef] [Green Version]
- Luo, L.; Li, D.-Q.; Doshi, A.; Farley, W.; Corrales, R.M.; Pflugfelder, S.C. Experimental Dry Eye Stimulates Production of Inflammatory Cytokines and MMP-9 and Activates MAPK Signaling Pathways on the Ocular Surface. Investig. Ophthalmol. Vis. Sci. 2004, 45, 4293–4301. [Google Scholar] [CrossRef]
- De Paiva, C.S.; Corrales, R.M.; Villarreal, A.L.; Farley, W.J.; Li, D.-Q.; Stern, M.E.; Pflugfelder, S.C. Corticosteroid and Doxycycline Suppress MMP-9 and Inflammatory Cytokine Expression, MAPK Activation in the Corneal Epithelium in Experimental Dry Eye. Exp. Eye Res. 2006, 83, 526–535. [Google Scholar] [CrossRef]
- Pam Theriot, O.D. Taming Inflammation in Dry Eye Disease. Available online: https://www.reviewofoptometry.com/article/taming-inflammation-in-dry-eye-disease (accessed on 15 April 2023).
- Shakoory, B.; Carcillo, J.A.; Chatham, W.W.; Amdur, R.L.; Zhao, H.; Dinarello, C.A.; Cron, R.Q.; Opal, S.M. Interleukin-1 Receptor Blockade Is Associated with Reduced Mortality in Sepsis Patients with Features of Macrophage Activation Syndrome: Reanalysis of a Prior Phase III Trial. Crit. Care Med. 2016, 44, 275–281. [Google Scholar] [CrossRef] [Green Version]
- Monteagudo, L.A.; Boothby, A.; Gertner, E. Continuous Intravenous Anakinra Infusion to Calm the Cytokine Storm in Macrophage Activation Syndrome. ACR Open Rheumatol. 2020, 2, 276–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huet, T.; Beaussier, H.; Voisin, O.; Jouveshomme, S.; Dauriat, G.; Lazareth, I.; Sacco, E.; Naccache, J.-M.; Bézie, Y.; Laplanche, S.; et al. Anakinra for Severe Forms of COVID-19: A Cohort Study. Lancet Rheumatol. 2020, 2, e393–e400. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, G.; De Luca, G.; Campochiaro, C.; Della-Torre, E.; Ripa, M.; Canetti, D.; Oltolini, C.; Castiglioni, B.; Tassan Din, C.; Boffini, N.; et al. Interleukin-1 Blockade with High-Dose Anakinra in Patients with COVID-19, Acute Respiratory Distress Syndrome, and Hyperinflammation: A Retrospective Cohort Study. Lancet Rheumatol. 2020, 2, e325–e331. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Donovan, K.A.; Kline, M.P.; Gornet, M.K.; Moon-Tasson, L.L.; Lacy, M.Q.; Dispenzieri, A.; Gertz, M.A.; Greipp, P.R.; Lust, J.A. Identification of Two Groups of Smoldering Multiple Myeloma Patients Who Are Either High or Low Producers of Interleukin-1. J. Interferon Cytokine Res. 2006, 26, 83–95. [Google Scholar] [CrossRef]
- Lust, J.A.; Lacy, M.Q.; Zeldenrust, S.R.; Dispenzieri, A.; Gertz, M.A.; Witzig, T.E.; Kumar, S.; Hayman, S.R.; Russell, S.J.; Buadi, F.K.; et al. Induction of a Chronic Disease State in Patients with Smoldering or Indolent Multiple Myeloma by Targeting Interleukin 1β-Induced Interleukin 6 Production and the Myeloma Proliferative Component. Mayo Clin. Proc. 2009, 84, 114–122. [Google Scholar] [CrossRef]
- Bresnihan, B.; Newmark, R.; Robbins, S.; Genant, H.K. Effects of Anakinra Monotherapy on Joint Damage in Patients with Rheumatoid Arthritis. Extension of a 24-Week Randomized, Placebo-Controlled Trial. J. Rheumatol. 2004, 31, 1103–1111. [Google Scholar]
- Meissner, F.; Molawi, K.; Zychlinsky, A. Mutant Superoxide Dismutase 1-Induced IL-1beta Accelerates ALS Pathogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 13046–13050. [Google Scholar] [CrossRef] [Green Version]
- Vigers, G.P.A.; Anderson, L.J.; Caffes, P.; Brandhuber, B.J. Crystal Structure of the Type-I Interleukin-1 Receptor Complexed with Interleukin-1β. Nature 1997, 386, 190–194. [Google Scholar] [CrossRef]
