The Effect of Plastic-Related Compounds on Transcriptome-Wide Gene Expression on CYP2C19-Overexpressing HepG2 Cells
Abstract
:1. Introduction
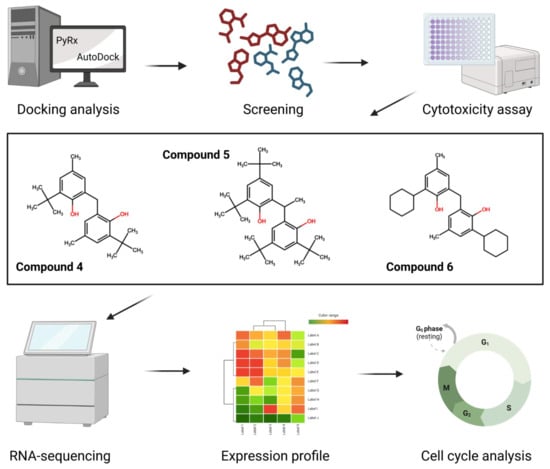
2. Results
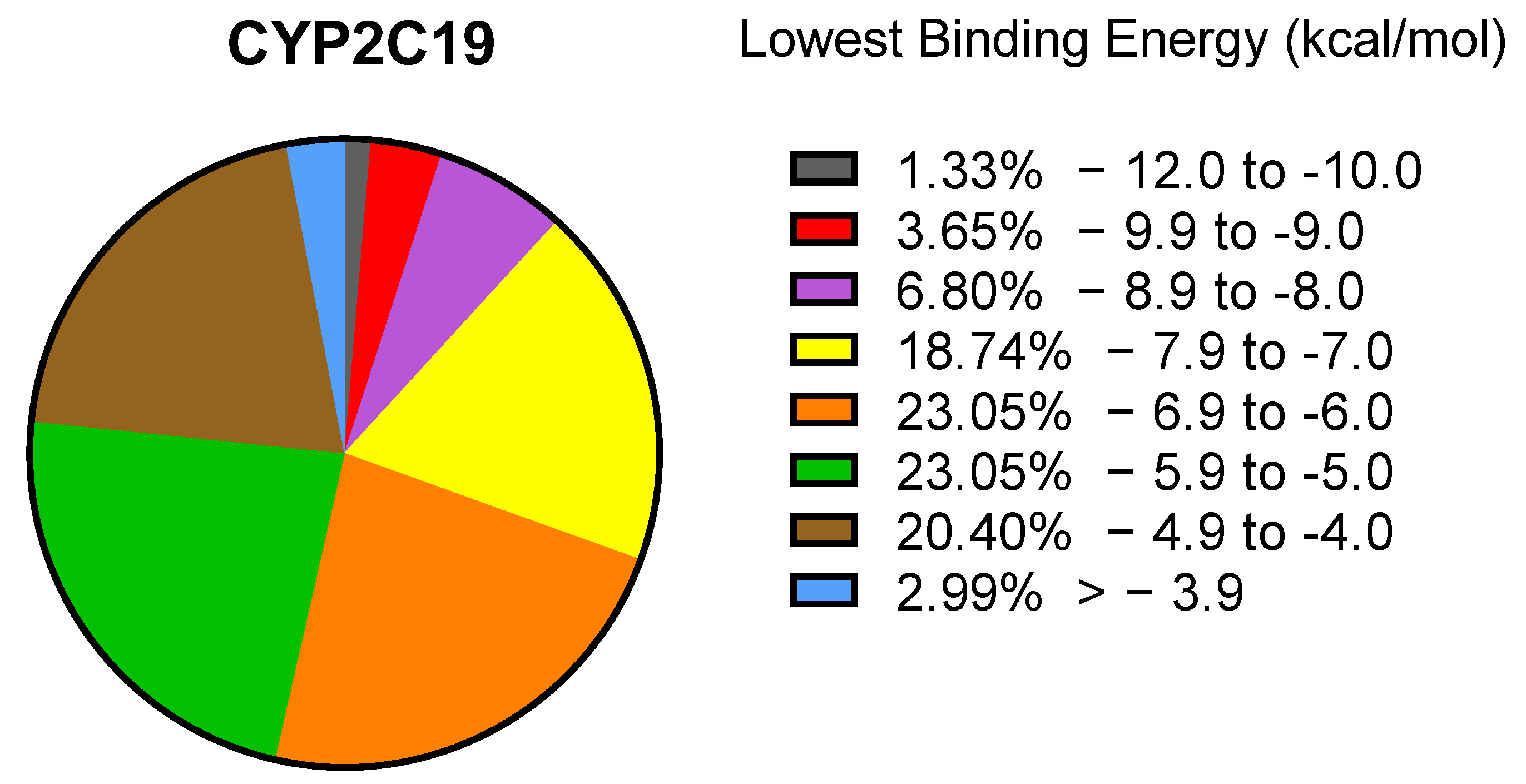
2.1. PyRx Screening Analyses
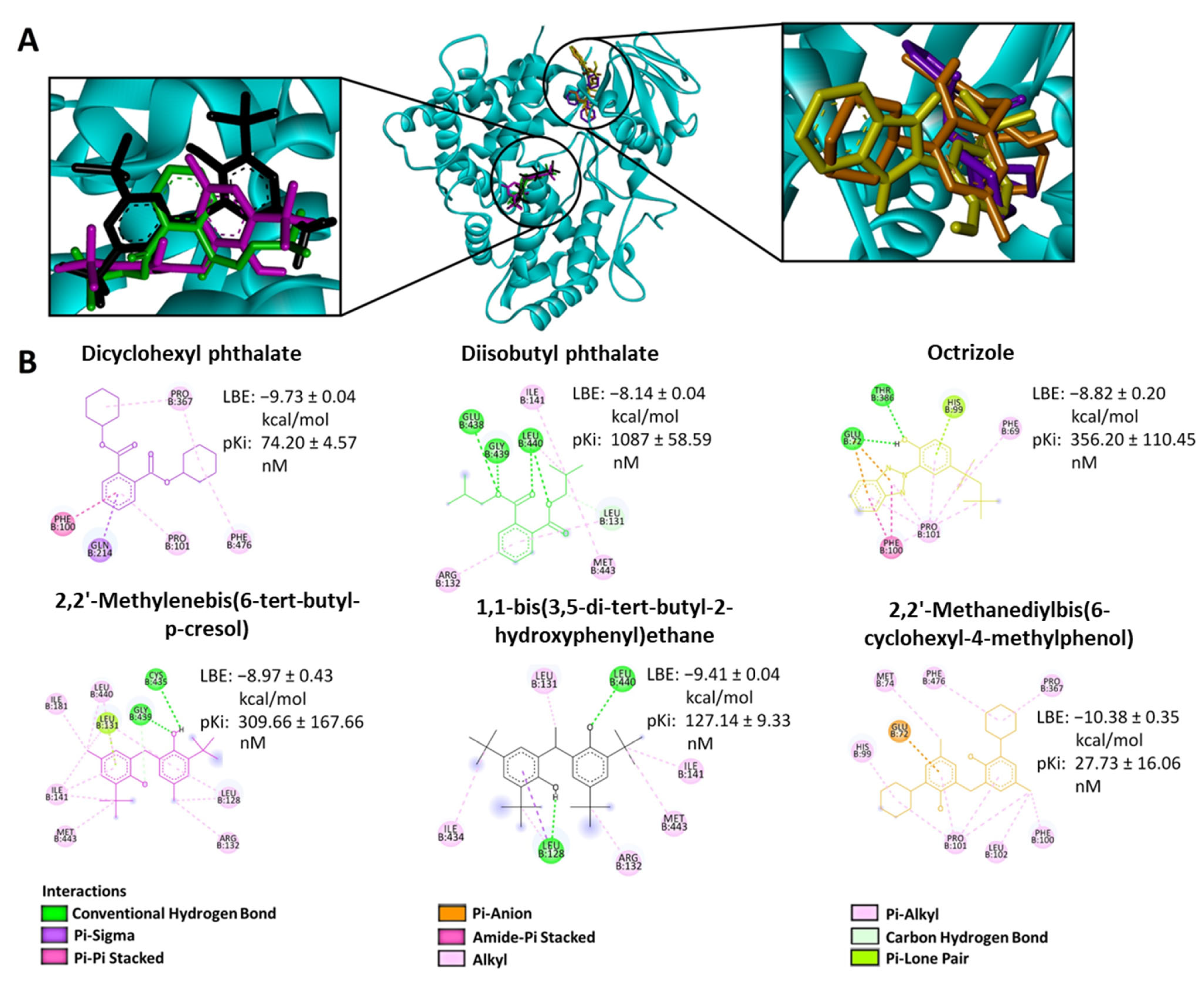
2.2. In Silico Binding of Plastic-Related Compounds to CYP2C19
2.3. Cytotoxicity
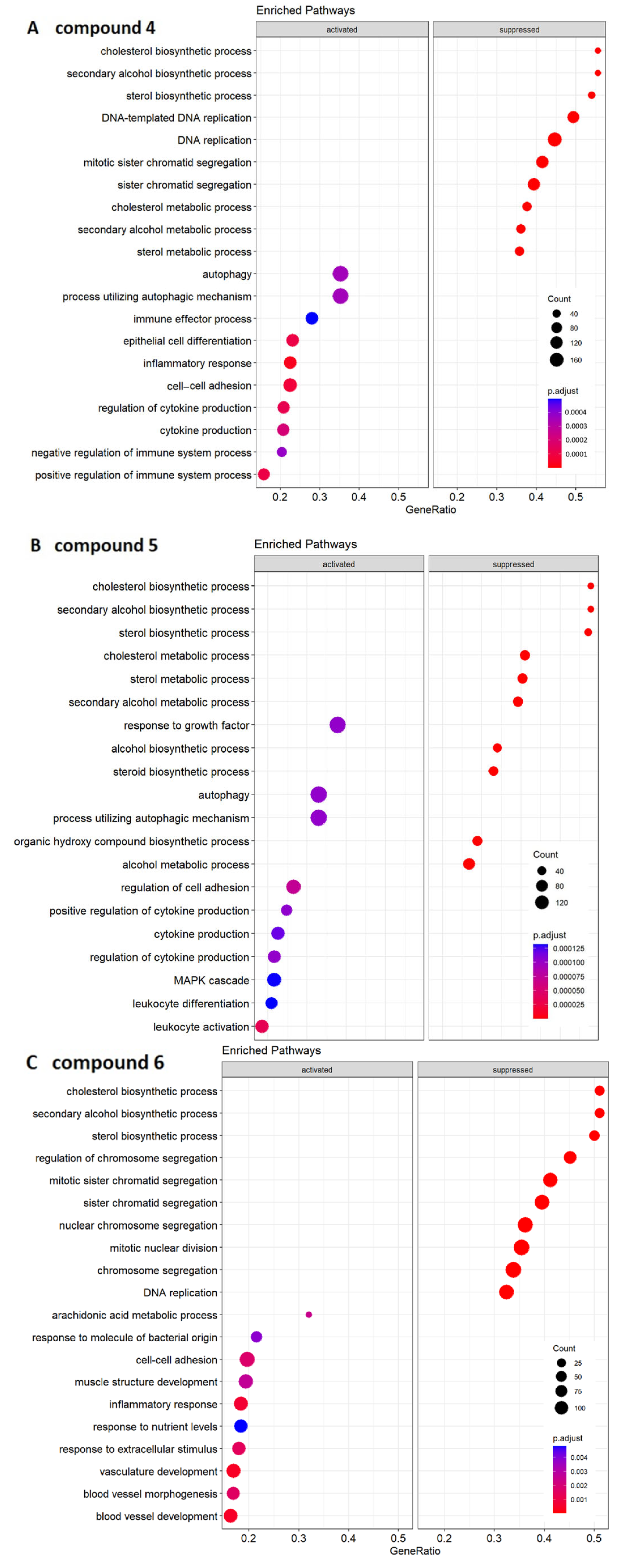
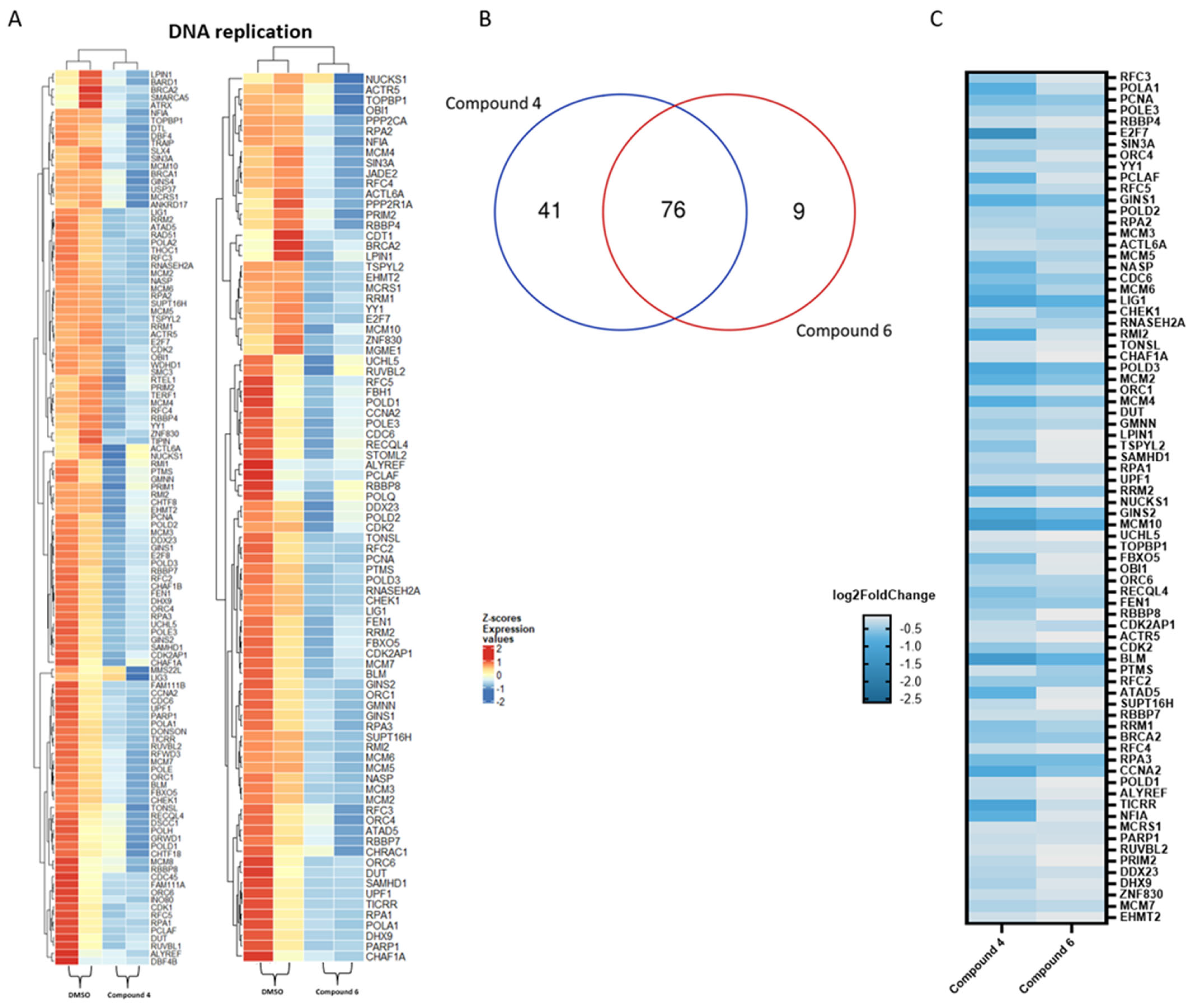
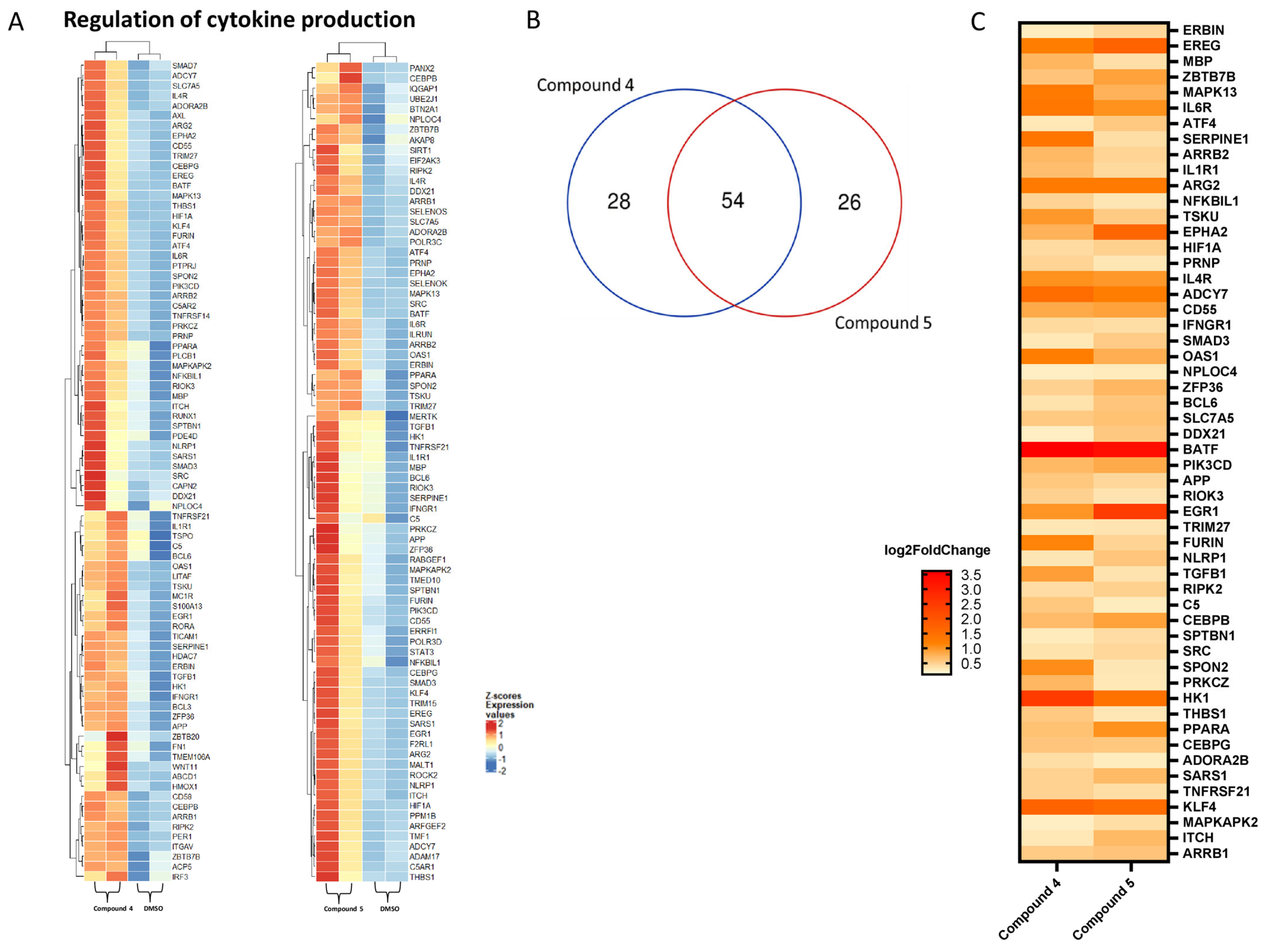
2.4. Transcriptomic Analysis and Deregulated Pathways upon Treatment with Compounds 4–6
2.5. Predicted Metabolism of Compounds 4–6
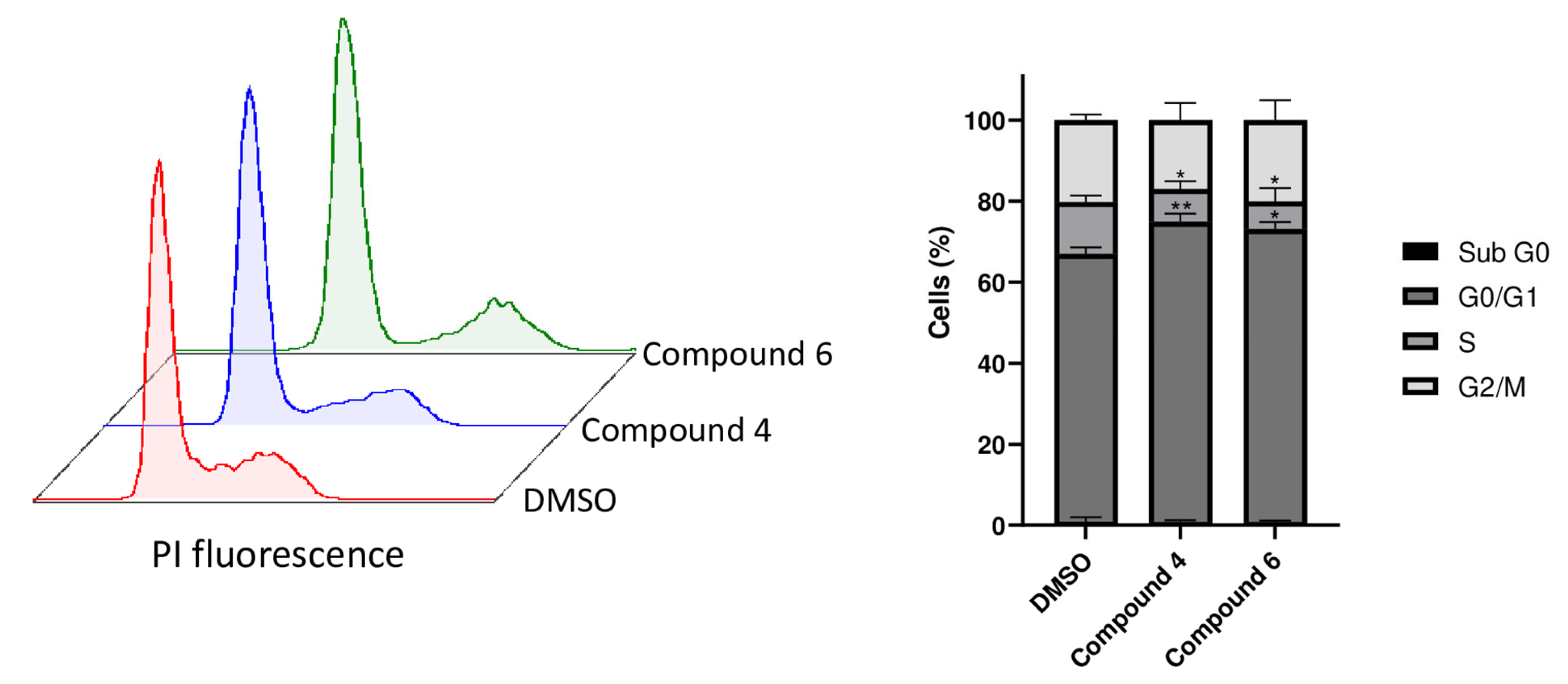
2.6. Compounds 4 and 6 Induce Cell Cycle Arrest in G0/G1 Phase
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Lines
4.3. PyRx Screening
4.4. Molecular Docking
4.5. Resazurin Reduction Assay
4.6. RNA Extraction
4.7. RNA Sequencing
4.8. Bioinformatics Analysis
4.9. Predicted Metabolites
4.10. Cell Cycle Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Cózar, A.; Echevarría, F.; González-Gordillo, J.I.; Irigoien, X.; Úbeda, B.; Hernández-León, S.; Palma, Á.T.; Navarro, S.; García-de-Lomas, J.; Ruiz, A.; et al. Plastic Debris in the Open Ocean. Proc. Natl. Acad. Sci. USA 2014, 111, 10239–10244. [Google Scholar] [CrossRef] [PubMed]
- Eriksen, M.; Lebreton, L.C.M.; Carson, H.S.; Thiel, M.; Moore, C.J.; Borerro, J.C.; Galgani, F.; Ryan, P.G.; Reisser, J. Plastic Pollution in the World’s Oceans: More than 5 Trillion Plastic Pieces Weighing over 250,000 Tons Afloat at Sea. PLoS ONE 2014, 9, e111913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Efferth, T.; Paul, N.W. Threats to Human Health by Great Ocean Garbage Patches. Lancet Planet. Health 2017, 1, e301–e303. [Google Scholar] [CrossRef] [PubMed]
- Vince, J.; Hardesty, B.D. Plastic Pollution Challenges in Marine and Coastal Environments: From Local to Global Governance. Restor. Ecol. 2017, 25, 123–128. [Google Scholar] [CrossRef]
- Alomar, C.; Sureda, A.; Capó, X.; Guijarro, B.; Tejada, S.; Deudero, S. Microplastic Ingestion by Mullus Surmuletus Linnaeus, 1758 Fish and Its Potential for Causing Oxidative Stress. Environ. Res. 2017, 159, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Anbumani, S.; Kakkar, P. Ecotoxicological Effects of Microplastics on Biota: A Review. Environ. Sci. Pollut. Res. 2018, 25, 14373–14396. [Google Scholar] [CrossRef] [PubMed]
- De Sá, L.C.; Oliveira, M.; Ribeiro, F.; Rocha, T.L.; Futter, M.N. Studies of the Effects of Microplastics on Aquatic Organisms: What Do We Know and Where Should We Focus Our Efforts in the Future? Sci. Total Environ. 2018, 645, 1029–1039. [Google Scholar] [CrossRef]
