Novel Design of an α-Amylase with an N-Terminal CBM20 in Aspergillus niger Improves Binding and Processing of a Broad Range of Starches
Abstract
:1. Introduction
2. Results and Discussion
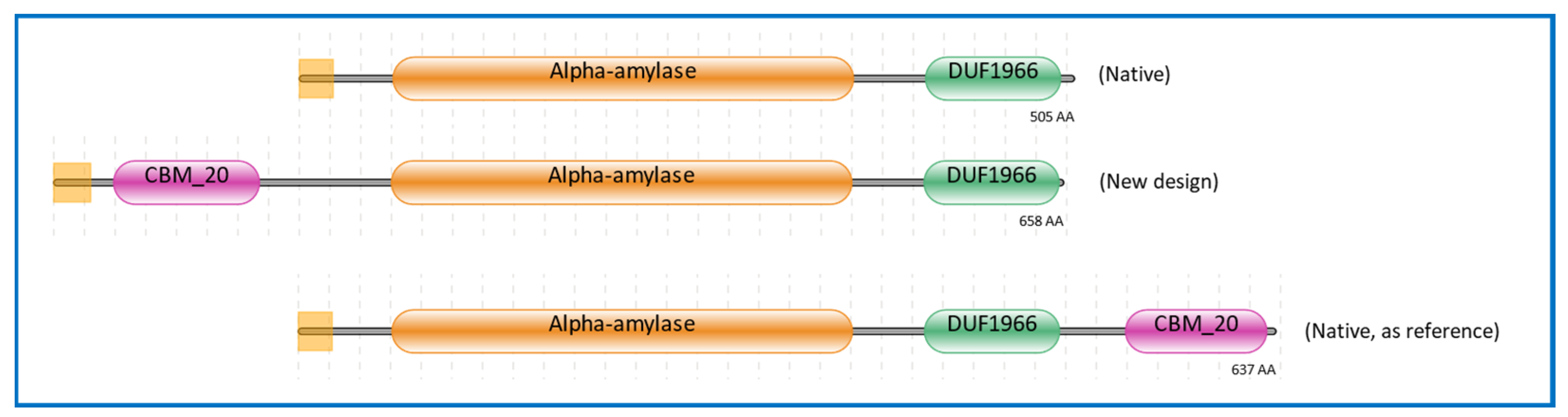
2.1. Domain Architecture of α-Amylase Design
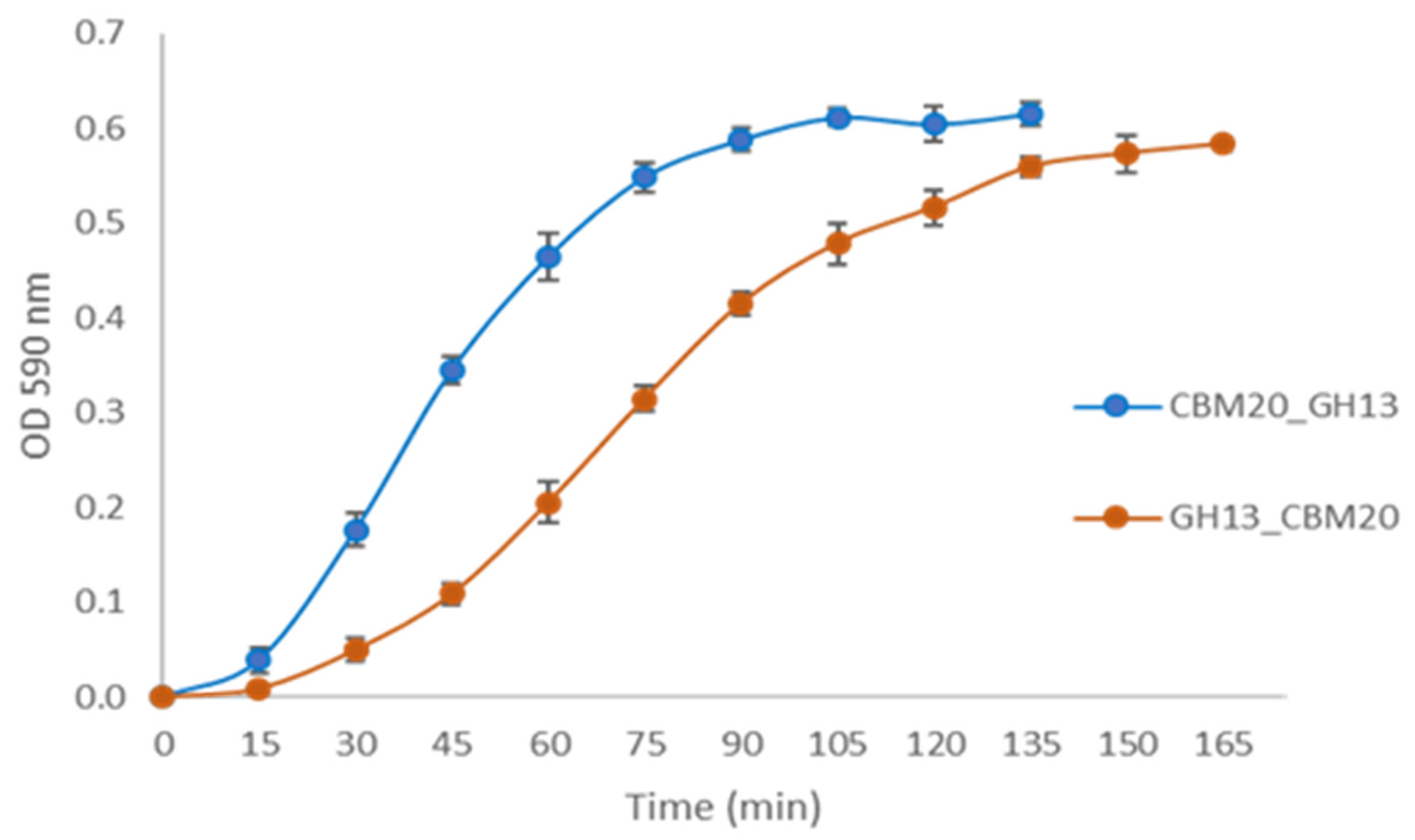
2.2. Expression of α-Amylase from A. niger Transformants
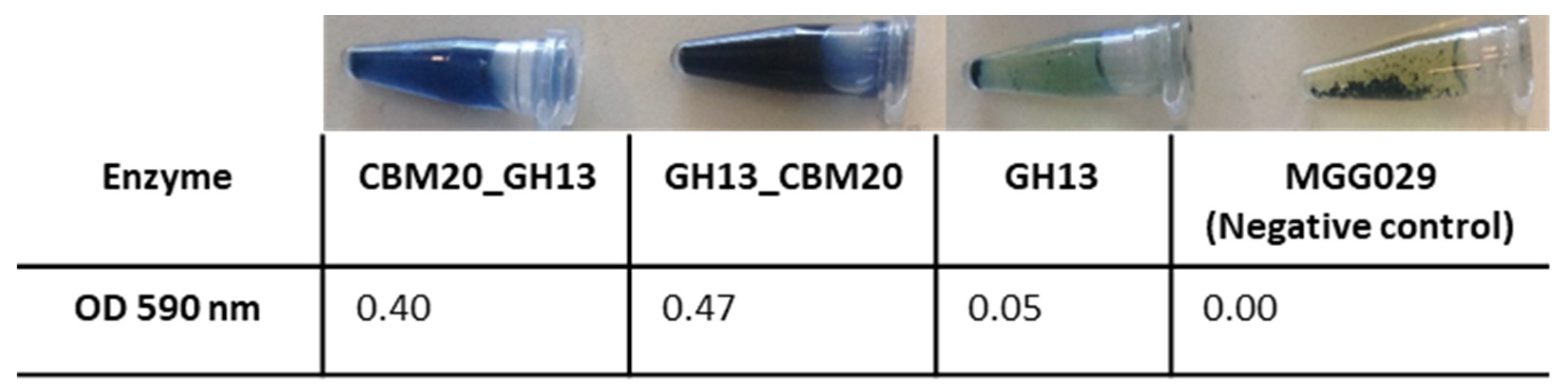
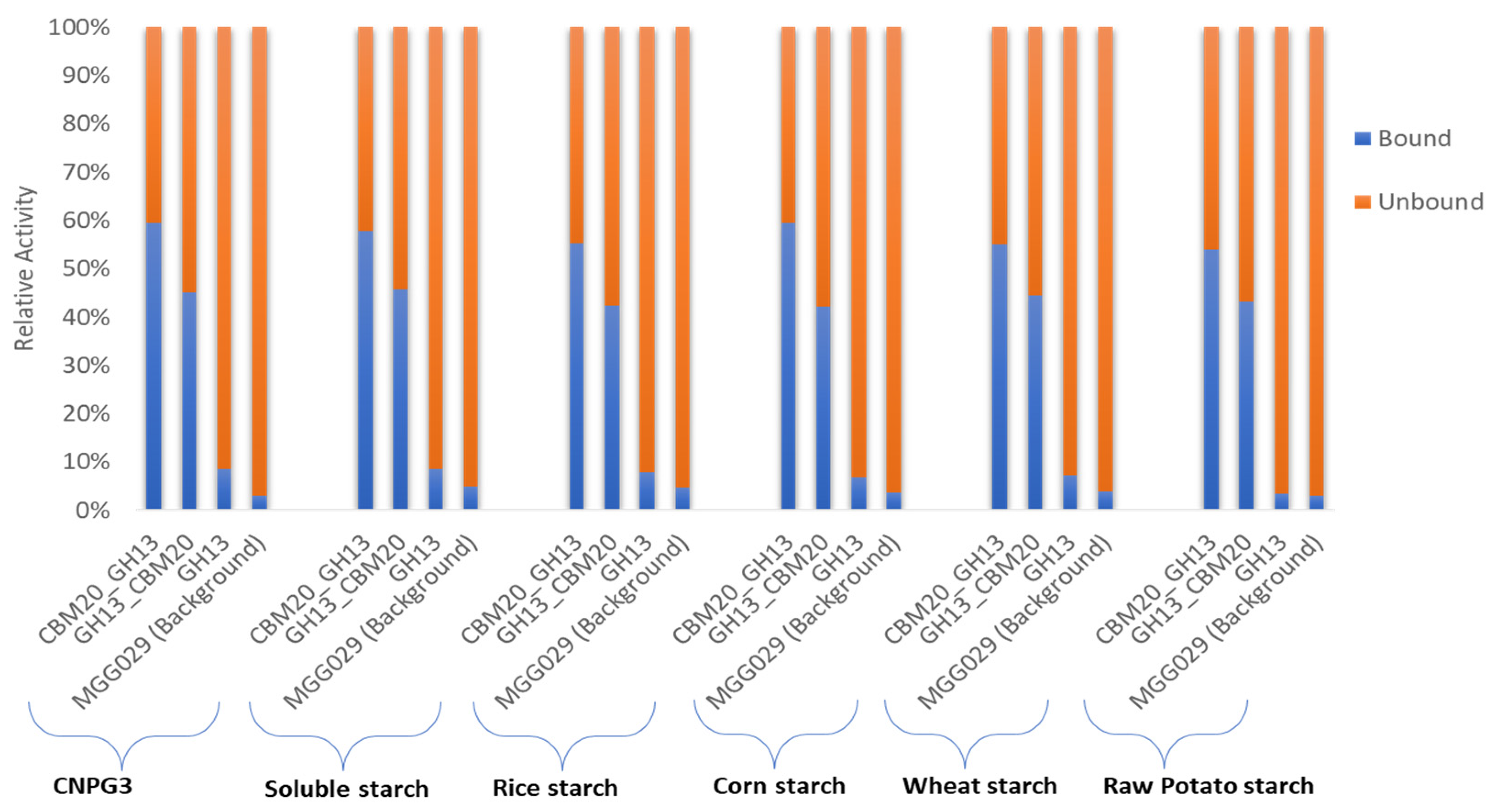
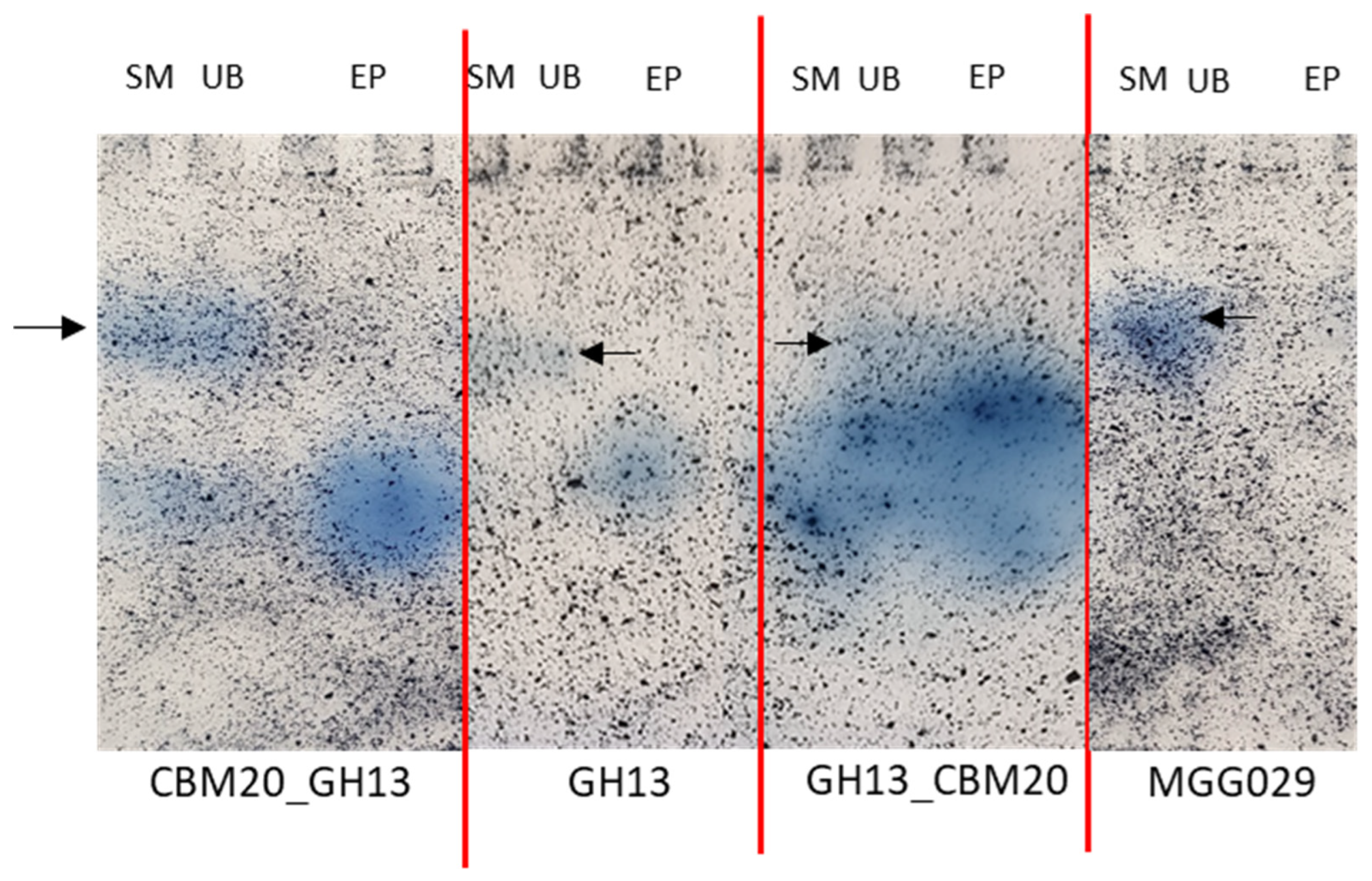
2.3. Purification of GH13 α-Amylase Variants Based on Starch Binding
2.4. Enzymatic Activity and Kinetic Parameter of Purified α-Amylases on Various Substrates
3. Material and Methods
3.1. Microbial Strains, Plasmid, Medium, and Substrates
3.2. Gene Design for Chimeric CBM20_GH13
3.3. Plasmid Construction, Transformation, Cultivation, and 3D Protein Modelling
3.4. Starch Binding Purification
3.5. Enzymatic Activity of α-Amylase
3.5.1. AZCL-Amylose
3.5.2. 2-Chloro-4-Nitrophenyl-α-D-Maltotrioside (CNPG3)
3.5.3. Dinitrosalicyclic Acid (DNS) Assay
3.6. SDS-PAGE and Zymogram Analysis, Protein Concentration
3.7. Kinetic Enzyme Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- de Souza, P.M.; de Oliveira Magalhães, P. Application of Microbial α-Amylase in Industry—A Review. Braz. J. Microbiol. 2010, 41, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.A.; Ali, S.; Hassan, A.; Tahir, H.M.; Mumtaz, S.; Mumtaz, S. Biosynthesis and Industrial Applications of α-Amylase: A Review. Arch. Microbiol. 2021, 203, 1281–1292. [Google Scholar] [CrossRef] [PubMed]
- Drula, E.; Garron, M.-L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The Carbohydrate-Active Enzyme Database: Functions and Literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef] [PubMed]
- Sidar, A.; Albuquerque, E.D.; Voshol, G.P.; Ram, A.F.J.; Vijgenboom, E.; Punt, P.J. Carbohydrate Binding Modules: Diversity of Domain Architecture in Amylases and Cellulases from Filamentous Microorganisms. Front. Bioeng. Biotechnol. 2020, 8, 871. [Google Scholar] [CrossRef]
- Janeček, Š.; Mareček, F.; MacGregor, E.A.; Svensson, B. Starch-Binding Domains as CBM Families–History, Occurrence, Structure, Function and Evolution. Biotechnol. Adv. 2019, 37, 107451. [Google Scholar] [CrossRef]
- Hayashida, S.; Kunisaki, S.; Nakao, M.; Flor, P.Q. Evidence for Raw Starch-Affinity Site on Aspergillus awamori Glucoamylase I. Agric. Biol. Chem. 1982, 46, 83–89. [Google Scholar] [CrossRef]
- Svensson, B.; Svendsen, T.G.; Svendsen, I.; Sakai, T.; Ottesen, M. Characterization of Two Forms of Glucoamylase from Aspergillus niger. Carlsberg Res. Commun. 1982, 47, 55–69. [Google Scholar] [CrossRef]
- Rodríguez-Sanoja, R.; Oviedo, N.; Sánchez, S. Microbial Starch-Binding Domain. Curr. Opin. Microbiol. 2005, 8, 260–267. [Google Scholar] [CrossRef]
