Four Undescribed Pyranones from the Scutellaria formosana-Derived Endophytic Fungi Ascomycota sp. FAE17
Abstract
:1. Introduction
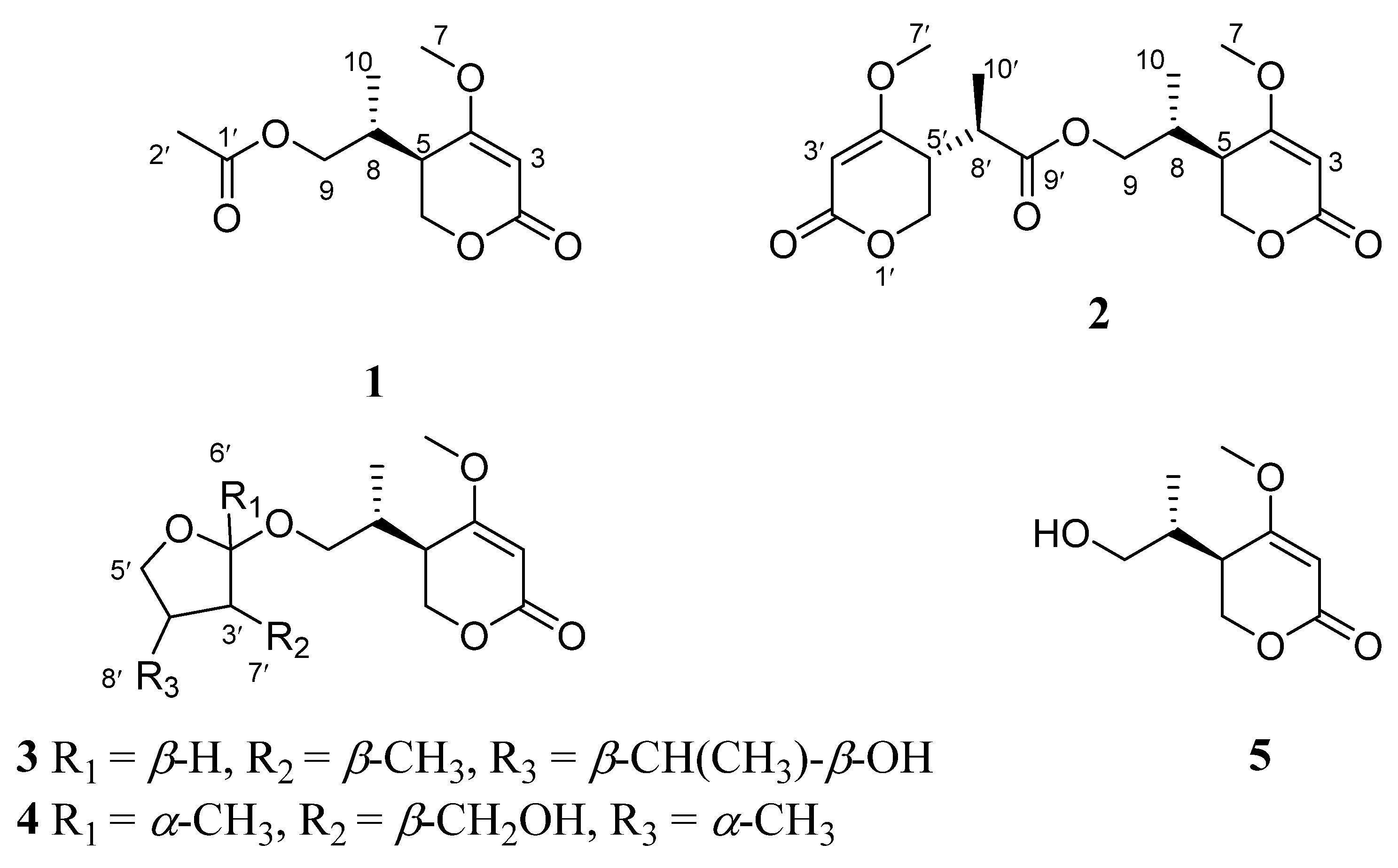
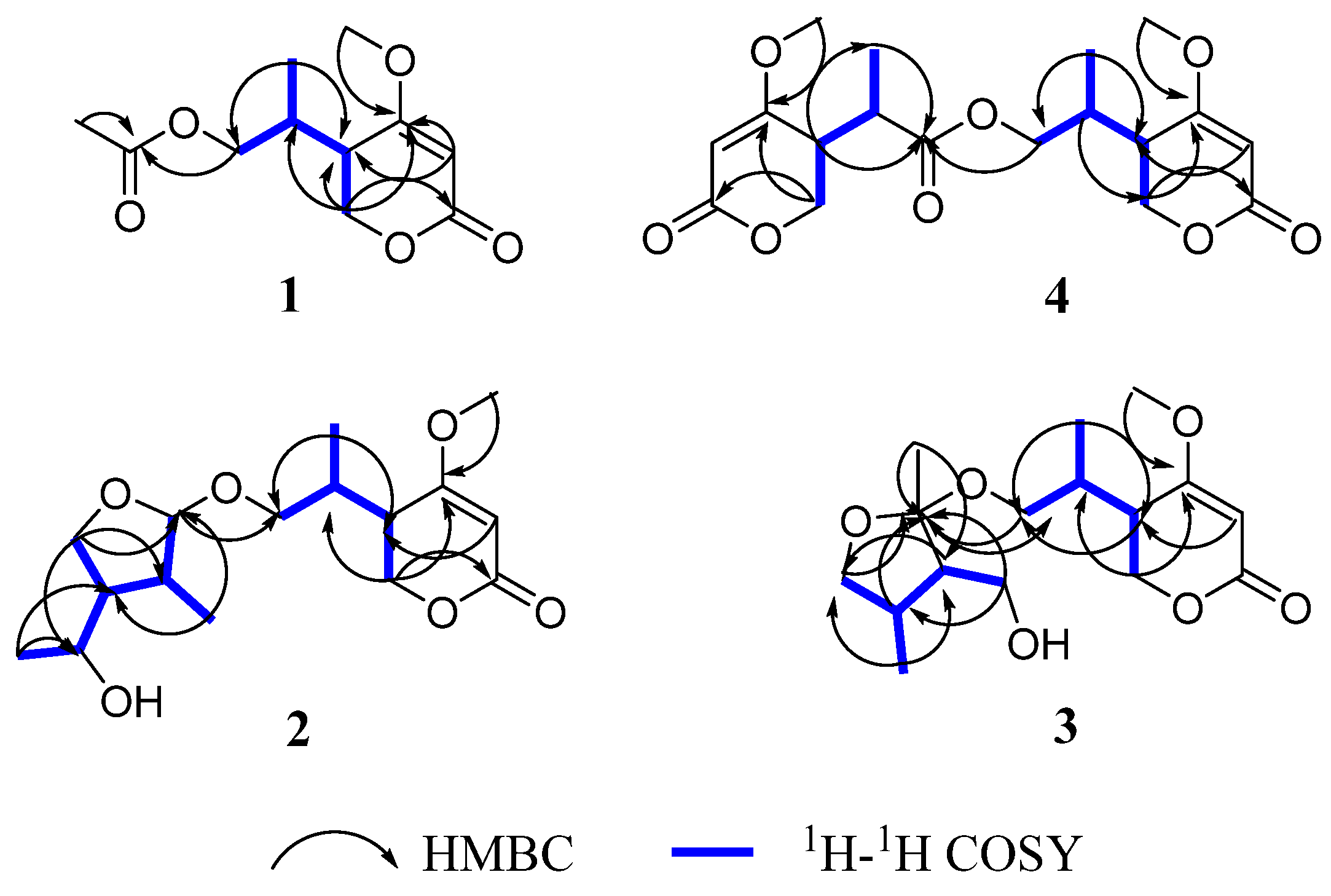
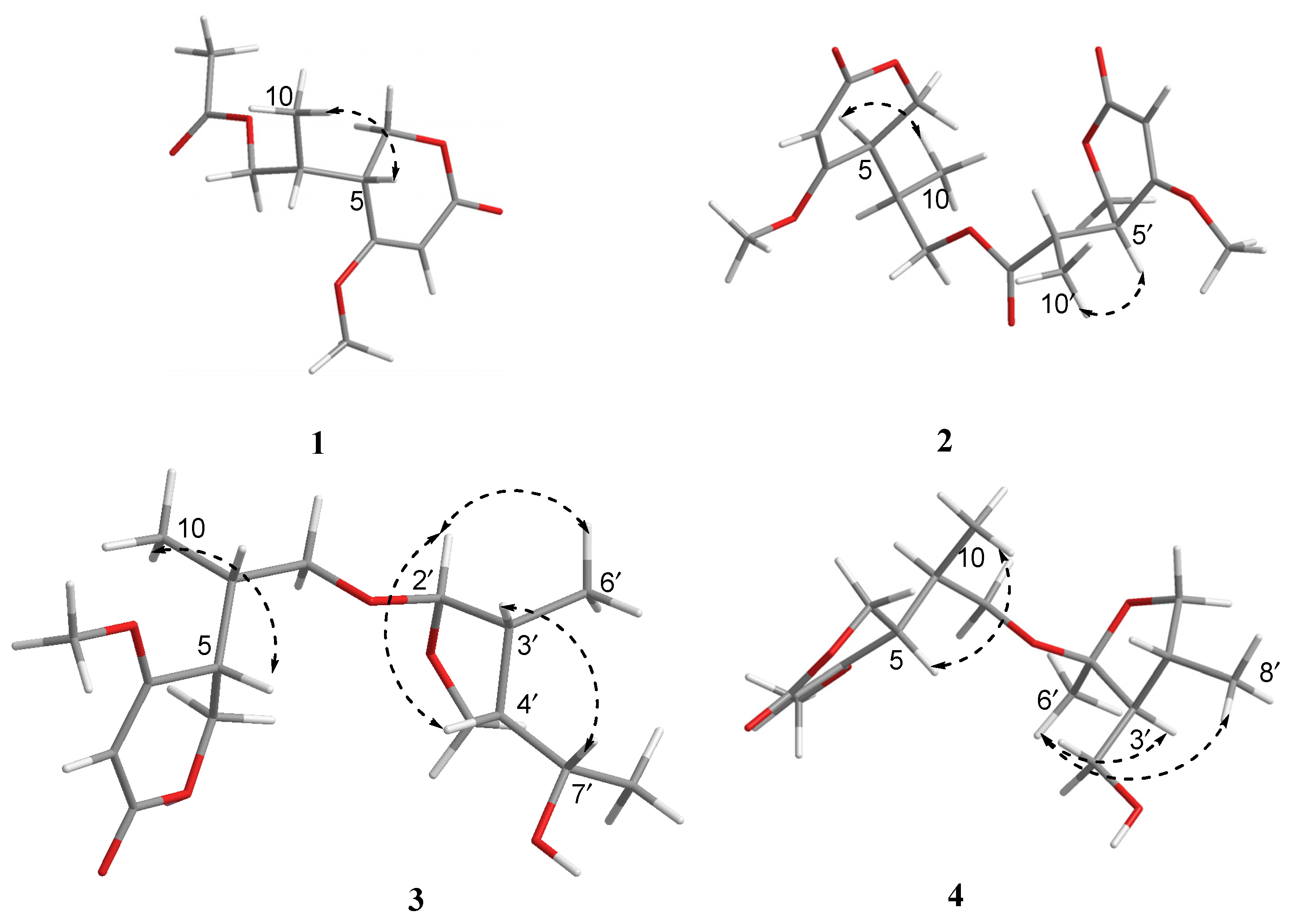
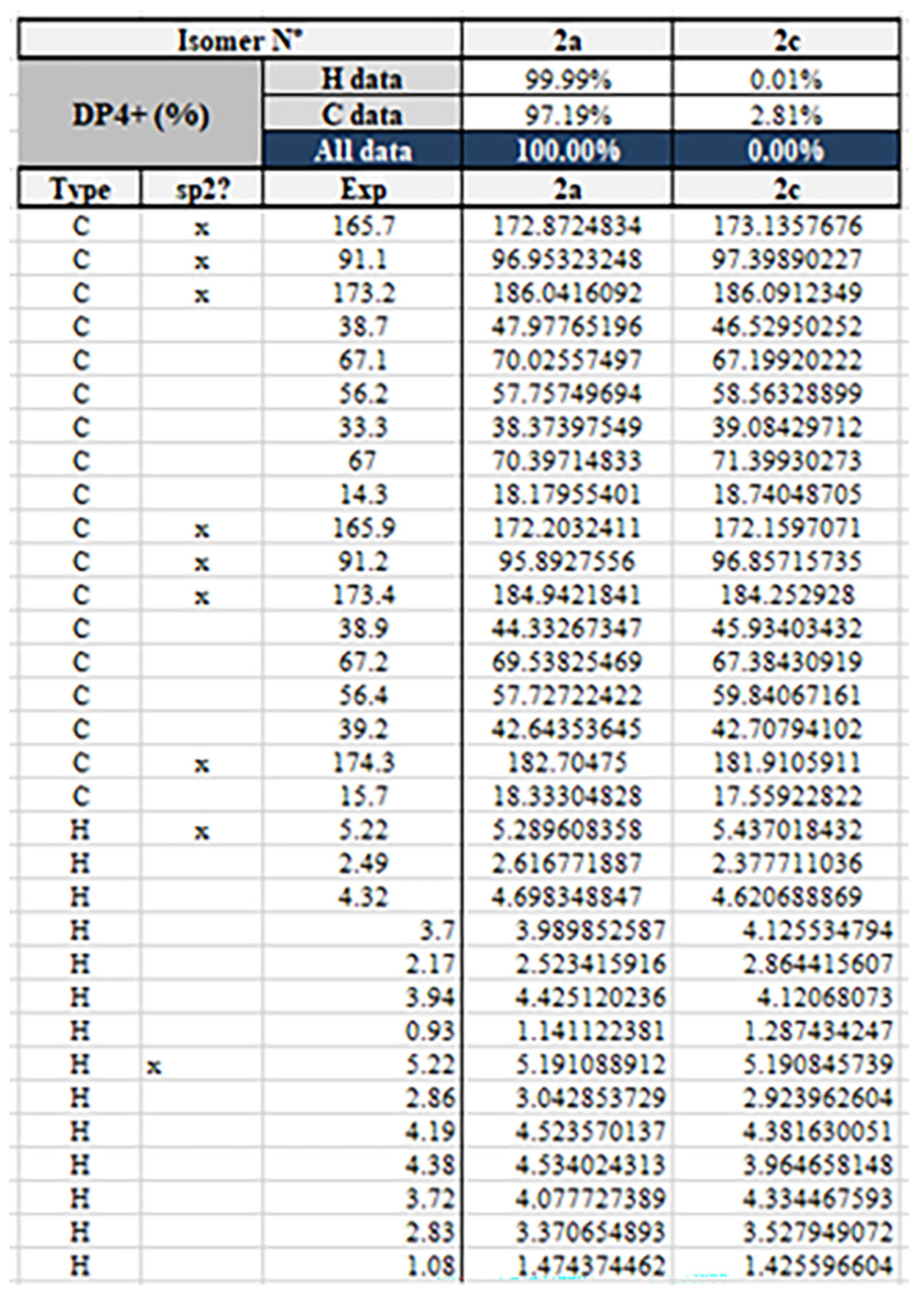
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Materials
3.3. Fermentation, Extraction, and Isolation
3.4. Biological Assays
3.4.1. Antibacterial Activity
3.4.2. Cytotoxic Activity
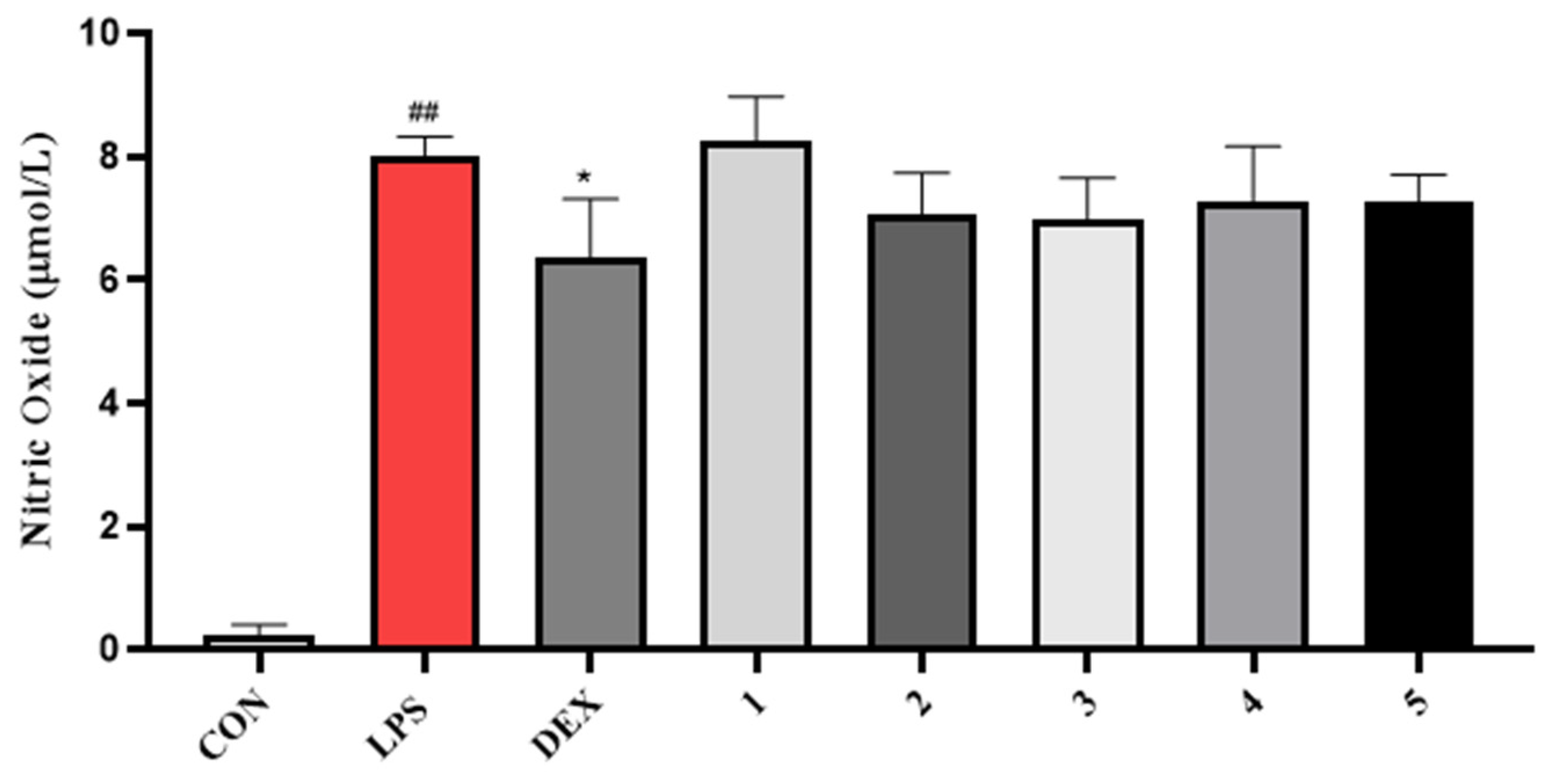
3.4.3. NO Inhibitory Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Guo, B.; Wang, Y.; Sun, X.; Tang, K. Bioactive natural products from endophytes: A review. Appl. Biochem. Micro. 2008, 44, 136–142. [Google Scholar] [CrossRef]
- Elsebai, M.F.; Tejesvi, M.V.; Pirttil€a, A.M. Endophytes as a novel source of bioactive new structures. In Advances in Endophytic Research; Springer: Berlin/Heidelberg, Germany, 2014; pp. 191–202. [Google Scholar]
- Strobel, G.; Daisy, B.; Castillo, U.; Harper, J. Natural products from endophytic microorganisms. J. Nat. Prod. 2004, 67, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, M.R.; Molina, G.; Dionísio, P.; Marostica, M.R.; Pastore, G.M. The use of endophytes to obtain bioactive compounds and their application in biotransformation process. Biotechnol. Res. Int. 2011, 2011, 576286. [Google Scholar] [CrossRef] [Green Version]
