Preliminary Study on Rapid and Simultaneous Detection of Viable Escherichia coli O157:H7, Staphylococcus aureus, and Salmonella by PMA-mPCR in Food
Abstract
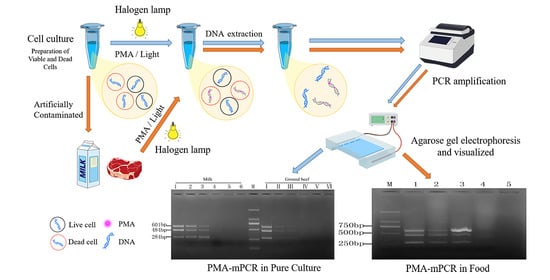
:1. Introduction
2. Results
2.1. Optimization of Concentration of PMA Treatment
2.2. Specificity and Sensitivity of PMA-mPCR in Pure Culture
2.3. Sensitivity of the PMA-mPCR Assay in Artificial Contaminated Food Products
2.4. Detection of Mixed Viable and Dead Cells
3. Materials and Methods
3.1. Bacterial Strains and Cultivation
3.2. Preparation of Viable and Dead Cells
3.3. PMA Treatment and DNA Extraction
3.4. Multiplex PCR Conditions
3.5. Specificity and Sensitivity of the PMA-mPCR
3.6. Detection of Viable Target Pathogens in Artificially Contaminated Food Products
3.7. Detection of Mixed Viable and Dead Cells
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Zhao, X.; Lin, C.W.; Wang, J.; Oh, D.H. Advances in rapid detection methods for foodborne pathogens. J. Microbiol. Biotechnol. 2014, 24, 297–312. [Google Scholar] [CrossRef] [Green Version]
- Shekar, A.; Babu, L.; Ramlal, S.; Sripathy, M.H.; Batra, H. Selective and concurrent detection of viable Salmonella spp., E. coli, Staphylococcus aureus, E. coli O157:H7, and Shigella spp., in low moisture food products by PMA-mPCR assay with internal amplification control. LWT—Food Sci. Technol. 2017, 86, 586–593. [Google Scholar] [CrossRef]
- Kim, J.H.; Rhim, S.R.; Kim, K.T.; Paik, H.D.; Lee, J.Y. Simultaneous Detection of Listeria monocytogenes, Escherichia coli O157:H7, Bacillus cereus, Salmonella spp., and Staphylococcus aureus in Low-fatted Milk by Multiplex PCR. Korean J. Food Sci. Anim. Resour. 2014, 34, 717–723. [Google Scholar] [CrossRef] [Green Version]
- Kourtis, A.P.; Hatfield, K.; Baggs, J.; Mu, Y.; See, I.; Epson, E.; Nadle, J.; Kainer, M.A.; Dumyati, G.; Petit, S.; et al. Vital Signs: Epidemiology and Recent Trends in Methicillin-Resistant and in Methicillin-Susceptible Staphylococcus aureus Bloodstream Infections—United States. Morb. Mortal. Wkly. Rep. 2019, 68, 214–219. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Ye, C.; Xu, H.; Aguilar, Z.P.; Xiong, Y.; Lai, W.; Wei, H. Development of an SD-PMA-mPCR assay with internal amplification control for rapid and sensitive detection of viable Salmonella spp., Shigella spp. and Staphylococcus aureus in food products. Food Control 2015, 57, 314–320. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, F.; Xu, H.; Aguilar, Z.P.; Niu, R.; Yuan, Y.; Sun, J.; You, X.; Lai, W.; Xiong, Y. Magnetic nano-beads based separation combined with propidium monoazide treatment and multiplex PCR assay for simultaneous detection of viable Salmonella Typhimurium, Escherichia coli O157:H7 and Listeria monocytogenes in food products. Food Microbiol. 2013, 34, 418–424. [Google Scholar] [CrossRef]
- Zhao, X.H.; Zhao, F.H.; Wang, J.; Zhong, N.J. Biofilm formation and control strategies of foodborne pathogens: Food safety perspectives. RSC Adv. 2017, 7, 36670–36683. [Google Scholar] [CrossRef] [Green Version]
- Dewey-Mattia, D.; Manikonda, K.; Hall, A.J.; Wise, M.E.; Crowe, S.J. Surveillance for Foodborne Disease Outbreaks—United States, 2009–2015. Mmwr Surveill. Summ. 2018, 67, 1–11. [Google Scholar] [CrossRef]
- Zhong, J.L.; Zhao, X.H. Isothermal Amplification Technologies for the Detection of Foodborne Pathogens. Food Anal. Methods 2018, 11, 1543–1560. [Google Scholar] [CrossRef]
- Cantekin, Z.; Ergun, Y.; Solmaz, H.; Özmen, G.Ö.; Demir, M.; Saidi, R. PCR assay with host specific internal control for Staphylococcus aureus from bovine milk samples. Maced. Vet. Rev. 2015, 38, 97–100. [Google Scholar] [CrossRef] [Green Version]