- Jia, X.; Guo, Y.; Lin, X.; You, M.; Lin, C.; Chen, L.; Chen, J. Fusion of a Family 20 Carbohydrate-Binding Module (CBM20) with Cyclodextrin Glycosyltransferase of Geobacillus sp. CHB1 Improves Catalytic Efficiency. J. Basic Microbiol. 2017, 57, 471–480. [Google Scholar] [CrossRef]
- Southall, S.M.; Simpson, P.J.; Gilbert, H.J.; Williamson, G.; Williamson, M.P. The Starch-Binding Domain from Glucoamylase Disrupts the Structure of Starch. FEBS Lett. 1999, 447, 58–60. [Google Scholar] [CrossRef]
- Morris, V.J.; Gunning, A.P.; Faulds, C.B.; Williamson, G.; Svensson, B. AFM Images of Complexes between Amylose and Aspergillus niger Glucoamylase Mutants, Native and Mutant Starch Binding Domains: A Model for the Action of Glucoamylase. Starch Stärke 2005, 57, 1–7. [Google Scholar] [CrossRef]
- Peng, H.; Li, R.; Li, F.; Zhai, L.; Zhang, X.; Xiao, Y.; Gao, Y. Extensive Hydrolysis of Raw Rice Starch by a Chimeric α-Amylase Engineered with α-Amylase (AmyP) and a Starch-Binding Domain from Cryptococcus sp. S-2. Appl. Microbiol. Biotechnol. 2018, 102, 743–750. [Google Scholar] [CrossRef]
- Ngo, S.T.; Tran-Le, P.D.; Ho, G.T.; Le, L.Q.; Bui, L.M.; Vu, B.K.; Thu Phung, H.T.; Nguyen, H.-D.; Vo, T.-S.; Vu, V.V. Interaction of Carbohydrate Binding Module 20 with Starch Substrates. RSC Adv. 2019, 9, 24833–24842. [Google Scholar] [CrossRef]
- You, H.; Liang, C.; Zhang, O.; Xu, H.; Xu, L.; Chen, Y.; Xiang, X. Variation of Resistant Starch Content in Different Processing Types and Their Starch Granules Properties in Rice. Carbohydr. Polym. 2022, 276, 118742. [Google Scholar] [CrossRef]
- Naguleswaran, S.; Li, J.; Vasanthan, T.; Bressler, D.; Hoover, R. Amylolysis of Large and Small Granules of Native Triticale, Wheat and Corn Starches Using a Mixture of α-Amylase and Glucoamylase. Carbohydr. Polym. 2012, 88, 864–874. [Google Scholar] [CrossRef]
- Janket, A.; Vorasoot, N.; Toomsan, B.; Kaewpradit, W.; Banterng, P.; Kesmala, T.; Theerakulpisut, P.; Jogloy, S. Seasonal Variation in Starch Accumulation and Starch Granule Size in Cassava Genotypes in a Tropical Savanna Climate. Agronomy 2018, 8, 297. [Google Scholar] [CrossRef]
- Lindeboom, N.; Chang, P.R.; Tyler, R.T. Analytical, Biochemical and Physicochemical Aspects of Starch Granule Size, with Emphasis on Small Granule Starches: A Review. Starch Stärke 2004, 56, 89–99. [Google Scholar] [CrossRef]
- Singh, J.; Colussi, R.; McCarthy, O.J.; Kaur, L. Potato Starch and Its Modification. In Advances in Potato Chemistry and Technology; Elsevier: Amsterdam, The Netherlands, 2016; pp. 195–247. [Google Scholar] [CrossRef]
- Li, M.; Daygon, V.D.; Solah, V.; Dhital, S. Starch Granule Size: Does It Matter? Crit. Rev. Food Sci. Nutr. 2021, 1–21. [Google Scholar] [CrossRef]
- Dhital, S.; Warren, F.J.; Butterworth, P.J.; Ellis, P.R.; Gidley, M.J. Mechanisms of Starch Digestion by α-Amylase-Structural Basis for Kinetic Properties. Crit. Rev. Food Sci. Nutr. 2017, 57, 875–892. [Google Scholar] [CrossRef]