- Chang, S.; Cai, M.; Xiao, T.; Chen, Y.; Zhao, W.; Yu, L.; Shao, R.G.; Jiang, W.; Zhang, T.; Gan, M.L.; et al. Prenylemestrins A and B: Two unexpected epipolythiodioxopiperazines with a thioethanothio bridge from Emericella sp. isolated by genomic analysis. Org. Lett. 2022, 24, 5941–5945. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.D.; Xu, H.B.; Deng, Z.W.; Yan, Y.J.; Yuan, Z.B.; Liu, X.Z.; Su, Z.P.; Yang, S.; Zhang, Y.; Rao, Y.J. Discovery of the biosynthetic pathway of beticolin 1 reveals a novel non-heme iron-dependent oxygenase for anthraquinone ring cleavage. Angew. Chem. Int. Ed. 2022, 37, e202208772. [Google Scholar]
- Xu, K.; Li, R.J.; Zhu, R.X.; Li, X.B.; Xu, Y.L.; He, Q.B.; Xie, F.; Qiao, Y.A.; Luan, X.Y.; Lou, H.X. Xylarins A–D, two pairs of diastereoisomeric isoindoline alkaloids from the endolichenic fungus Xylaria sp. Org. Lett. 2021, 23, 7751–7754. [Google Scholar] [CrossRef]
- Yan, L.H.; Li, P.H.; Li, X.M.; Yang, S.Q.; Liu, K.C.; Wang, B.G.; Li, X. Chevalinulins A and B, proangiogenic alkaloids with a spiro [bicyclo [2.2.2] octane-diketopiperazine] skeleton from deep-sea cold-seep-derived fungus Aspergillus chevalieri CS-122. Org. Lett. 2022, 24, 2684–2688. [Google Scholar] [CrossRef]