- Sardar, M.; Zia, K.; Ashraf, S.; Malik, H.N.; Jabeen, A.; Khan, K.M.; Ul-Haq, Z. Interface Inhibitory Action on Interleukin-1β Using Selected Anti-Inflammatory Compounds to Mitigate the Depression: A Computational Investigation. Comput. Biol. Chem. 2022, 101, 107774. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.-Y.; Zhang, Y.-Q.; Liu, Z.-M.; Chen, T.; Lv, C.-Y.; Tang, S.-H.; Zhang, X.-B.; Zhang, W.; Li, Z.-Y.; Zhou, R.-R.; et al. ETCM: An Encyclopaedia of Traditional Chinese Medicine. Nucleic Acids Res. 2019, 47, D976–D982. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-J.; Zhang, T. Integration of Traditional Chinese Medicine and Western Medicine in the Era of Precision Medicine. J. Integr. Med. 2017, 15, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yang, Y.; Feng, H.; Wang, H.; Zhao, R.; Liu, H. Baicalein Inhibits Interleukin-1β-Induced Proliferation of Human Rheumatoid Arthritis Fibroblast-like Synoviocytes. Inflammation 2014, 37, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Farghali, H.; Kutinová Canová, N.; Lekić, N. Resveratrol and Related Compounds as Antioxidants with an Allosteric Mechanism of Action in Epigenetic Drug Targets. Physiol. Res. 2013, 62, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Jurenka, J.S. Anti-Inflammatory Properties of Curcumin, a Major Constituent of Curcuma Longa: A Review of Preclinical and Clinical Research. Altern. Med. Rev. 2009, 14, 141–153. [Google Scholar]
- Li, E.; Zhang, J. Therapeutic Effects of Triptolide from Tripterygium Wilfordii Hook. f. on Interleukin-1-Beta-Induced Osteoarthritis in Rats. Eur. J. Pharmacol. 2020, 883, 173341. [Google Scholar] [CrossRef]
- Stierle, D.B.; Stierle, A.A.; Patacini, B.; McIntyre, K.; Girtsman, T.; Bolstad, E. Berkeleyones and Related Meroterpenes from a Deep Water Acid Mine Waste Fungus That Inhibit the Production of Interleukin 1-β from Induced Inflammasomes. J. Nat. Prod. 2011, 74, 2273–2277. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.Y.-C. TCM Database@Taiwan: The World’s Largest Traditional Chinese Medicine Database for Drug Screening In Silico. PLoS ONE 2011, 6, e15939. [Google Scholar] [CrossRef] [Green Version]
- Narayanaswamy, R.; Veeraragavan, V. Chapter 8—Natural Products as Antiinflammatory Agents. In Studies in Natural Products Chemistry; Rahman, A., Ed.; Bioactive Natural Products; Elsevier: Amsterdam, The Netherlands, 2020; Volume 67, pp. 269–306. [Google Scholar]
- Blevins, H.M.; Xu, Y.; Biby, S.; Zhang, S. The NLRP3 Inflammasome Pathway: A Review of Mechanisms and Inhibitors for the Treatment of Inflammatory Diseases. Front. Aging Neurosci. 2022, 14, 879021. [Google Scholar] [CrossRef] [PubMed]
- Chemical Computing Group. Molecular Operating Environment (MOE) Software; Chemical Computing Group: Montreal, QC, Canada, 2018. [Google Scholar]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminformatics 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An Overview of the Amber Biomolecular Simulation Package. WIREs Comput. Mol. Sci. 2013, 3, 198–210. [Google Scholar] [CrossRef]



| Compound Code | Structure | Binding Score (kcal/mol) |
|---|---|---|
| Compound 1 | 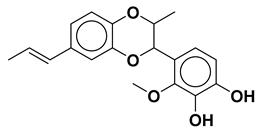 | −10.10 |
| Compound 2 |  | −11.12 |
| Compound 3 |  | −10.53 |
| Compound 4 |  | −14.19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, T.-t.; Chen, Y.-k.; Adil, M.; Almehmadi, M.; Alshabrmi, F.M.; Allahyani, M.; Alsaiari, A.A.; Liu, P.; Khan, M.R.; Peng, Q. In Silico Identification of Natural Product-Based Inhibitors Targeting IL-1β/IL-1R Protein–Protein Interface. Molecules 2023, 28, 4885. https://doi.org/10.3390/molecules28134885
Liu T-t, Chen Y-k, Adil M, Almehmadi M, Alshabrmi FM, Allahyani M, Alsaiari AA, Liu P, Khan MR, Peng Q. In Silico Identification of Natural Product-Based Inhibitors Targeting IL-1β/IL-1R Protein–Protein Interface. Molecules. 2023; 28(13):4885. https://doi.org/10.3390/molecules28134885
Chicago/Turabian StyleLiu, Ting-ting, Yan-kun Chen, Muhammad Adil, Mazen Almehmadi, Fahad M. Alshabrmi, Mamdouh Allahyani, Ahad Amer Alsaiari, Pei Liu, Muhammad Raheel Khan, and Qinghua Peng. 2023. "In Silico Identification of Natural Product-Based Inhibitors Targeting IL-1β/IL-1R Protein–Protein Interface" Molecules 28, no. 13: 4885. https://doi.org/10.3390/molecules28134885





