Coptisine Inhibits Influenza Virus Replication by Upregulating p21
Abstract
1. Introduction
2. Results
2.1. Antiviral Activity of Coptisine against H1N1 In Vitro
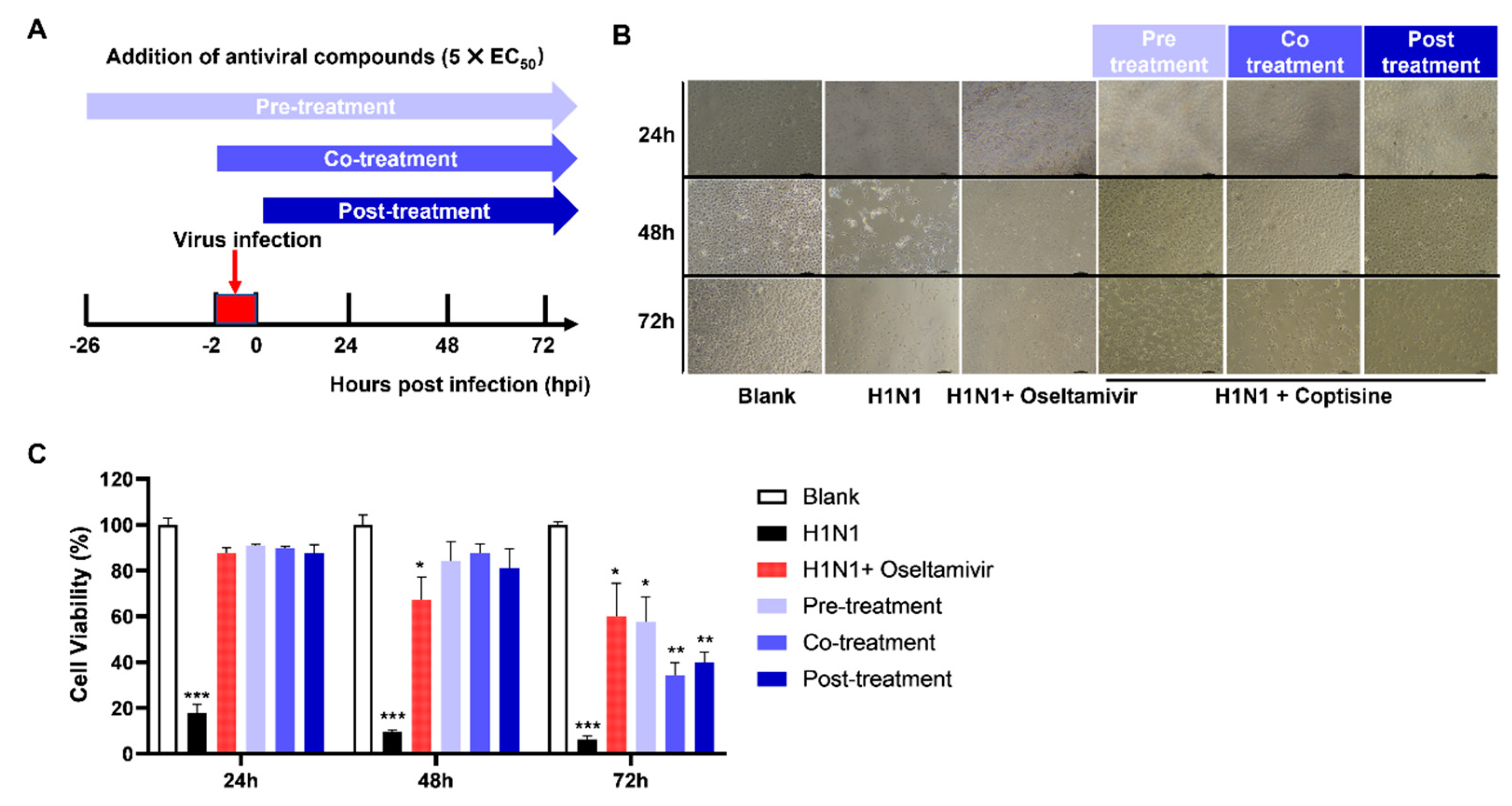
2.2. Administering Coptisine before Infection Is More Effective in Exerting Its Antiviral Effects
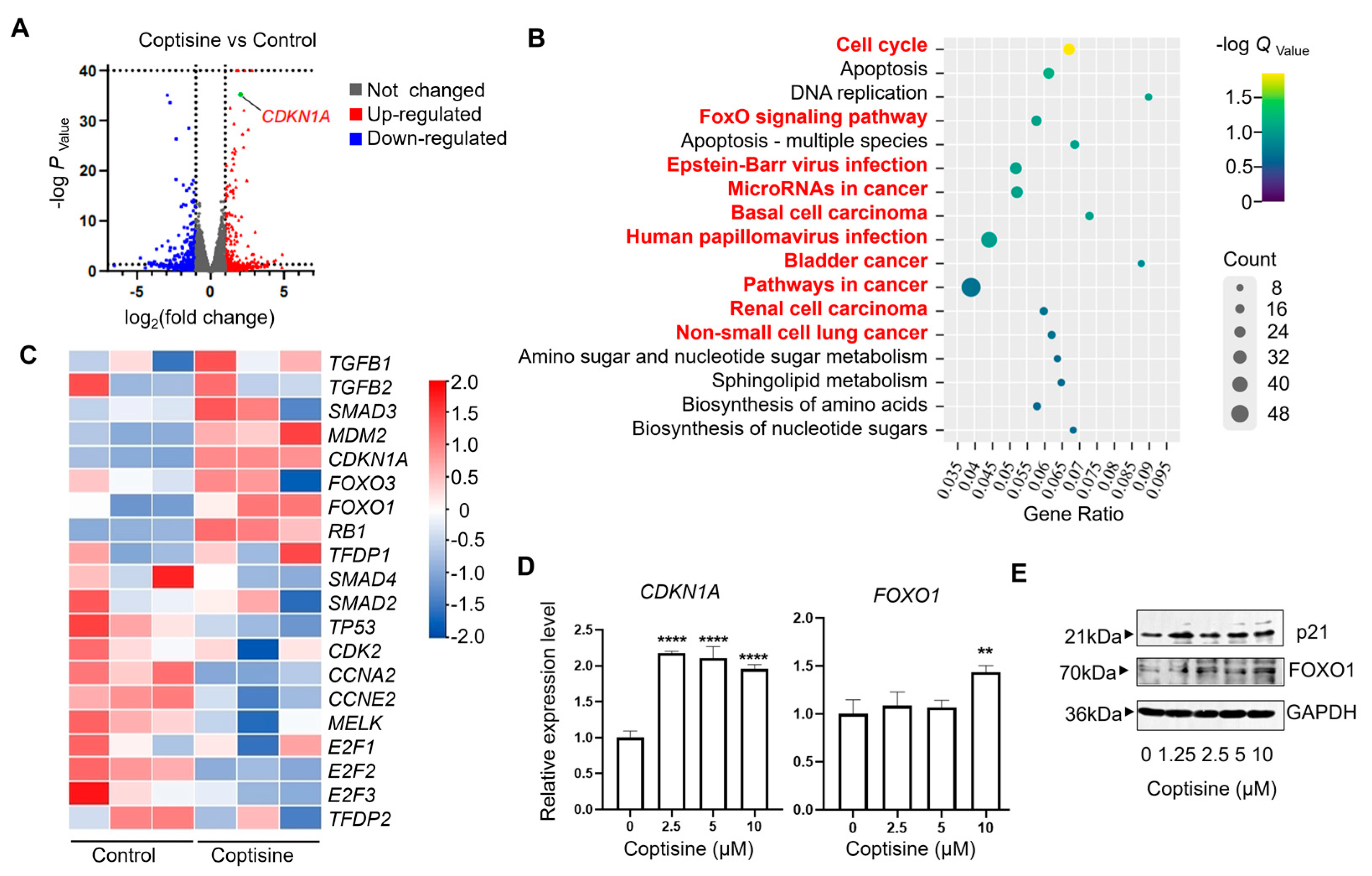
2.3. Coptisine Upregulates the Expression of CDKNI1A and FOXO1
2.4. Coptisine Might Target against MELK to Inhibit Influenza Virus
3. Discussion
4. Materials and Methods
4.1. Cells and Viruses
4.2. Cytotoxicity Assay
4.3. Cytopathic Effect Assay
4.4. Plaque Reduction Assay
4.5. RNA Sequencing
4.6. Western Blot Analysis
4.7. RNA Extraction and RT-qPCR
4.8. Molecular Docking
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Krammer, F.; Smith, G.J.D.; Fouchier, R.A.M.; Peiris, M.; Kedzierska, K.; Doherty, P.C.; Palese, P.; Shaw, M.L.; Treanor, J.; Webster, R.G.; et al. Influenza. Nat. Rev. Dis. Prim. 2018, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Petrova, V.N.; Russell, C.A. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018, 16, 47–60. [Google Scholar] [CrossRef]
- Su, S.; Fu, X.; Li, G.; Kerlin, F.; Veit, M. Novel Influenza D virus: Epidemiology, pathology, evolution and biological characteristics. Virulence 2017, 8, 1580–1591. [Google Scholar] [CrossRef] [PubMed]
- Cheung, T.K.; Poon, L.L. Biology of influenza a virus. Ann. N. Y. Acad. Sci. 2007, 1102, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Shao, W.; Li, X.; Goraya, M.U.; Wang, S.; Chen, J.L. Evolution of Influenza A Virus by Mutation and Re-Assortment. Int. J. Mol. Sci. 2017, 18, 1650. [Google Scholar] [CrossRef] [PubMed]
- Hayden, F.G. Antiviral resistance in influenza viruses—Implications for management and pandemic response. N. Engl. J. Med. 2006, 354, 785–788. [Google Scholar] [CrossRef]
- Ison, M.G. Antivirals and resistance: Influenza virus. Curr. Opin. Virol. 2011, 1, 563–573. [Google Scholar] [CrossRef]