- Aleem, B.; Rashid, M.H.; Zeb, N.; Saqib, A.; Ihsan, A.; Iqbal, M.; Ali, H. Random Mutagenesis of Super Koji (Aspergillus oryzae): Improvement in Production and Thermal Stability of α-Amylases for Maltose Syrup Production. BMC Microbiol. 2018, 18, 200. [Google Scholar] [CrossRef]
- Tong, L.; Huang, H.; Zheng, J.; Wang, X.; Bai, Y.; Wang, X.; Wang, Y.; Tu, T.; Yao, B.; Qin, X.; et al. Engineering a Carbohydrate-Binding Module to Increase the Expression Level of Glucoamylase in Pichia Pastoris. Microb. Cell Factories 2022, 21, 95. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Tu, T.; Wang, Y.; Li, Y.; Luo, X.; Zheng, F.; Wang, X.; Bai, Y.; Huang, H.; Su, X.; et al. Improving the Catalytic Performance of a Talaromyces leycettanus α-Amylase by Changing the Linker Length. J. Agric. Food Chem. 2017, 65, 5041–5048. [Google Scholar] [CrossRef] [PubMed]
- Miao, H.; Zhao, Y.; Ma, Y.; Han, N.; Zhe, Y.; Tang, X.; Huang, Z. Improving the Thermostability of Endo-β-1,4-Glucanase by the Fusion of a Module Subdivided from Hyperthermophilic CBM9_1-2. Process Biochem. 2022, 114, 147–155. [Google Scholar] [CrossRef]
- Kuchtová, A.; Janeček, Š. Domain Evolution in Enzymes of the Neopullulanase Subfamily. Microbiology 2016, 162, 2099–2115. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Morimoto, N.; Saburi, W.; Mukai, A.; Imoto, K.; Takehana, T.; Koike, S.; Mori, H.; Matsui, H. Purification and Characterization of a Liquefying α-Amylase from Alkalophilic Thermophilic Bacillus sp. AAH-31. Biosci. Biotechnol. Biochem. 2012, 76, 1378–1383. [Google Scholar] [CrossRef]
- Saburi, W.; Morimoto, N.; Mukai, A.; Kim, D.H.; Takehana, T.; Koike, S.; Matsui, H.; Mori, H. A Thermophilic Alkalophilic α-Amylase from Bacillus sp. AAH-31 Shows a Novel Domain Organization among Glycoside Hydrolase Family 13 Enzymes. Biosci. Biotechnol. Biochem. 2013, 77, 1867–1873. [Google Scholar] [CrossRef]
- Oslancová, A.; Janeček, Š. Oligo-1,6-Glucosidase and Neopullulanase Enzyme Subfamilies from the α-Amylase Family Defined by the Fifth Conserved Sequence Region. Cell. Mol. Life Sci. CMLS 2002, 59, 1945–1959. [Google Scholar] [CrossRef]
- Chen, W.; Xie, T.; Shao, Y.; Chen, F. Phylogenomic Relationships between Amylolytic Enzymes from 85 Strains of Fungi. PLoS ONE 2012, 7, e49679. [Google Scholar] [CrossRef]
- Lin, S.-C.; Lin, I.-P.; Chou, W.-I.; Hsieh, C.-A.; Liu, S.-H.; Huang, R.-Y.; Sheu, C.-C.; Chang, M.D.-T. CBM21 Starch-Binding Domain: A New Purification Tag for Recombinant Protein Engineering. Protein Expr. Purif. 2009, 65, 261–266. [Google Scholar] [CrossRef]
- Xu, W.; Liu, Y.; Ye, Y.; Liu, M.; Han, L.; Song, A.; Liu, L. C-Terminal Carbohydrate-Binding Module 9_2 Fused to the N-Terminus of GH11 Xylanase from Aspergillus niger. Biotechnol. Lett. 2016, 38, 1739–1745. [Google Scholar] [CrossRef]