- Zhai, Y.J.; Huo, G.M.; Zhang, Q.; Li, D.; Wang, D.C.; Qi, J.Z.; Han, W.B.; Gao, J.M. Phaeosphaones: Tyrosinase inhibitory thiodiketopiperazines from an endophytic Phaeosphaeria fuckelii. J. Nat. Prod. 2020, 83, 1592–1597. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, Z.; Fukaya, K.; Urabe, D.; Harunari, E.; Oku, N.; Igarashi, Y. Catellatolactams A–C, plant growth-promoting ansamacrolactams from a rare actinomycete of the genus Catellatospora. J. Nat. Prod. 2022, 85, 1993–1999. [Google Scholar] [CrossRef]
- Chen, X.; Chen, W.H.; Chen, G.Y.; Han, C.R.; He, J.J.; Zhou, X.M.; Yu, Z.X.; Dai, C.Y.; Song, X.P. Neo-clerodane diterpenoids from the whole plants of Scutellaria formosana. Phytochemistry 2018, 145, 1–9. [Google Scholar] [CrossRef]
- Wang, P.X.; Liu, F.; Yang, X.Y.; Liang, Y.; Li, S.; Su, G.C.; Jin, D.Q.; Ohizumi, Y.; Xu, J.; Guo, Y.Q. Clerodane diterpenoids from Scutellaria formosana with inhibitory effects on NO production and interactions with iNOS protein. Phytochemistry 2017, 144, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.X.; Yang, X.Y.; Liu, F.; Liang, Y.; Su, G.C.; Tuerhong, M.; Jin, D.Q.; Xu, J.; Lee, D.H.; Guo, Y.Q. Nitric oxide inhibitors with a spiro diterpenoid skeleton from Scutellaria formosana: Structures, NO inhibitory effects, and interactions with iNOS. Bioorg. Chem. 2018, 76, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, G.Y.; Chen, W.H.; Han, C.R.; Song, X.P.; Pan, Y. Study on flavone constituents from Scutellaria formosana and their biological activities. J. Chin. Med. Mater. 2016, 39, 2270–2273. [Google Scholar]
