In Silico Modeling and Structural Analysis of Soluble Epoxide Hydrolase Inhibitors for Enhanced Therapeutic Design
Abstract
1. Introduction
2. Results
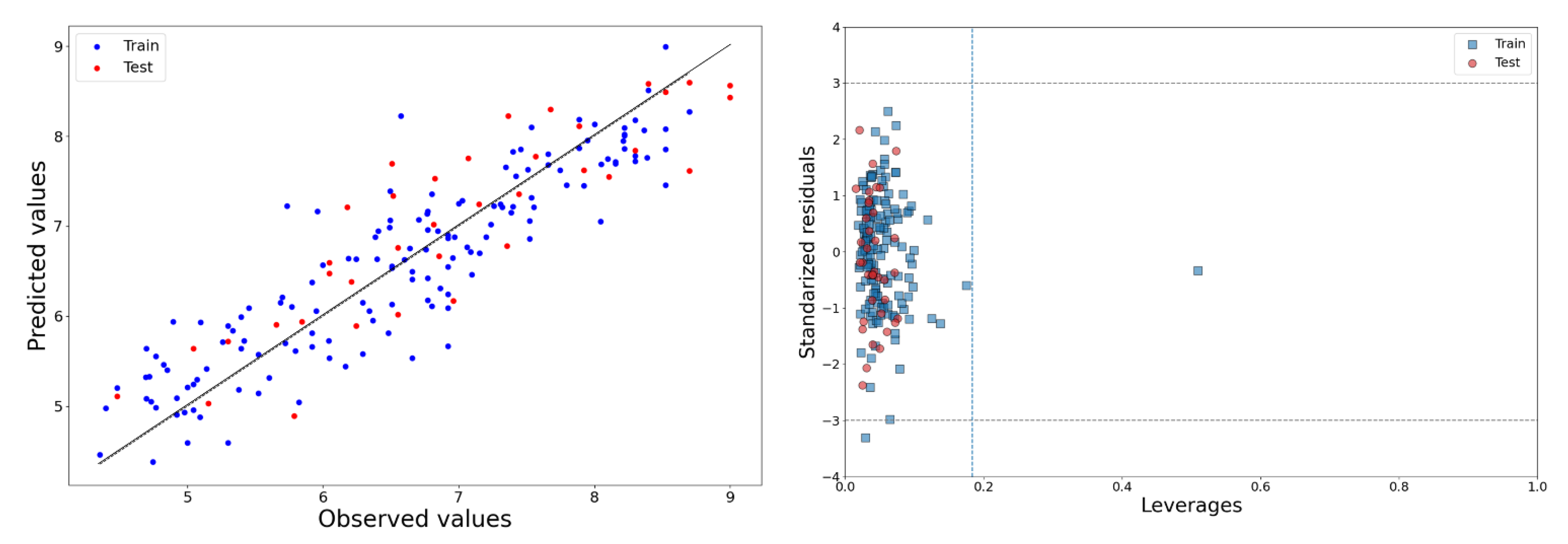
2.1. 2D-QSAR Model
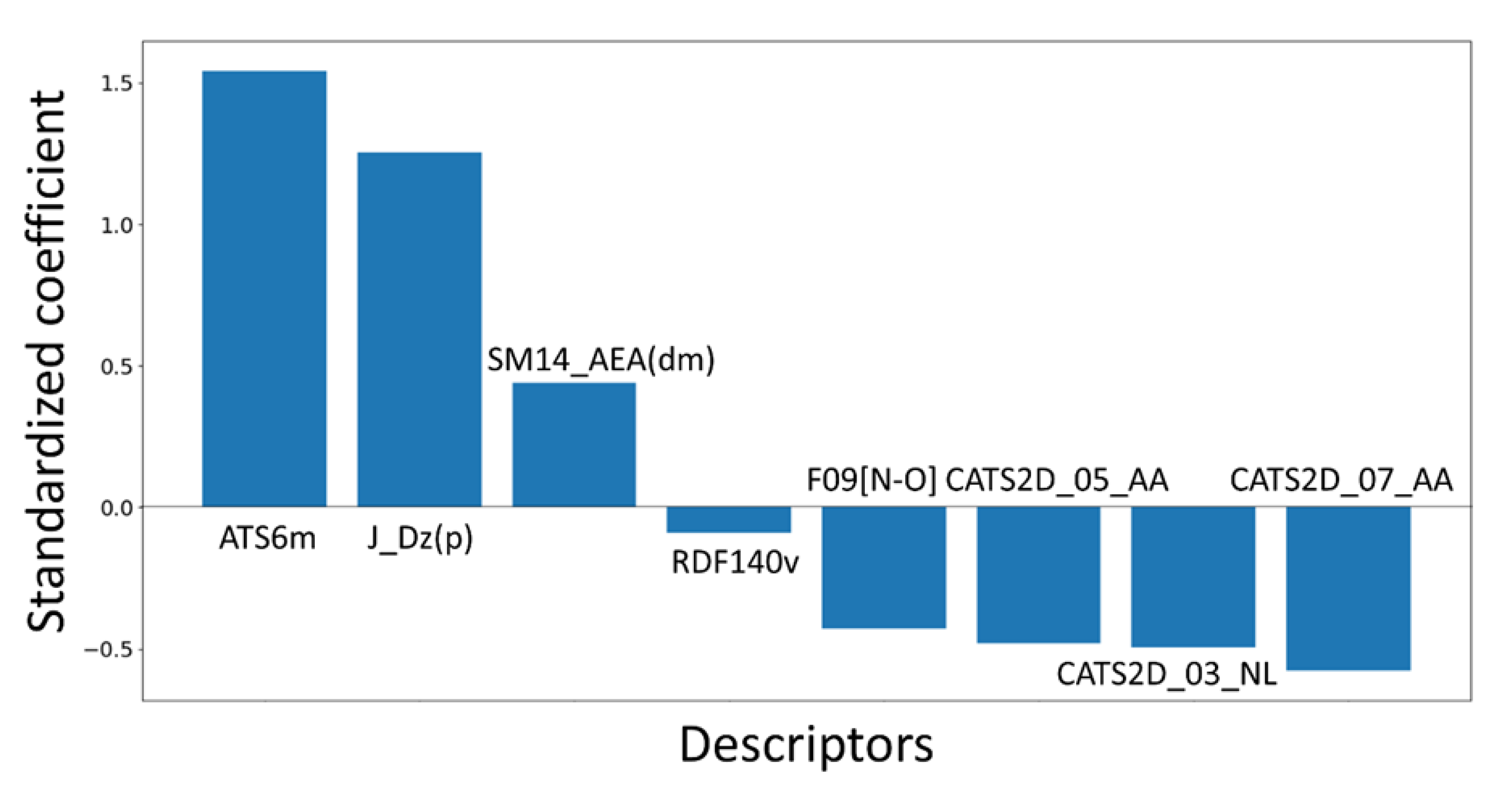
− 0.48(±0.043) CATS2D_05_AA + 0.44(±0.07)*SM14_AEA(dm)