- Juge, N.; Nøhr, J.; Le Gal-Coëffet, M.-F.; Kramhøft, B.; Furniss, C.S.M.; Planchot, V.; Archer, D.B.; Williamson, G.; Svensson, B. The Activity of Barley Alpha-Amylase on Starch Granules Is Enhanced by Fusion of a Starch Binding Domain from Aspergillus niger Glucoamylase. Biochim. Biophys. Acta 2006, 1764, 275–284. [Google Scholar] [CrossRef]
- Vu, V.V.; Hangasky, J.A.; Detomasi, T.C.; Henry, S.J.W.; Ngo, S.T.; Span, E.A.; Marletta, M.A. Substrate Selectivity in Starch Polysaccharide Monooxygenases. J. Biol. Chem. 2019, 294, 12157–12166. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Y.; Christensen, S.J.; Janeček, Š.; Bai, Y.; Møller, M.S.; Svensson, B. Impact of Starch Binding Domain Fusion on Activities and Starch Product Structure of 4-α-Glucanotransferase. Molecules 2023, 28, 1320. [Google Scholar] [CrossRef]
- Ruiz, D.M.; Turowski, V.R.; Murakami, M.T. Effects of the Linker Region on the Structure and Function of Modular GH5 Cellulases. Sci. Rep. 2016, 6, 28504. [Google Scholar] [CrossRef]
- Setter-Lamed, E.; Moraïs, S.; Stern, J.; Lamed, R.; Bayer, E.A. Modular Organization of the Thermobifida fusca Exoglucanase Cel6B Impacts Cellulose Hydrolysis and Designer Cellulosome Efficiency. Biotechnol. J. 2017, 12, 1700205. [Google Scholar] [CrossRef]
- Srivastava, A.; Nagar, P.; Rathore, S.; Adlakha, N. The Linker Region Promotes Activity and Binding Efficiency of Modular LPMO towards Polymeric Substrate. Microbiol. Spectr. 2022, 10, e0269721. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, Y.-H.; Liu, T.-Q.; Zhang, H.; Zhang, H.; Li, J. The GlaA Signal Peptide Substantially Increases the Expression and Secretion of α-Galactosidase in Aspergillus niger. Biotechnol. Lett. 2018, 40, 949–955. [Google Scholar] [CrossRef]
- Jørgensen, A.D.; Nøhr, J.; Kastrup, J.S.; Gajhede, M.; Sigurskjold, B.W.; Sauer, J.; Svergun, D.I.; Svensson, B.; Vestergaard, B. Small Angle X-Ray Studies Reveal That Aspergillus niger Glucoamylase Has a Defined Extended Conformation and Can Form Dimers in Solution. J. Biol. Chem. 2008, 283, 14772–14780. [Google Scholar] [CrossRef]
- Boel, E.; Hjort, I.; Svensson, B.; Norris, F.; Norris, K.E.; Fiil, N.P. Glucoamylases G1 and G2 from Aspergillus niger Are Synthesized from Two Different but Closely Related MRNAs. EMBO J. 1984, 3, 1097–1102. [Google Scholar] [CrossRef]
- Sorimachi, K.; Le Gal-Coëffet, M.F.; Williamson, G.; Archer, D.B.; Williamson, M.P. Solution Structure of the Granular Starch Binding Domain of Aspergillus niger Glucoamylase Bound to Beta-Cyclodextrin. Structure 1997, 5, 647–661. [Google Scholar] [CrossRef] [PubMed]
- Paldi, T.; Levy, I.; Shoseyov, O. Glucoamylase Starch-Binding Domain of Aspergillus niger B1: Molecular Cloning and Functional Characterization. Biochem. J. 2003, 372 Pt 3, 905–910. [Google Scholar] [CrossRef]
- Lee, J.; Paetzel, M. Structure of the Catalytic Domain of Glucoamylase from Aspergillus niger. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011, 67 Pt 2, 188–192. [Google Scholar] [CrossRef] [PubMed]