- da Costa, J.P.; Santos, P.S.M.; Duarte, A.C.; Rocha-Santos, T. (Nano)Plastics in the Environment—Sources, Fates and Effects. Sci. Total Environ. 2016, 566–567, 15–26. [Google Scholar] [CrossRef]
- Prata, J.C. Airborne Microplastics: Consequences to Human Health? Environ. Pollut. 2018, 234, 115–126. [Google Scholar] [CrossRef]
- Jambeck, J.; Hardesty, B.D.; Brooks, A.L.; Friend, T.; Teleki, K.; Fabres, J.; Beaudoin, Y.; Bamba, A.; Francis, J.; Ribbink, A.J.; et al. Challenges and Emerging Solutions to the Land-Based Plastic Waste Issue in Africa. Mar. Policy 2018, 96, 256–263. [Google Scholar] [CrossRef]
- Koelmans, A.A.; Redondo-Hasselerharm, P.E.; Nor, N.H.M.; de Ruijter, V.N.; Mintenig, S.M.; Kooi, M. Risk Assessment of Microplastic Particles. Nat. Rev. Mater. 2022, 7, 138–152. [Google Scholar] [CrossRef]
- Leslie, H.A.; van Velzen, M.J.M.; Brandsma, S.H.; Vethaak, A.D.; Garcia-Vallejo, J.J.; Lamoree, M.H. Discovery and Quantification of Plastic Particle Pollution in Human Blood. Environ. Int. 2022, 163, 107199. [Google Scholar] [CrossRef] [PubMed]
- Ragusa, A.; Svelato, A.; Santacroce, C.; Catalano, P.; Notarstefano, V.; Carnevali, O.; Papa, F.; Rongioletti, M.C.A.; Baiocco, F.; Draghi, S.; et al. Plasticenta: First Evidence of Microplastics in Human Placenta. Environ. Int. 2021, 146, 106274. [Google Scholar] [CrossRef]
- Gregory, M.R. Environmental Implications of Plastic Debris in Marine Settings- Entanglement, Ingestion, Smothering, Hangers-on, Hitch-Hiking and Alien Invasions. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 2013–2025. [Google Scholar] [CrossRef] [Green Version]
- Waring, R.H.; Harris, R.M.; Mitchell, S.C. Plastic Contamination of the Food Chain: A Threat to Human Health? Maturitas 2018, 115, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Wesch, C.; Bredimus, K.; Paulus, M.; Klein, R. Towards the Suitable Monitoring of Ingestion of Microplastics by Marine Biota: A Review. Environ. Pollut. 2016, 218, 1200–1208. [Google Scholar] [CrossRef]
- Danopoulos, E.; Twiddy, M.; Rotchell, J.M. Microplastic Contamination of Drinking Water: A Systematic Review. PLoS ONE 2020, 15, e0236838. [Google Scholar] [CrossRef]
- Oliveri Conti, G.; Ferrante, M.; Banni, M.; Favara, C.; Nicolosi, I.; Cristaldi, A.; Fiore, M.; Zuccarello, P. Micro- and Nano-Plastics in Edible Fruit and Vegetables. The First Diet Risks Assessment for the General Population. Environ. Res. 2020, 187, 109677. [Google Scholar] [CrossRef]
- Wright, S.L.; Kelly, F.J. Plastic and Human Health: A Micro Issue? Environ. Sci. Technol. 2017, 51, 6634–6647. [Google Scholar] [CrossRef]
- Amato-Lourenço, L.F.; Carvalho-Oliveira, R.; Júnior, G.R.; dos Santos Galvão, L.; Ando, R.A.; Mauad, T. Presence of Airborne Microplastics in Human Lung Tissue. J. Hazard. Mater. 2021, 416, 126124. [Google Scholar] [CrossRef]