− 0.495(±0.061) CATS2D_03_NL − 0.091(±0.036) RDF140v
− 0.577(±0.097) CATS2D_07_AA grama + 1.255(±0.296) J_Dz(p)
∆rm2LOO = 0.147, KXX = 0.371, ΔK = 0.03. Ntest = 37, R2Pred/Q2(F1) = 0.792, Q2(F2) = 0.769,
Q2(F3) = 0.763, RMSEP = 0.558, rm2test = 0.685, ∆rm2test = 0.167
2.2. Transformer-CNN-Based QSAR Model
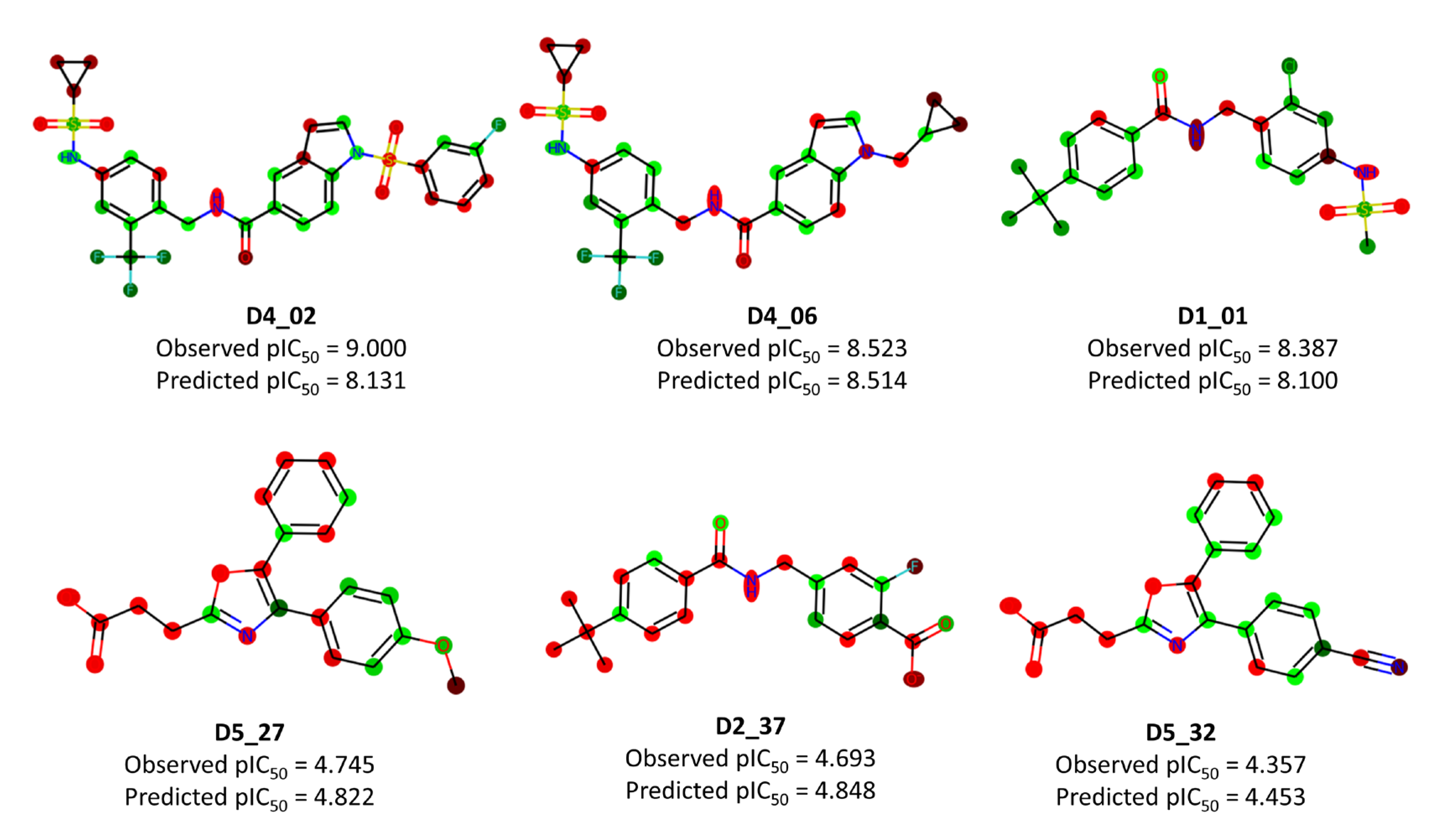