- Tanackovic, V.; Rydahl, M.G.; Pedersen, H.L.; Motawia, M.S.; Shaik, S.S.; Mikkelsen, M.D.; Krunic, S.L.; Fangel, J.U.; Willats, W.G.T.; Blennow, A. High Throughput Screening of Starch Structures Using Carbohydrate Microarrays. Sci. Rep. 2016, 6, 30551. [Google Scholar] [CrossRef] [PubMed]
- Suyama, Y.; Muraki, N.; Kusunoki, M.; Miyake, H. Crystal Structure of the Starch-Binding Domain of Glucoamylase from Aspergillus niger. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2017, 73 Pt 10, 550–554. [Google Scholar] [CrossRef]
- Zhong, Y.; Sagnelli, D.; Topbjerg, H.B.; Hasler-Sheetal, H.; Andrzejczak, O.A.; Hooshmand, K.; Gislum, R.; Jiang, D.; Møller, I.M.; Blennow, A.; et al. Expression of Starch-Binding Factor CBM20 in Barley Plastids Controls the Number of Starch Granules and the Level of CO2 Fixation. J. Exp. Bot. 2020, 71, 234–246. [Google Scholar] [CrossRef]
- Pel, H.J.; de Winde, J.H.; Archer, D.B.; Dyer, P.S.; Hofmann, G.; Schaap, P.J.; Turner, G.; de Vries, R.P.; Albang, R.; Albermann, K.; et al. Genome Sequencing and Analysis of the Versatile Cell Factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 2007, 25, 221–231. [Google Scholar] [CrossRef]
- van der Kaaij, R.M.; Yuan, X.-L.; Franken, A.; Ram, A.F.J.; Punt, P.J.; van der Maarel, M.J.E.C.; Dijkhuizen, L. Two Novel, Putatively Cell Wall-Associated and Glycosylphosphatidylinositol-Anchored Alpha-Glucanotransferase Enzymes of Aspergillus niger. Eukaryot. Cell 2007, 6, 1178–1188. [Google Scholar] [CrossRef]
- Yuan, X.-L.; van der Kaaij, R.M.; van den Hondel, C.A.M.J.J.; Punt, P.J.; van der Maarel, M.J.E.C.; Dijkhuizen, L.; Ram, A.F.J. Aspergillus niger Genome-Wide Analysis Reveals a Large Number of Novel Alpha-Glucan Acting Enzymes with Unexpected Expression Profiles. Mol. Genet. Genom. MGG 2008, 279, 545–561. [Google Scholar] [CrossRef]
- Wang, J.; Li, Y.; Lu, F. Molecular Cloning and Biochemical Characterization of an α-Amylase Family from Aspergillus niger. Electron. J. Biotechnol. 2018, 32, 55–62. [Google Scholar] [CrossRef]
- Vujičić-Žagar, A.; Dijkstra, B.W. Monoclinic Crystal Form of Aspergillus niger α-Amylase in Complex with Maltose at 1.8 Å Resolution. Acta Crystallogr. Sect. F 2006, 62, 716–721. [Google Scholar] [CrossRef]
- Boel, E.; Brady, L.; Brzozowski, A.M.; Derewenda, Z.; Dodson, G.G.; Jensen, V.J.; Petersen, S.B.; Swift, H.; Thim, L.; Woldike, H.F. Calcium Binding in Alpha-Amylases: An X-Ray Diffraction Study at 2.1-Å Resolution of Two Enzymes from Aspergillus. Biochemistry 1990, 29, 6244–6249. [Google Scholar] [CrossRef]
- Williamson, M.P. The Structure and Function of Proline-Rich Regions in Proteins. Biochem. J. 1994, 297 Pt 2, 249–260. [Google Scholar] [CrossRef]
- Skaf, M.S.; Polikarpov, I.; Stanković, I.M. A Linker of the Proline-Threonine Repeating Motif Sequence Is Bimodal. J. Mol. Model. 2020, 26, 178. [Google Scholar] [CrossRef]