- Meeker, J.D.; Sathyanarayana, S.; Swan, S.H. Phthalates and Other Additives in Plastics: Human Exposure and Associated Health Outcomes. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 2097–2113. [Google Scholar] [CrossRef] [Green Version]
- Koch, H.M.; Calafat, A.M. Human Body Burdens of Chemicals Used in Plastic Manufacture. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 2063–2078. [Google Scholar] [CrossRef] [PubMed]
- Organisation for Economic Co-operation and Development. Global Plastics Outlook; OECD: Paris, France, 2022. [Google Scholar]
- Böckers, M.; Paul, N.W.; Efferth, T. Bisphenolic Compounds Alter Gene Expression in MCF-7 Cells through Interaction with Estrogen Receptor α. Toxicol. Appl. Pharmacol. 2020, 399, 115030. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Y. Non-Coplanar and Coplanar Polychlorinated Biphenyls Potentiate Genotoxicity of Aflatoxin B1 in a Human Hepatocyte Line by Enhancing CYP1A2 and CYP3A4 Expression. Environ. Pollut. 2019, 246, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Manikandan, P.; Nagini, S. Cytochrome P450 Structure, Function and Clinical Significance: A Review. Curr. Drug Targets 2017, 19, 38–54. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P. Cytochrome P450 and Chemical Toxicology. Chem. Res. Toxicol. 2008, 21, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Esteves, F.; Rueff, J.; Kranendonk, M. The Central Role of Cytochrome P450 in Xenobiotic Metabolism—A Brief Review on a Fascinating Enzyme Family. J. Xenobiotics 2021, 11, 94–114. [Google Scholar] [CrossRef]
- Guengerich, F.P.; Waterman, M.R.; Egli, M. Recent Structural Insights into Cytochrome P450 Function. Trends Pharmacol. Sci. 2016, 37, 625–640. [Google Scholar] [CrossRef] [Green Version]
- Sevior, D.K.; Pelkonen, O.; Ahokas, J.T. Hepatocytes: The Powerhouse of Biotransformation. Int. J. Biochem. Cell Biol. 2012, 44, 257–261. [Google Scholar] [CrossRef]
- McGraw, J.; Waller, D. Cytochrome P450 Variations in Different Ethnic Populations. Expert Opin. Drug Metab. Toxicol. 2012, 8, 371–382. [Google Scholar] [CrossRef]
- Zanger, U.M.; Schwab, M. Cytochrome P450 Enzymes in Drug Metabolism: Regulation of Gene Expression, Enzyme Activities, and Impact of Genetic Variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef] [PubMed]
- The Lancet Planetary Health. Microplastics and Human Health—An Urgent Problem. Lancet Planet. Health 2017, 1, e254. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Kauffman, A.E.; Li, L.; McFee, W.; Cai, B.; Weinstein, J.; Lead, J.R.; Chatterjee, S.; Scott, G.I.; Xiao, S. Health Impacts of Environmental Contamination of Micro- And Nanoplastics: A Review. Environ. Health Prev. Med. 2020, 25, 29. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Marini, F.; Ludt, A.; Linke, J.; Strauch, K. GeneTonic: An R/Bioconductor Package for Streamlining the Interpretation of RNA-Seq Data. BMC Bioinform. 2021, 22, 610. [Google Scholar] [CrossRef]