- Tian, Y.Q.; Lin, X.P.; Liu, J.; Kaliyaperumal, K.; Ai, W.; Ju, Z.R.; Yang, B.; Wang, J.F.; Yang, X.W.; Liu, Y.H. Ascomycotin A, a new citromycetin analogue produced by Ascomycota sp. Ind19F07 isolated from deep sea sediment. Nat. Prod. Res. 2015, 29, 820–826. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.M.; Chen, S.H.; Qiu, P.; Tan, C.B.; Long, Y.H.; Lu, Y.J.; She, Z.G. (+)- and (-)-Ascomlactone A: A pair of novel dimeric polyketides from a mangrove endophytic fungus Ascomycota sp. SK2YWS-L. Org. Biomol. Chem. 2017, 15, 10276–10280. [Google Scholar] [CrossRef]
- Tan, C.B.; Liu, Z.M.; Chen, S.H.; Huang, X.S.; Cui, H.; Long, Y.H.; Lu, Y.J.; She, Z.G. Antioxidative polyketones from the mangrove-derived fungus Ascomycota sp. SK2YWS-L. Sci. Rep. 2016, 6, 36609–36618. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.M.; Qiu, P.J.; Chen, G.Y.; Chen, Y.; Liu, H.J.; She, Z.G. Anti-inflammatory polyketides from the mangrove-derived fungus Ascomycota sp. SK2YWS-L. Tetrahedron 2018, 74, 746–751. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Z.M.; Liu, H.J.; Pan, Y.P.; Li, J.; Liu, L.; She, Z.G. Dichloroisocoumarins with potential anti-inflammatory activity from the mangrove endophytic fungus Ascomycota sp. CYSK-4. Mar. Drugs 2018, 16, 54. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.; Nong, X.H.; Yang, J.N.; Li, C.; Han, C.R.; Song, X.P.; Sun, Z.F.; Hui, Y.; Chen, W.H. Study on the secondary metabolites of the endophytic fungus Aspergillus terreus HQ100X-1 in Scutellaria formosana. Chin. J. Org. Chem. 2022, 42, 2961–2966. [Google Scholar] [CrossRef]
- Jian Li, J.; Wu, X.F.; Ding, G.; Feng, Y.; Jiang, X.J.; Guo, L.D.; Che, Y.S. α-pyrones and pyranes from the plant pathogenic fungus Pestalotiopsis scirpina. Eur. J. Org. Chem. 2012, 12, 2445–2452. [Google Scholar]
- Praphatsorn, S.; Vatcharin, R.; Souwalak, P.; Sita, P.; Jariya, S.; Suparerk, B.; Sawinee, S.; Chatchai, M. Depsidones and an α-pyrone derivative from Simpilcillium sp. PSU-H41, an endophytic fungus from Hevea brasiliensis leaf. Phytochemistry 2017, 143, 115–123. [Google Scholar]
- Li, H.L.; Li, X.M.; Ying, Z.; Li, H.Y.; Wang, B.G. Bisabolane sesquiterpene and cyclopentene derivatives from the marine algal-derived endophytic fungus Trichoderma asperellum EN-764. Phytochemistry 2023, 210, 113644. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A. Gaussian 09W (Revision A.02); Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Bruhn, T.; Schaumloffel, A.; Hemberger, Y.; Bringmann, G. SpecDis: Quantifying the comparison of calculated and experimental electronic circular dichroism spectra. Chirality 2013, 25, 243–249. [Google Scholar] [CrossRef]