- Gilkes, N.R.; Henrissat, B.; Kilburn, D.G.; Miller, R.C.; Warren, R.A. Domains in Microbial Beta-1, 4-Glycanases: Sequence Conservation, Function, and Enzyme Families. Microbiol. Rev. 1991, 55, 303–315. [Google Scholar] [CrossRef]
- Rizk, M.; Antranikian, G.; Elleuche, S. End-to-End Gene Fusions and Their Impact on the Production of Multifunctional Biomass Degrading Enzymes. Biochem. Biophys. Res. Commun. 2012, 428, 1–5. [Google Scholar] [CrossRef]
- Sørensen, C.S.; Kjaergaard, M. Effective Concentrations Enforced by Intrinsically Disordered Linkers Are Governed by Polymer Physics. Proc. Natl. Acad. Sci. USA 2019, 116, 23124–23131. [Google Scholar] [CrossRef]
- Samuel, D.; Kumar, T.K.; Ganesh, G.; Jayaraman, G.; Yang, P.W.; Chang, M.M.; Trivedi, V.D.; Wang, S.L.; Hwang, K.C.; Chang, D.K.; et al. Proline Inhibits Aggregation during Protein Refolding. Protein Sci. Publ. Protein Soc. 2000, 9, 344–352. [Google Scholar] [CrossRef]
- Sammond, D.W.; Payne, C.M.; Brunecky, R.; Himmel, M.E.; Crowley, M.F.; Beckham, G.T. Cellulase Linkers Are Optimized Based on Domain Type and Function: Insights from Sequence Analysis, Biophysical Measurements, and Molecular Simulation. PLoS ONE 2012, 7, e48615. [Google Scholar] [CrossRef]
- Gräwe, A.; Stein, V. Linker Engineering in the Context of Synthetic Protein Switches and Sensors. Trends Biotechnol. 2021, 39, 731–744. [Google Scholar] [CrossRef] [PubMed]
- Sujka, M.; Jamroz, J. α-Amylolysis of Native Potato and Corn Starches—SEM, AFM, Nitrogen and Iodine Sorption Investigations. LWT Food Sci. Technol. 2009, 42, 1219–1224. [Google Scholar] [CrossRef]
- Huang, H.-B.; Chi, M.-C.; Hsu, W.-H.; Liang, W.-C.; Lin, L.-L. Construction and One-Step Purification of Bacillus Kaustophilus Leucine Aminopeptidase Fused to the Starch-Binding Domain of Bacillus sp. Strain TS-23 Alpha-Amylase. Bioprocess Biosyst. Eng. 2005, 27, 389. [Google Scholar] [CrossRef]
- Mendu, D.R.; Ratnam, B.V.V.; Purnima, A.; Ayyanna, C. Affinity Chromatography of α-Amylase from Bacillus Licheniformis. Enzyme Microb. Technol. 2005, 37, 712–717. [Google Scholar] [CrossRef]
- Warren, F.J.; Royall, P.G.; Gaisford, S.; Butterworth, P.J.; Ellis, P.R. Binding Interactions of α-Amylase with Starch Granules: The Influence of Supramolecular Structure and Surface Area. Carbohydr. Polym. 2011, 86, 1038–1047. [Google Scholar] [CrossRef]
- Aller, E.E.J.G.; Abete, I.; Astrup, A.; Martinez, J.A.; van Baak, M.A. Starches, Sugars and Obesity. Nutrients 2011, 3, 341–369. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Oh, S.-K.; Chung, H.-J.; Park, H.-J. Structural and Physicochemical Properties of Native Starches and Non-Digestible Starch Residues from Korean Rice Cultivars with Different Amylose Contents. Food Hydrocoll. 2020, 102, 105544. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Delano, W. The PyMOL Molecular Graphics System (2002); Delano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Rosignoli, S.; Paiardini, A. Boosting the Full Potential of PyMOL with Structural Biology Plugins. Biomolecules 2022, 12, 1764. [Google Scholar] [CrossRef]