- Rydberg, P.; Gloriam, D.E.; Zaretzki, J.; Breneman, C.; Olsen, L. SMARTCyp: A 2D Method for Prediction of Cytochrome P450-Mediated Drug Metabolism. ACS Med. Chem. Lett. 2010, 1, 96–100. [Google Scholar] [CrossRef] [Green Version]
- Rydberg, P.; Gloriam, D.E.; Olsen, L. The SMARTCyp Cytochrome P450 Metabolism Prediction Server. Bioinformatics 2010, 26, 2988–2989. [Google Scholar] [CrossRef] [Green Version]
- Cooper, J.E.; Kendig, E.L.; Belcher, S.M. Assessment of Bisphenol A Released from Reusable Plastic, Aluminium and Stainless Steel Water Bottles. Chemosphere 2011, 85, 943–947. [Google Scholar] [CrossRef] [Green Version]
- Kwan, C.S.; Takada, H. Release of Additives and Monomers from Plastic Wastes. In Handbook of Environmental Chemistry; Springer: Cham, Switzerland, 2019; Volume 78, pp. 51–70. [Google Scholar]
- Ellen MacArthur Foundation. Ellen MacArthur Foundation and World Economic Forum The New Plastics Economy: Rethinking the Future of Plastics. 2016. Available online: https://ellenmacarthurfoundation.org/the-new-plastics-economy-rethinking-the-future-of-plastics (accessed on 3 April 2022).
- Hodgson, E.; Rose, R.L. The Importance of Cytochrome P450 2B6 in the Human Metabolism of Environmental Chemicals. Pharmacol. Ther. 2007, 113, 420–428. [Google Scholar] [CrossRef]
- Saintot, M.; Malaveille, C.; Hautefeuille, A.; Gerber, M. Interaction between Genetic Polymorphism of Cytochrome P450-1B1 and Environmental Pollutants in Breast Cancer Risk. Eur. J. Cancer Prev. 2004, 13, 83–86. [Google Scholar] [CrossRef]
- Lagunas-Rangel, F.A.; Linnea-Niemi, J.V.; Kudłak, B.; Williams, M.J.; Jönsson, J.; Schiöth, H.B. Role of the Synergistic Interactions of Environmental Pollutants in the Development of Cancer. GeoHealth 2022, 6, e2021GH000552. [Google Scholar] [CrossRef]
- Abdelfatah, S.; Naß, J.; Knorz, C.; Klauck, S.M.; Küpper, J.H.; Efferth, T. Pyrrolizidine Alkaloids Cause Cell Cycle and DNA Damage Repair Defects as Analyzed by Transcriptomics in Cytochrome P450 3A4-Overexpressing HepG2 Clone 9 Cells. Cell Biol. Toxicol. 2022, 38, 325–345. [Google Scholar] [CrossRef]
- Medina, M.W.; Gao, F.; Naidoo, D.; Rudel, L.L.; Temel, R.E.; McDaniel, A.L.; Marshall, S.M.; Krauss, R.M. Coordinately Regulated Alternative Splicing of Genes Involved in Cholesterol Biosynthesis and Uptake. PLoS ONE 2011, 6, e19420. [Google Scholar] [CrossRef] [Green Version]