- Izzedine, H.; Launay-Vacher, V.; Deray, G. Antiviral drug-induced nephrotoxicity. Am. J. Kidney Dis. Off. J. Natl. Kidney Found. 2005, 45, 804–817. [Google Scholar] [CrossRef]
- McHutchison, J.G.; Manns, M.P.; Longo, D.L. Definition and management of anemia in patients infected with hepatitis C virus. Liver Int. Off. J. Int. Assoc. Study Liver 2006, 26, 389–398. [Google Scholar] [CrossRef]
- Tao, J.; Wang, H.; Wang, W.; Mi, N.; Zhang, W.; Wen, Q.; Ouyang, J.; Liang, X.; Chen, M.; Guo, W.; et al. Binding mechanism of oseltamivir and influenza neuraminidase suggests perspectives for the design of new anti-influenza drugs. PLoS Comput. Biol. 2022, 18, e1010343. [Google Scholar] [CrossRef] [PubMed]
- Kausar, S.; Said Khan, F.; Ishaq Mujeeb Ur Rehman, M.; Akram, M.; Riaz, M.; Rasool, G.; Hamid Khan, A.; Saleem, I.; Shamim, S.; Malik, A. A review: Mechanism of action of antiviral drugs. Int. J. Immunopathol. Pharmacol. 2021, 35, 20587384211002621. [Google Scholar] [CrossRef] [PubMed]
- Świerczyńska, M.; Mirowska-Guzel, D.M.; Pindelska, E. Antiviral Drugs in Influenza. Int. J. Environ. Res. Public Health 2022, 19, 3018. [Google Scholar] [CrossRef] [PubMed]
- Runfeng, L.; Yunlong, H.; Jicheng, H.; Weiqi, P.; Qinhai, M.; Yongxia, S.; Chufang, L.; Jin, Z.; Zhenhua, J.; Haiming, J.; et al. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2). Pharmacol. Res. 2020, 156, 104761. [Google Scholar] [CrossRef]
- Lee, D.Y.W.; Li, Q.Y.; Liu, J.; Efferth, T. Traditional Chinese herbal medicine at the forefront battle against COVID-19: Clinical experience and scientific basis. Phytomedicine 2021, 80, 153337. [Google Scholar] [CrossRef]
- Mohammed, F.S.; Uysal, İ.; Sevindik, M. A Review on Antiviral Plants Effective against Different Virus Types. Prospect. Pharm. Sci. 2023, 21, 1–21. [Google Scholar] [CrossRef]