- Van den Hondel, C.A.M.J.J.; Punt, P.J.; Van Gorcom, R.F.M. 18—Heterologous Gene Expression in Filamentous Fungi. In More Gene Manipulations in Fungi; Bennett, J.W., Lasure, L.L., Eds.; Academic Press: San Diego, CA, USA, 1991; pp. 396–428. [Google Scholar] [CrossRef]
- Yuan, X.-L.; Roubos, J.A.; van den Hondel, C.A.M.J.J.; Ram, A.F.J. Identification of InuR, a New Zn(II)2Cys6 Transcriptional Activator Involved in the Regulation of Inulinolytic Genes in Aspergillus niger. Mol. Genet. Genom. 2008, 279, 11–26. [Google Scholar] [CrossRef]
- Arentshorst, M.; Ram, A.F.J.; Meyer, V. Using Non-Homologous End-Joining-Deficient Strains for Functional Gene Analyses in Filamentous Fungi. In Plant Fungal Pathogens; Bolton, M.D., Thomma, B.P.H.J., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2012; Volume 835, pp. 133–150. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam Protein Families Database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Shimokawa, T.; Yamaguchi, M.; Murata, H. Agar Plate Assays Using Dye-Linked Substrates Differentiate Members of Tricholoma sect. Caligata, Ectomycorrhizal Symbionts Represented by Tricholoma matsutake. Mycoscience 2017, 58, 432–437. [Google Scholar] [CrossRef]
- Timalsina, D.; Bhusal, D.; Devkota, H.P.; Pokhrel, K.P.; Sharma, K.R. α-Amylase Inhibitory Activity of Catunaregam spinosa (Thunb.) Tirveng.: In Vitro and In Silico Studies. BioMed Res. Int. 2021, 2021, e4133876. [Google Scholar] [CrossRef]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Elia, G. Cell Surface Protein Biotinylation for SDS-PAGE Analysis. In Electrophoretic Separation of Proteins: Methods and Protocols; Kurien, B.T., Scofield, R.H., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; pp. 449–459. [Google Scholar] [CrossRef]
- Sahnoun, M.; Bejar, S.; Sayari, A.; Triki, M.A.; Kriaa, M.; Kammoun, R. Production, Purification and Characterization of Two α-Amylase Isoforms from a Newly Isolated Aspergillus Oryzae Strain S2. Process Biochem. 2012, 47, 18–25. [Google Scholar] [CrossRef]
- Yi, Z.; Fang, Y.; He, K.; Liu, D.; Luo, H.; Zhao, D.; He, H.; Jin, Y.; Zhao, H. Directly Mining a Fungal Thermostable α-Amylase from Chinese Nong-Flavor Liquor Starter. Microb. Cell Factories 2018, 17, 30. [Google Scholar] [CrossRef]
- Lineweaver, H.; Burk, D. The Determination of Enzyme Dissociation Constants. J. Am. Chem. Soc. 1934, 56, 658–666. [Google Scholar] [CrossRef]
- Whitaker, J.R. Principles of Enzymology for the Food Sciences; CRC Press LLC: Boca Raton, FL, USA, 2018. [Google Scholar]

| α-Amylase | Linker Composition |
|---|---|
| Chimeric CBM20_GH13 | VSQEQWWCSEDDPAAVAASQAARVYMDCHPKPRHPRKPIPVFVPD |