- Moebius, F.F.; Fitzky, B.U.; Glossmann, H. Genetic Defects in Postsqualene Cholesterol Biosynthesis. Trends Endocrinol. Metab. 2000, 11, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Sacks, F.M. The Apolipoprotein Story. Atheroscler. Suppl. 2006, 7, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Bell, F.P. Effects of Phthalate Esters on Lipid Metabolism in Various Tissues, Cells and Organelles in Mammals. Environ. Health Perspect. 1982, 45, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Ganning, A.E.; Brunk, U.; Dallner, G. Phthalate Esters and Their Effect on the Liver. Hepatology 1984, 4, 541–547. [Google Scholar] [CrossRef]
- Mes, J.; Coffin, D.E.; Campbell, D.S. Di-n-Butyl-and Di-2-Ethylhexyl Phthalate in Human Adipose Tissue. Bull. Environ. Contam. Toxicol. 1974, 12, 721–725. [Google Scholar] [CrossRef]
- Sánchez, A.; Rodríguez-Viso, P.; Domene, A.; Orozco, H.; Vélez, D.; Devesa, V. Dietary Microplastics: Occurrence, Exposure and Health Implications. Environ. Res. 2022, 212, 113150. [Google Scholar] [CrossRef]
- Koelmans, A.A.; Mohamed Nor, N.H.; Hermsen, E.; Kooi, M.; Mintenig, S.M.; De France, J. Microplastics in Freshwaters and Drinking Water: Critical Review and Assessment of Data Quality. Water Res. 2019, 155, 410–422. [Google Scholar] [CrossRef]
- Yin, J.; Ju, Y.; Qian, H.; Wang, J.; Miao, X.; Zhu, Y.; Zhou, L.; Ye, L. Nanoplastics and Microplastics May Be Damaging Our Livers. Toxics 2022, 10, 586. [Google Scholar] [CrossRef]
- Pereira, C.; Mapuskar, K.; Rao, C.V. Chronic Toxicity of Diethyl Phthalate in Male Wistar Rats-A Dose-Response Study. Regul. Toxicol. Pharmacol. 2006, 45, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Horvatits, T.; Tamminga, M.; Liu, B.; Sebode, M.; Carambia, A.; Fischer, L.; Püschel, K.; Huber, S.; Fischer, E.K. Microplastics Detected in Cirrhotic Liver Tissue. eBioMedicine 2022, 82, 104147. [Google Scholar] [CrossRef]
- Bartels, Ä.; O’Donoghue, K. Cholesterol in Pregnancy: A Review of Knowns and Unknowns. Obstet. Med. 2011, 4, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Kapourchali, F.R.; Surendiran, G.; Goulet, A.; Moghadasian, M.H. The Role of Dietary Cholesterol in Lipoprotein Metabolism and Related Metabolic Abnormalities: A Mini-Review. Crit. Rev. Food Sci. Nutr. 2016, 56, 2408–2415. [Google Scholar] [CrossRef] [PubMed]
- Wilson, P.W.F.; Hoeg, J.M.; D’Agostino, R.B.; Silbershatz, H.; Belanger, A.M.; Poehlmann, H.; O’Leary, D.; Wolf, P.A. Cumulative Effects of High Cholesterol Levels, High Blood Pressure, and Cigarette Smoking on Carotid Stenosis. N. Engl. J. Med. 1997, 337, 516–522. [Google Scholar] [CrossRef]
- LaRosa, J.C. Understanding Risk in Hypercholesterolemia. Clin. Cardiol. 2003, 26, 3–6. [Google Scholar] [CrossRef]
- Falk, E. Pathogenesis of Atherosclerosis. J. Am. Coll. Cardiol. 2006, 47, C7–C12. [Google Scholar] [CrossRef] [Green Version]
- Yeagle, P.L. Cholesterol and the Cell Membrane. Biochim. Biophys. Acta BBA-Rev. Biomembr. 1985, 822, 267–287. [Google Scholar] [CrossRef]