- Rolf, J.; Sakshi, S.; Volker, H.; Brigitte, K.; Marc, S.; Rolf, M. Pyrronazols, metabolites from the Myxobacteria Nannocystis pusilla and N. exedens, are unusual chlorinated pyrone-oxazole-pyrroles. J. Nat. Prod. 2014, 77, 320–326. [Google Scholar]
- Meyer, B.N.; Ferrigni, N.R.; Putnam, J.E.; Jacobsen, L.B.; Nichols, D.E. Brine shrimp: A convenient general bioassay foractive plant constituents. Planta Med. 1982, 45, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Fei, K.Y.; Ming, L.C.; Li, K.C.; Chen, C.Y. Antioxidant and anticancer constituents from the leaves of Liriodendron tulipifera. Molecules 2014, 19, 4234–4245. [Google Scholar]
- Cheon, S.Y.; Chung, K.S.; Jeon, E.; Nugroho, A.; Park, H.J.; An, H.J. Anti-inflammatory activity of saxifragin via inhibition of NF-κB involves caspase-1 activation. J. Nat. Prod. 2015, 78, 1579–1585. [Google Scholar] [CrossRef]


| Position | 1 | 2 | ||
|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | |
| 2 | 165.8 | 165.7 | ||
| 3 | 91.0 | 5.22, s | 91.1 | 5.22, s |
| 4 | 174.2 | 173.2 | ||
| 5 | 38.7 | 2.49, m | 38.7 | 2.49, m |
| 6 | 67.1 | 4.32, d (3.6) | 67.1 | 4.32, d (3.2) |
| 7 | 56.2 | 3.73, s | 56.2 | 3.70, s |
| 8 | 33.3 | 2.17, m | 33.3 | 2.17, m |
| 9 | 66.4 | 3.96, dd (11.2, 6.4); 3.91, dd (11.2, 6.4) | 67.0 | 4.00, dd (11.2, 6.0); 3.92, dd (11.2, 6.0) |
| 10 | 14.3 | 0.93, d (7.2) | 14.3 | 0.93, d (7.2) |
| 1′ | 170.3 | |||
| 2′ | 20.6 | 2.00, s | 165.9 | |
| 3′ | 91.2 | 5.22, s | ||