- Yu, S.; Yan, L.; Wu, X.; Li, F.; Wang, D.; Xu, H. Multiplex PCR coupled with propidium monoazide for the detection of viable Cronobacter sakazakii, Bacillus cereus, and Salmonella spp. in milk and milk products. J. Dairy Sci. 2017, 100, 7874–7882. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Wang, L.; Xu, H.; Aguilar, Z.P.; Liu, C.; Gan, B.; Xiong, Y.; Lai, W.; Xu, F.; Wei, H. Detection of non-emetic and emetic Bacillus cereus by propidium monoazide multiplex PCR (PMA-mPCR) with internal amplification control. Food Control 2014, 35, 401–406. [Google Scholar] [CrossRef]
- Zhao, X.H.; Li, M.; Xu, Z.B. Detection of Foodborne Pathogens by Surface Enhanced Raman Spectroscopy. Front. Microbiol. 2018, 9, 1236. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Mustapha, A. Detection of viable Escherichia coli O157:H7 in ground beef by propidium monoazide real-time PCR. Int. J. Food Microbiol. 2014, 170, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Y.; Zhou, R.; Li, L.; Peters, B.M.; Li, B.; Lin, C.W.; Chuang, T.L.; Chen, D.Q.; Zhao, X.H.; Xiong, Z.Y.; et al. Viable but non-culturable state and toxin gene expression of enterohemorrhagic Escherichia coli 0157 under cryopreservation. Res. Microbiol. 2017, 168, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Nocker, A.; Cheung, C.-Y.; Camper, A.K. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 2006, 67, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Rudi, K.; Moen, B.; Drømtorp, S.M.; Holck, A.L. Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl. Environ. Microbiol. 2005, 71, 1018–1024. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Breidt, F., Jr. Enumeration of viable Listeria monocytogenes cells by real-time PCR with propidium monoazide and ethidium monoazide in the presence of dead cells. Appl. Environ. Microbiol. 2008, 73, 8028–8031. [Google Scholar] [CrossRef] [Green Version]
- Zhu, R.G.; Li, T.P.; Jia, Y.F.; Song, L.F. Quantitative study of viable Vibrio parahaemolyticus cells in raw seafood using propidium monoazide in combination with quantitative PCR. J. Microbiol. Methods 2012, 90, 262–266. [Google Scholar] [CrossRef]
- Gerard, À.; Marta, G.; Sergio, I.; Vanessa, B.; Rubén, L. Method to quantify live and dead cells in multi-species oral biofilm by real-time PCR with propidium monoazide. Amb Express 2013, 3, 1. [Google Scholar]
- Zhao, X.; Zhong, J.; Wei, C.; Lin, C.-W.; Ding, T. Current Perspectives on Viable but Non-culturable State in Foodborne Pathogens. Front. Microbiol. 2017, 8, 580. [Google Scholar] [CrossRef] [Green Version]
- Zhao, F.; Bi, X.; Hao, Y.; Liao, X. Induction of viable but nonculturable Escherichia coli O157: H7 by high pressure CO2 and its characteristics. PLoS ONE 2013, 8, e62388. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Gilchrist, A.; Zhang, J.; Li, X.F. Detection of Viable but Nonculturable Escherichia coli O157:H7 Bacteria in Drinking Water and River Water. Appl. Environ. Microbiol. 2008, 74, 1502–1507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, J.L.; Zhao, X.H. Detection of viable but non-culturable Escherichia coli O157:H7 by PCR in combination with propidium monoazide. 3 Biotech 2017, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, J.; Forghani, F.; Park, J.H.; Park, M.S.; Seo, K.H.; Oh, D.H. Rapid detection of viable Escherichia coli O157 by coupling propidium monoazide with loop-mediated isothermal amplification. J. Microbiol. Biotechnol. 2013, 23, 1708–1716. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Zhong, J.; Hu, T.; Zhao, X. Simultaneous detection of Escherichia coli O157: H7, Staphylococcus aureus and Salmonella by multiplex PCR in milk. 3 Biotech 2018, 8, 76. [Google Scholar] [CrossRef]
- Xu, Y.; Cheng, W.; Chen, F. Detection of Salmonella spp., Escherichia coli and Staphylococcus aureus by Multiplex PCR. J. Microbiol. 2006, 33, 89–94. [Google Scholar]
- Li, F.; Xie, G.; Zhou, B.; Yu, P.; Yu, S.; Aguilar, Z.P.; Wei, H.; Xu, H. Rapid and simultaneous detection of viable Cronobacter sakazakii, Staphylococcus aureus, and Bacillus cereus in infant food products by PMA-mPCR assay with internal amplification control. LWT—Food Sci. Technol. 2016, 74, 176–182. [Google Scholar] [CrossRef]