- Luo, J.; Yang, H.; Song, B.L. Mechanisms and Regulation of Cholesterol Homeostasis. Nat. Rev. Mol. Cell Biol. 2020, 21, 225–245. [Google Scholar] [CrossRef]
- Hu, J.; Zhang, Z.; Shen, W.J.; Azhar, S. Cellular Cholesterol Delivery, Intracellular Processing and Utilization for Biosynthesis of Steroid Hormones. Nutr. Metab. 2010, 7, 7–9. [Google Scholar] [CrossRef] [Green Version]
- Morgan, A.E.; Mooney, K.M.; Wilkinson, S.J.; Pickles, N.A.; Mc Auley, M.T. Cholesterol Metabolism: A Review of How Ageing Disrupts the Biological Mechanisms Responsible for Its Regulation. Ageing Res. Rev. 2016, 27, 108–124. [Google Scholar] [CrossRef] [PubMed]
- Subczynski, W.K.; Pasenkiewicz-Gierula, M.; Widomska, J.; Mainali, L.; Raguz, M. High Cholesterol/Low Cholesterol: Effects in Biological Membranes: A Review. Cell Biochem. Biophys. 2017, 75, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Su, T. Cholesterol Binding and Cholesterol Transport Proteins; Springer: Dordrecht, The Netherlands, 2010; Volume 51, ISBN 978-90-481-8621-1. [Google Scholar]
- Schade, D.S.; Shey, L.; Eaton, R.P. Cholesterol Review: A Metabolically Important Molecule. Endocr. Pract. 2020, 26, 1514–1523. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Jannatun, N.; Zeng, Y.; Liu, T.; Zhang, G.; Chen, C.; Li, Y. Impacts of Microplastics on Immunity. Front. Toxicol. 2022, 4, 956885. [Google Scholar] [CrossRef]
- Jin, Y.; Xia, J.; Pan, Z.; Yang, J.; Wang, W.; Fu, Z. Polystyrene Microplastics Induce Microbiota Dysbiosis and Inflammation in the Gut of Adult Zebrafish. Environ. Pollut. 2018, 235, 322–329. [Google Scholar] [CrossRef]
- Murata, M.; Kang, J.H. Bisphenol A (BPA) and Cell Signaling Pathways. Biotechnol. Adv. 2018, 36, 311–327. [Google Scholar] [CrossRef]
- Sorci, G.; Loiseau, C. Should We Worry about the Accumulation of Microplastics in Human Organs? eBioMedicine 2022, 82, 104191. [Google Scholar] [CrossRef]
- Kumari, A.; Arora, S.; Kaur, R. Comparative Cytotoxic and Genotoxic Potential of Benzyl-Butyl Phthalate and Di-n-Butyl Phthalate Using Allium Cepa Assay. Energy Ecol. Environ. 2021, 6, 244–257. [Google Scholar] [CrossRef]
- Erkekoglu, P.; Rachidi, W.; Yuzugullu, O.G.; Giray, B.; Favier, A.; Ozturk, M.; Hincal, F. Evaluation of Cytotoxicity and Oxidative DNA Damaging Effects of Di(2-Ethylhexyl)-Phthalate (DEHP) and Mono(2-Ethylhexyl)-Phthalate (MEHP) on MA-10 Leydig Cells and Protection by Selenium. Toxicol. Appl. Pharmacol. 2010, 248, 52–62. [Google Scholar] [CrossRef]
- Erkekoglu, P.; Kocer-Gumusel, B. Genotoxicity of Phthalates. Toxicol. Mech. Methods 2014, 24, 616–626. [Google Scholar] [CrossRef] [PubMed]
- Rider, C.F.; Carlsten, C. Air Pollution and DNA Methylation: Effects of Exposure in Humans. Clin. Epigenetics 2019, 11, 131. [Google Scholar] [CrossRef] [Green Version]