| 4′ | 173.4 | |||
| 5′ | 38.9 | 2.86, m | ||
| 6′ | 67.2 | 4.38, m; 4.19, dd (11.2, 3.2) | ||
| 7′ | 56.4 | 3.72, s | ||
| 8′ | 39.2 | 2.83, m | ||
| 9′ | 174.3 | |||
| 10′ | 15.7 | 1.08, d (6.8) | ||
| Position | 3 | 4 | ||
|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | |
| 2 | 166.0 | 165.9 | ||
| 3 | 90.7 | 5.18, s | 90.6 | 5.18, s |
| 4 | 174.9 | 175.0 | ||
| 5 | 39.0 | 2.44, m | 39.0 | 2.44, m |
| 6 | 67.3 | 4.30, d (3.6) | 67.5 | 4.31, d (3.6) |
| 7 | 56.1 | 3.72, s | 56.0 | 3.72, s |
| 8 | 34.2 | 2.05, m | 34.1 | 2.05, m |
| 9 | 69.8 | 3.44, dd (9.6, 6.8); 3.25, dd (9.6, 6.8) | 73.6 | 3.27, m |
| 10 | 14.8 | 0.89, d (7.2) | 14.8 | 0.91, d (6.8) |
| 1′ | ||||
| 2′ | 110.8 | 4.58, d (2.4) | 104.2 | |
| 3′ | 42.0 | 1.80, dt (6.8, 2.4) | 54.8 | 1.57, ddd (10.2, 7.6, 6.0) |
| 4′ | 53.9 | 1.59, m | 35.8 | 2.08, m |
| 5′ | 69.2 | 3.90, t (8.0); 3.66, t (8.8) | 71.9 | 3.91, t (8.0) 3.19, m |
| 6′ | 18.0 | 0.97, d (7.2) | 26.6 | 1.34, s |
| 7′ | 67.4 | 3.54, t (6.8) | 70.4 | 3.52, m; 3.33, d (3.2) |
| 8′ | 22.6 | 1.07, d (6.4) | 17.5 | 0.99, d (6.8) |
| MIC (μg/mL) | ||||
|---|---|---|---|---|
| Compounds | S. aureus | E. faecalis | E. coli | S. maltophilia |
| 1 | >64 | >64 | >64 | >64 |
| 2 | >64 | >64 | >64 | >64 |
| 3 | >64 | >64 | >64 | >64 |
| 4 | >64 | >64 | >64 | >64 |
| 5 | >64 | >64 | >64 | >64 |
| vancomycin | 1.25 | 1.25 | ||
| meropenem | 0.03 | 0.03 | ||