- Vugia, D.; Cronquist, A.; Cartter, M.; Tobin-D’Angelo, M.; Blythe, D.; Smith, K.; Lathrop, S.; Morse, D.; Cieslak, P.; Dunn, J. Preliminary FoodNet Data on the incidence of infection with pathogens transmitted commonly through food—10 States, 2008. Mmwr Morb. Mortal. Wkly. Rep. 2010, 59, 418–422. [Google Scholar]
- Saeki, E.K.; Alves, J.; Bonfante, R.C.; Hirooka, E.Y. Multiplex PCR (mPCR) for the Detection of Salmonella spp. and the Differentiation of the Typhimurium and Enteritidis Serovars in Chicken Meat. J. Food Saf. 2013, 33, 25–29. [Google Scholar] [CrossRef]
- Park, Y.S.; Lee, S.R.; Kim, Y.G. Detection of Escherichia coli O157:H7, Salmonella spp., Staphylococcus aureus and Listeria monocytogenes in kimchi by multiplex polymerase chain reaction (mPCR). J. Microbiol. 2006, 44, 92–97. [Google Scholar] [PubMed]
- Forghani, F.; Langaee, T.; Eskandari, M.; Seo, K.H.; Chung, M.J.; Oh, D.H. Rapid detection of viable Bacillus cereus emetic and enterotoxic strains in food by coupling propidium monoazide and multiplex PCR (PMA-mPCR). Food Control 2015, 55, 151–157. [Google Scholar] [CrossRef]
- Chen, J.; Tang, J.; Liu, J.; Cai, Z.; Bai, X. Development and evaluation of a multiplex PCR for simultaneous detection of five foodborne pathogens. J. Appl. Microbiol. 2012, 112, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.U.; Batule, B.S.; Mun, H.; Shim, W.B.; Kim, M.G. Ultrasensitive colorimetric detection of Salmonella enterica Typhimurium on lettuce leaves by HRPzyme-Integrated polymerase chain reaction. Food Control 2018, 84, 522–528. [Google Scholar] [CrossRef]
- Ahn, H.; Batule, B.S.; Seok, Y.; Kim, M.G. Single-Step Recombinase Polymerase Amplification Assay Based on a Paper Chip for Simultaneous Detection of Multiple Foodborne Pathogens. Anal. Chem. 2018, 90, 10211–10216. [Google Scholar] [CrossRef]
- Batule, B.S.; Kim, S.U.; Mun, H.; Shim, W.-B.; Kim, M.-G. Development of HRPzyme-Integrated PCR Platform for Colorimetric Detection of Foodborne Pathogens. In Biosensing Technologies for the Detection of Pathogens; Rinken, T., Ed.; IntechOpen Limited: London, UK, 2018. [Google Scholar]
- Li, L.; Mendis, N.; Trigui, H.; Oliver, J.D.; Faucher, S.P. The importance of the viable but non-culturable state in human bacterial pathogens. Front. Microbiol. 2014, 5, 258. [Google Scholar] [CrossRef] [Green Version]









| Microorganism | Target Gene | Primer Sequence (5′-3′) | G + C (%) | Tm (°C) | Amplicon Length (bp) | Reference |
|---|---|---|---|---|---|---|
| E. coli O157:H7 | rfbE (S83460) | F:GCCACCCCCATTTTCGTTG R:TCCTCTCTTTCCTCTGCGGT | 57.9 47.4 | 63.2 51.7 | 601 | [26] |
| S. aureus | nuc (AP017922) | F:TACAGGTGACTGCGGGCTTATC R:CTTACCGGGCAATACACTCACTA | 50 45.4 | 60.2 58.3 | 484 | [27] |
| Salmonella | invA (M90846) | F:CTTTAGCCAAGCCTTGACGAAC R:AAAGGCAATACGCAAAGAGGT | 54.5 47.8 | 62.1 60.6 | 284 | [27] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Wei, C.; Wan, H.; Sarengaowa; Liang, X.; Jiang, T.; Dong, Y.; Zhao, X.; Zhong, T. Preliminary Study on Rapid and Simultaneous Detection of Viable Escherichia coli O157:H7, Staphylococcus aureus, and Salmonella by PMA-mPCR in Food. Molecules 2023, 28, 5835. https://doi.org/10.3390/molecules28155835
Liu Y, Wei C, Wan H, Sarengaowa, Liang X, Jiang T, Dong Y, Zhao X, Zhong T. Preliminary Study on Rapid and Simultaneous Detection of Viable Escherichia coli O157:H7, Staphylococcus aureus, and Salmonella by PMA-mPCR in Food. Molecules. 2023; 28(15):5835. https://doi.org/10.3390/molecules28155835
Chicago/Turabian StyleLiu, Yao, Caijiao Wei, Hui Wan, Sarengaowa, Xiaoping Liang, Tao Jiang, Yuhe Dong, Xihong Zhao, and Tian Zhong. 2023. "Preliminary Study on Rapid and Simultaneous Detection of Viable Escherichia coli O157:H7, Staphylococcus aureus, and Salmonella by PMA-mPCR in Food" Molecules 28, no. 15: 5835. https://doi.org/10.3390/molecules28155835