- Gillings, M.R.; Westoby, M.; Ghaly, T.M. Pollutants That Replicate: Xenogenetic DNAs. Trends Microbiol. 2018, 26, 975–977. [Google Scholar] [CrossRef] [PubMed]
- Somers, C.M.; Yauk, C.L.; White, P.A.; Parfett, C.L.J.; Quinn, J.S. Air Pollution Induces Heritable DNA Mutations. Proc. Natl. Acad. Sci. USA 2002, 99, 15904–15907. [Google Scholar] [CrossRef] [PubMed]
- Bilancio, A.; Bontempo, P.; Di Donato, M.; Conte, M.; Giovannelli, P.; Altucci, L.; Migliaccio, A.; Castoria, G. Bisphenol a Induces Cell Cycle Arrest in Primary and Prostate Cancer Cells through EGFR/ERK/P53 Signaling Pathway Activation. Oncotarget 2017, 8, 115620–115631. [Google Scholar] [CrossRef] [Green Version]
- Zu, M.L.; Duan, Y.; Xie, J.B.; Qi, Y.S.; Xie, P.; Borjigidai, A.; Piao, X.L. Gypenoside LI Arrests the Cell Cycle of Breast Cancer in G0/G1 Phase by down-Regulating E2F1. J. Ethnopharmacol. 2021, 273, 114017. [Google Scholar] [CrossRef]
- Fausto, N.; Campbell, J.S.; Riehle, K.J. Liver Regeneration. Hepatology 2006, 43, 45–53. [Google Scholar] [CrossRef]
- Jeon, M.S.; Kim, J.W.; Han, Y.B.; Jeong, M.H.; Kim, H.R.; Sik Kim, H.; Park, Y.J.; Chung, K.H. Polystyrene Microplastic Particles Induce Autophagic Cell Death in BEAS-2B Human Bronchial Epithelial Cells. Environ. Toxicol. 2023, 38, 359–367. [Google Scholar] [CrossRef]
- Toshima, T.; Shirabe, K.; Fukuhara, T.; Ikegami, T.; Yoshizumi, T.; Soejima, Y.; Ikeda, T.; Okano, S.; Maehara, Y. Suppression of Autophagy during Liver Regeneration Impairs Energy Charge and Hepatocyte Senescence in Mice. Hepatology 2014, 60, 290–300. [Google Scholar] [CrossRef]
- Jang, M.; Kim, H.; Park, R.; Jo, D.; Lee, E.J.; Oh, W.K.; Park, J. 2,2′-Methylenebis (6-Tert-Butyl 4-Methylphenol) Enhances the Antitumor Efficacy of Belotecan, a Derivative of Camptothecin, by Inducing Autophagy. Oncotarget 2017, 8, 115068–115078. [Google Scholar] [CrossRef] [Green Version]
- Cox, K.D.; Covernton, G.A.; Davies, H.L.; Dower, J.F.; Juanes, F.; Dudas, S.E. Human Consumption of Microplastics. Environ. Sci. Technol. 2019, 53, 7068–7074. [Google Scholar] [CrossRef] [Green Version]
- Guengerich, F.P.; Liebler, D.C.; Reed, D.L. Enzymatic Activation of Chemicals to Toxic Metabolites. CRC Crit. Rev. Toxicol. 1985, 14, 259–307. [Google Scholar] [CrossRef] [PubMed]
- Steinbrecht, S.; König, R.; Schmidtke, K.U.; Herzog, N.; Scheibner, K.; Krüger-Genge, A.; Jung, F.; Kammerer, S.; Küpper, J.H. Metabolic Activity Testing Can Underestimate Acute Drug Cytotoxicity as Revealed by HepG2 Cell Clones Overexpressing Cytochrome P450 2C19 and 3A4. Toxicology 2019, 412, 37–47. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New Data Content and Improved Web Interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M. The Protein Data Bank: A Historical Perspective. Acta Crystallogr. Sect. A Found. Crystallogr. 2008, 64, 88–95. [Google Scholar] [CrossRef]