- Liu, Y.; Song, X.; Li, C.; Hu, H.; Li, W.; Wang, L.; Hu, J.; Liao, C.; Liang, H.; He, Z.; et al. Chrysin Ameliorates Influenza Virus Infection in the Upper Airways by Repressing Virus-Induced Cell Cycle Arrest and Mitochondria-Dependent Apoptosis. Front. Immunol. 2022, 13, 872958. [Google Scholar] [CrossRef]
- Zhu, X.; Hu, Z.; Yu, T.; Hu, H.; Zhao, Y.; Li, C.; Zhu, Q.; Wang, M.; Zhai, P.; He, L.; et al. The Antiviral Effects of Jasminin via Endogenous TNF-alpha and the Underlying TNF-alpha-Inducing Action. Molecules 2022, 27, 1598. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.H.; Chathuranga, K.; Uddin, M.B.; Weeratunga, P.; Kim, M.S.; Cho, W.K.; Kim, H.I.; Ma, J.Y.; Lee, J.S. Coptidis Rhizoma extract inhibits replication of respiratory syncytial virus in vitro and in vivo by inducing antiviral state. J. Microbiol. 2017, 55, 488–498. [Google Scholar] [CrossRef]
- Zhao, J.; Tian, S.; Lu, D.; Yang, J.; Zeng, H.; Zhang, F.; Tu, D.; Ge, G.; Zheng, Y.; Shi, T.; et al. Systems pharmacological study illustrates the immune regulation, anti-infection, anti-inflammation, and multi-organ protection mechanism of Qing-Fei-Pai-Du decoction in the treatment of COVID-19. Phytomedicine 2021, 85, 153315. [Google Scholar] [CrossRef]
- Balomenos, D.; Martín-Caballero, J.; García, M.I.; Prieto, I.; Flores, J.M.; Serrano, M.; Martínez, A.C. The cell cycle inhibitor p21 controls T-cell proliferation and sex-linked lupus development. Nat. Med. 2000, 6, 171–176. [Google Scholar] [CrossRef]
- Chen, H.; Li, C.; Huang, J.; Cung, T.; Seiss, K.; Beamon, J.; Carrington, M.F.; Porter, L.C.; Burke, P.S.; Yang, Y.; et al. CD4+ T cells from elite controllers resist HIV-1 infection by selective upregulation of p21. J. Clin. Investig. 2011, 121, 1549–1560. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Li, Y.; Zong, Y.; Velkov, T.; Wang, C.; Yang, X.; Zhang, M.; Jiang, Z.; Sun, H.; Tong, Q.; et al. p21 restricts influenza A virus by perturbing the viral polymerase complex and upregulating type I interferon signaling. PLoS Pathog. 