| Native GH13_CBM20 | GSNSSTTTTTTATSSSTATSKSASTSSTSTACTATST |
| α-Amylase | Rice Starch | Soluble Starch | Corn Starch | Wheat Starch | Raw/Native Potato Starch | CNPG3 | |
|---|---|---|---|---|---|---|---|
| Specific activity a | CBM20_GH13 | 1752 ± 18 | 1126 ± 12 | 684 ± 3 | 370 ± 11 | 177 ± 1 | 4815 ± 16 |
| GH13_CBM20 | 1426 ± 27 | 902 ± 14 | 538 ± 6 | 275 ± 7 | 84 ± 0.8 | 3886 ± 33 | |
| Relative activity | CBM20_GH13 | 100 ± 1 | 100 ± 1 | 100 ± 0.4 | 100 ± 3 | 100 ± 0.6 | 100 ± 0.3 |
| GH13_CBM20 | 81 ± 2 | 80 ± 2 | 78 ± 1 | 74 ± 2.5 | 47 ± 1 | 81 ± 0.8 |
| Substrate | Vmax (µmol/min) a | Km (mg/mL) a | ||
|---|---|---|---|---|
| CBM20_GH13 | GH13_CBM20 | CBM20_GH13 | GH13_CBM20 | |
| CNPG3 | 90.9 | 71.4 | 4.2 | 4.9 |
| Soluble starch | 20.3 | 15.6 | 3.5 | 4.5 |
| Rice starch | 30.2 | 22.4 | 4.0 | 6.4 |
| Corn Starch | 10.3 | 8.1 | 2.9 | 4.6 |
| Wheat starch | 6.2 | 4.2 | 1.2 | 2.2 |
| Raw Potato starch | 3.5 | 1.5 | 0.8 | 1.6 |
| List of Primer Name | Sequence 5′ to 3′ | Targeted Site |
|---|---|---|
| AamA_2 (forward) | TGGCGGACACAATCCATC | GH13 aamA gene of A. niger CBS 513.88 from the plasmid containing CBM20_GH13 |
| GH13AamARev (reverse) | GGCCAGACCTGTGCAGAC | GH13 aamA gene of A. niger CBS 513.88 including glaA signal sequence from the plasmid containing CBM20_GH13 |
| OriAamAf (forward) | ACATGTCGAGACTATCGACTTCA | Native GH13_CBM20 from A. niger AB4.1 |
| OriAamA_CBM20r (reverse) | GGATCCCTACCTCCAAGTATCAACCACC | Native GH13_CBM20 from A. niger AB4.1 |
| MBL852 (forward) | GCTACATCCATACTCCA | GPD Promoter until the gene of insert for confirming correct construct |
| MBL858 (reverse) | ATATCCAGATTCGTCAAGCTG | trpC terminator until the gene of insert for confirming correct construct |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sidar, A.; Voshol, G.P.; Vijgenboom, E.; Punt, P.J. Novel Design of an α-Amylase with an N-Terminal CBM20 in Aspergillus niger Improves Binding and Processing of a Broad Range of Starches. Molecules 2023, 28, 5033. https://doi.org/10.3390/molecules28135033
Sidar A, Voshol GP, Vijgenboom E, Punt PJ. Novel Design of an α-Amylase with an N-Terminal CBM20 in Aspergillus niger Improves Binding and Processing of a Broad Range of Starches. Molecules. 2023; 28(13):5033. https://doi.org/10.3390/molecules28135033
Chicago/Turabian StyleSidar, Andika, Gerben P. Voshol, Erik Vijgenboom, and Peter J. Punt. 2023. "Novel Design of an α-Amylase with an N-Terminal CBM20 in Aspergillus niger Improves Binding and Processing of a Broad Range of Starches" Molecules 28, no. 13: 5033. https://doi.org/10.3390/molecules28135033
APA StyleSidar, A., Voshol, G. P., Vijgenboom, E., & Punt, P. J. (2023). Novel Design of an α-Amylase with an N-Terminal CBM20 in Aspergillus niger Improves Binding and Processing of a Broad Range of Starches. Molecules, 28(13), 5033. https://doi.org/10.3390/molecules28135033