- Reynald, R.L.; Sansen, S.; Stout, C.D.; Johnson, E.F. Structural Characterization of Human Cytochrome P450 2C19: Active Site Differences between P450s 2C8, 2C9, and 2C19. J. Biol. Chem. 2012, 287, 44581–44591. [Google Scholar] [CrossRef] [Green Version]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Soneson, C.; Hickey, P.F.; Johnson, L.K.; Tessa Pierce, N.; Shepherd, L.; Morgan, M.; Patro, R. Tximeta: Reference Sequence Checksums for Provenance Identification in RNA-Seq. PLoS Comput. Biol. 2020, 16, e1007664. [Google Scholar] [CrossRef] [Green Version]
- Ludt, A.; Ustjanzew, A.; Binder, H.; Strauch, K.; Marini, F. Interactive and Reproducible Workflows for Exploring and Modeling RNA-Seq Data with PcaExplorer, Ideal, and GeneTonic. Curr. Protoc. 2022, 2, e411. [Google Scholar] [CrossRef]
- Marini, F.; Binder, H. PcaExplorer: An R/Bioconductor Package for Interacting with RNA-Seq Principal Components. BMC Bioinform. 2019, 20, 331. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zitovsky, J.P.; Love, M.I. Fast Effect Size Shrinkage Software for Beta- Binomial Models of Allelic Imbalance [Version 2 ; Peer Review: 3 Approved with Reservations]. F1000Research 2021, 8, 2024. [Google Scholar] [CrossRef] [PubMed]
- Alexa, A.; Rahnenführer, J.; Lengauer, T. Improved Scoring of Functional Groups from Gene Expression Data by Decorrelating GO Graph Structure. Bioinformatics 2006, 22, 1600–1607. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosellini, M.; Schulze, A.; Omer, E.A.; Ali, N.T.; Marini, F.; Küpper, J.-H.; Efferth, T. The Effect of Plastic-Related Compounds on Transcriptome-Wide Gene Expression on CYP2C19-Overexpressing HepG2 Cells. Molecules 2023, 28, 5952. https://doi.org/10.3390/molecules28165952
Rosellini M, Schulze A, Omer EA, Ali NT, Marini F, Küpper J-H, Efferth T. The Effect of Plastic-Related Compounds on Transcriptome-Wide Gene Expression on CYP2C19-Overexpressing HepG2 Cells. Molecules. 2023; 28(16):5952. https://doi.org/10.3390/molecules28165952
Chicago/Turabian StyleRosellini, Matteo, Alicia Schulze, Ejlal A. Omer, Nadeen T. Ali, Federico Marini, Jan-Heiner Küpper, and Thomas Efferth. 2023. "The Effect of Plastic-Related Compounds on Transcriptome-Wide Gene Expression on CYP2C19-Overexpressing HepG2 Cells" Molecules 28, no. 16: 5952. https://doi.org/10.3390/molecules28165952
APA StyleRosellini, M., Schulze, A., Omer, E. A., Ali, N. T., Marini, F., Küpper, J.-H., & Efferth, T. (2023). The Effect of Plastic-Related Compounds on Transcriptome-Wide Gene Expression on CYP2C19-Overexpressing HepG2 Cells. Molecules, 28(16), 5952. https://doi.org/10.3390/molecules28165952