2022, 18, e1010295. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.Q.; Zhang, Y.; Xia, T.; Jiang, L.Q.; Zhong, B.; Shu, H.B. FoxO1 negatively regulates cellular antiviral response by promoting degradation of IRF3. J. Biol. Chem. 2013, 288, 12596–12604. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Zhang, D. Maternal embryonic leucine zipper kinase (MELK): A novel regulator in cell cycle control, embryonic development, and cancer. Int. J. Mol. Sci. 2013, 14, 21551–21560. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, T.; Kato, T.; Kiyotani, K.; Tarhan, Y.E.; Saloura, V.; Chung, S.; Ueda, K.; Nakamura, Y.; Park, J.H. p53-independent p21 induction by MELK inhibition. Oncotarget 2017, 8, 57938–57947. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, M.-F.; Liang, J.-H.; Shen, Y.-N.; Zhang, C.-W.; Yang, K.-Y.; Liu, L.-C.; Xie, Q.; Hu, C.; Song, X.; Wang, Y. Coptisine Inhibits Influenza Virus Replication by Upregulating p21. Molecules 2023, 28, 5398. https://doi.org/10.3390/molecules28145398
He M-F, Liang J-H, Shen Y-N, Zhang C-W, Yang K-Y, Liu L-C, Xie Q, Hu C, Song X, Wang Y. Coptisine Inhibits Influenza Virus Replication by Upregulating p21. Molecules. 2023; 28(14):5398. https://doi.org/10.3390/molecules28145398
Chicago/Turabian StyleHe, Ming-Feng, Jian-Hui Liang, Yan-Ni Shen, Chao-Wei Zhang, Kuang-Yang Yang, Li-Chu Liu, Qian Xie, Chun Hu, Xun Song, and Yan Wang. 2023. "Coptisine Inhibits Influenza Virus Replication by Upregulating p21" Molecules 28, no. 14: 5398. https://doi.org/10.3390/molecules28145398
APA StyleHe, M.-F., Liang, J.-H., Shen, Y.-N., Zhang, C.-W., Yang, K.-Y., Liu, L.-C., Xie, Q., Hu, C., Song, X., & Wang, Y. (2023). Coptisine Inhibits Influenza Virus Replication by Upregulating p21. Molecules, 28(14), 5398. https://doi.org/10.3390/molecules28145398