| Inhibition Rate (%) | |||
|---|---|---|---|
| Compounds | MCF-7 | A549 | Hela |
| 1 | 13.21 | 21.04 | 8.13 |
| 2 | 8.19 | 10.23 | 13.11 |
| 3 | 16.28 | 31.22 | 24.56 |
| 4 | 5.36 | 2.79 | 6.35 |
| 5 | 20.13 | 12.65 | 17.03 |
| adriamycin | 93.25 | 95.42 | 92.16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Hui, Y.; Chen, Z.; Chen, G.; Song, X.; Sun, Z.; Han, C.; Chen, W. Four Undescribed Pyranones from the Scutellaria formosana-Derived Endophytic Fungi Ascomycota sp. FAE17. Molecules 2023, 28, 5388. https://doi.org/10.3390/molecules28145388
Yang J, Hui Y, Chen Z, Chen G, Song X, Sun Z, Han C, Chen W. Four Undescribed Pyranones from the Scutellaria formosana-Derived Endophytic Fungi Ascomycota sp. FAE17. Molecules. 2023; 28(14):5388. https://doi.org/10.3390/molecules28145388
Chicago/Turabian StyleYang, Jianni, Yang Hui, Zhaoxia Chen, Guangying Chen, Xiaoping Song, Zhenfan Sun, Changri Han, and Wenhao Chen. 2023. "Four Undescribed Pyranones from the Scutellaria formosana-Derived Endophytic Fungi Ascomycota sp. FAE17" Molecules 28, no. 14: 5388. https://doi.org/10.3390/molecules28145388
APA StyleYang, J., Hui, Y., Chen, Z., Chen, G., Song, X., Sun, Z., Han, C., & Chen, W. (2023). Four Undescribed Pyranones from the Scutellaria formosana-Derived Endophytic Fungi Ascomycota sp. FAE17. Molecules, 28(14), 5388. https://doi.org/10.3390/molecules28145388