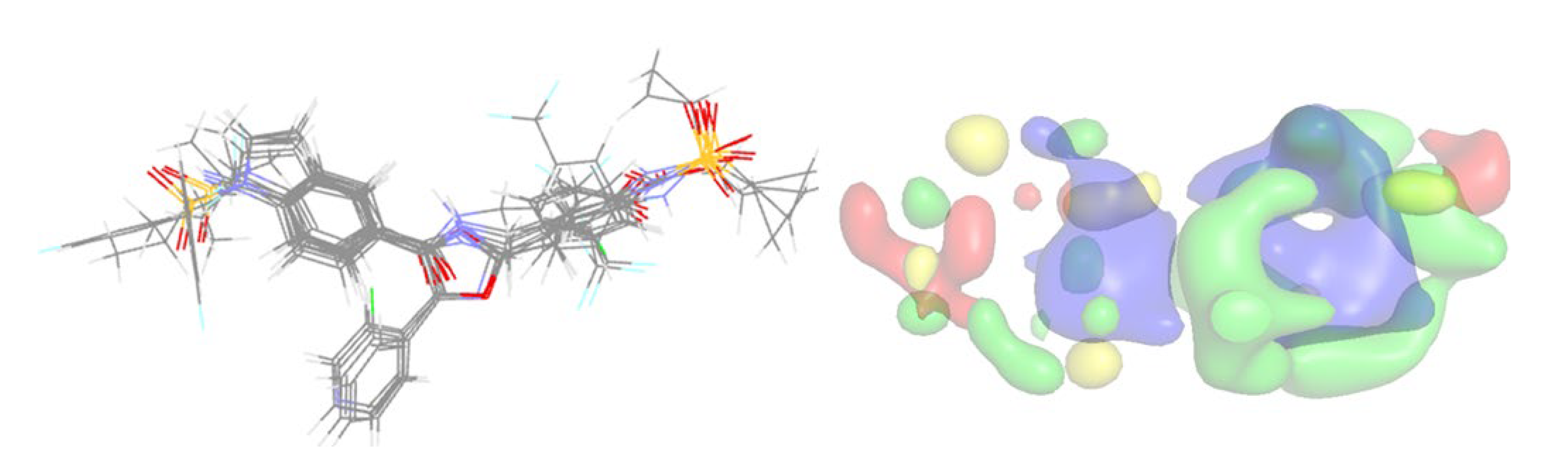
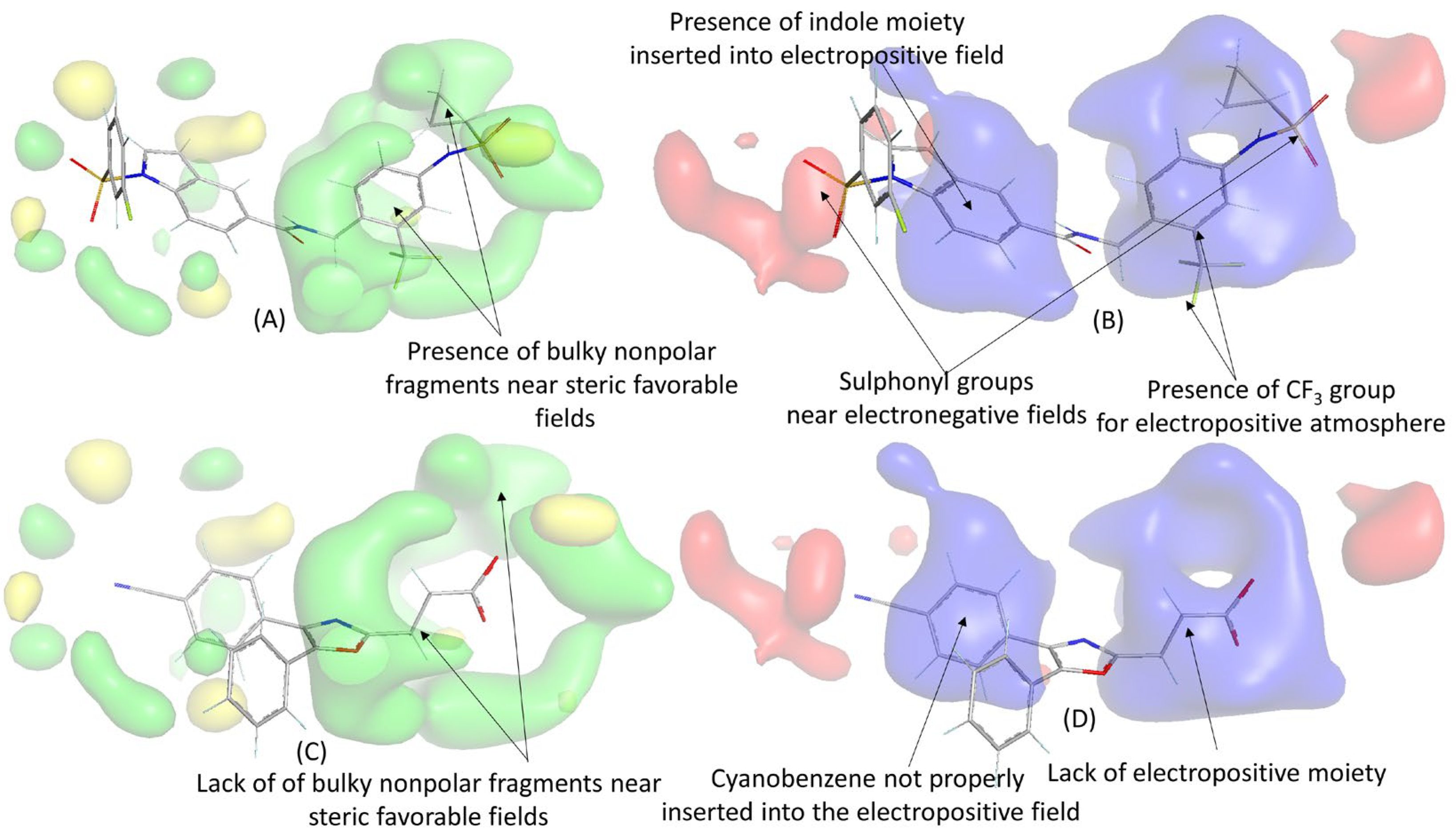
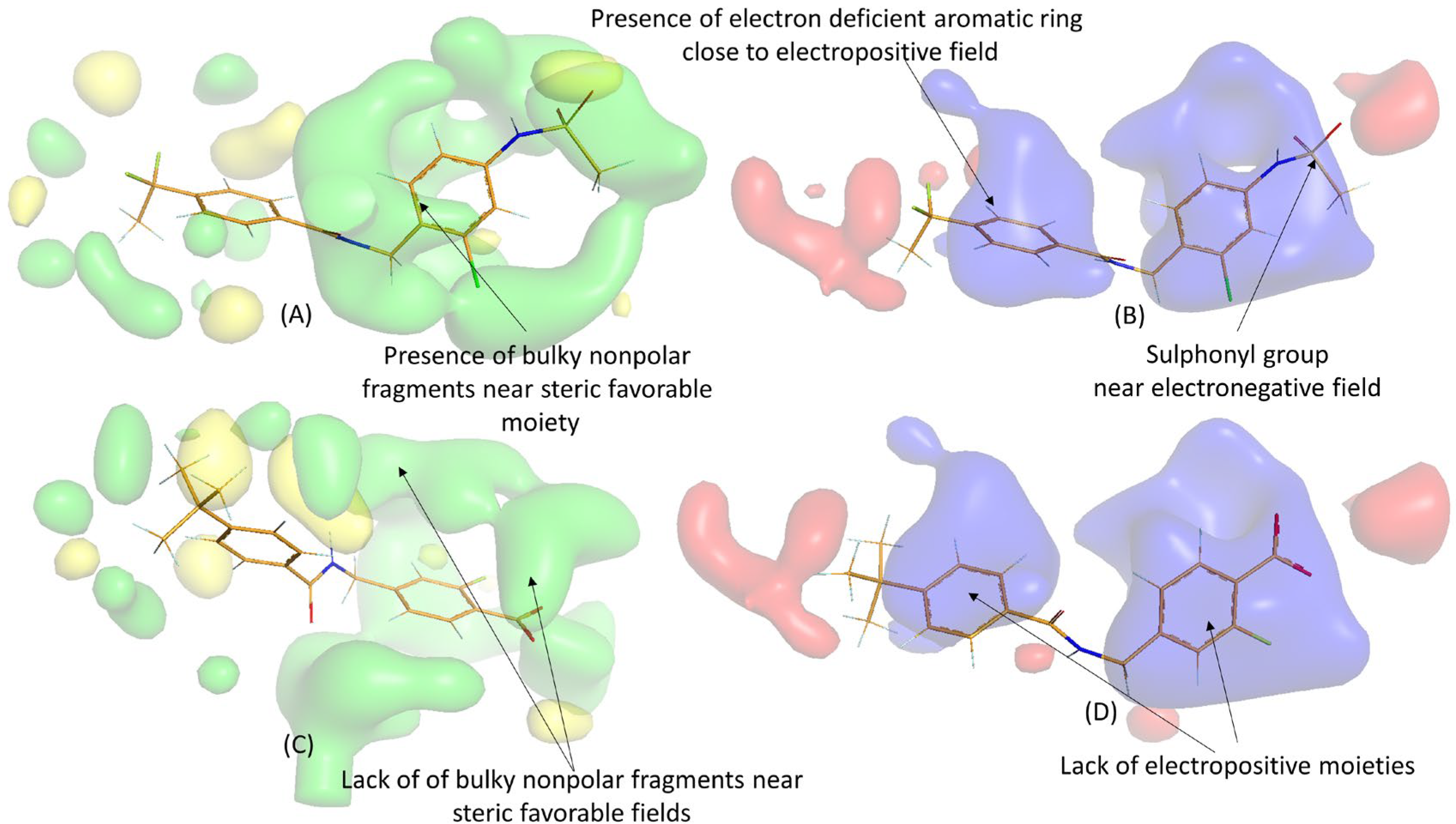
2.3. 3D-QSAR Analysis
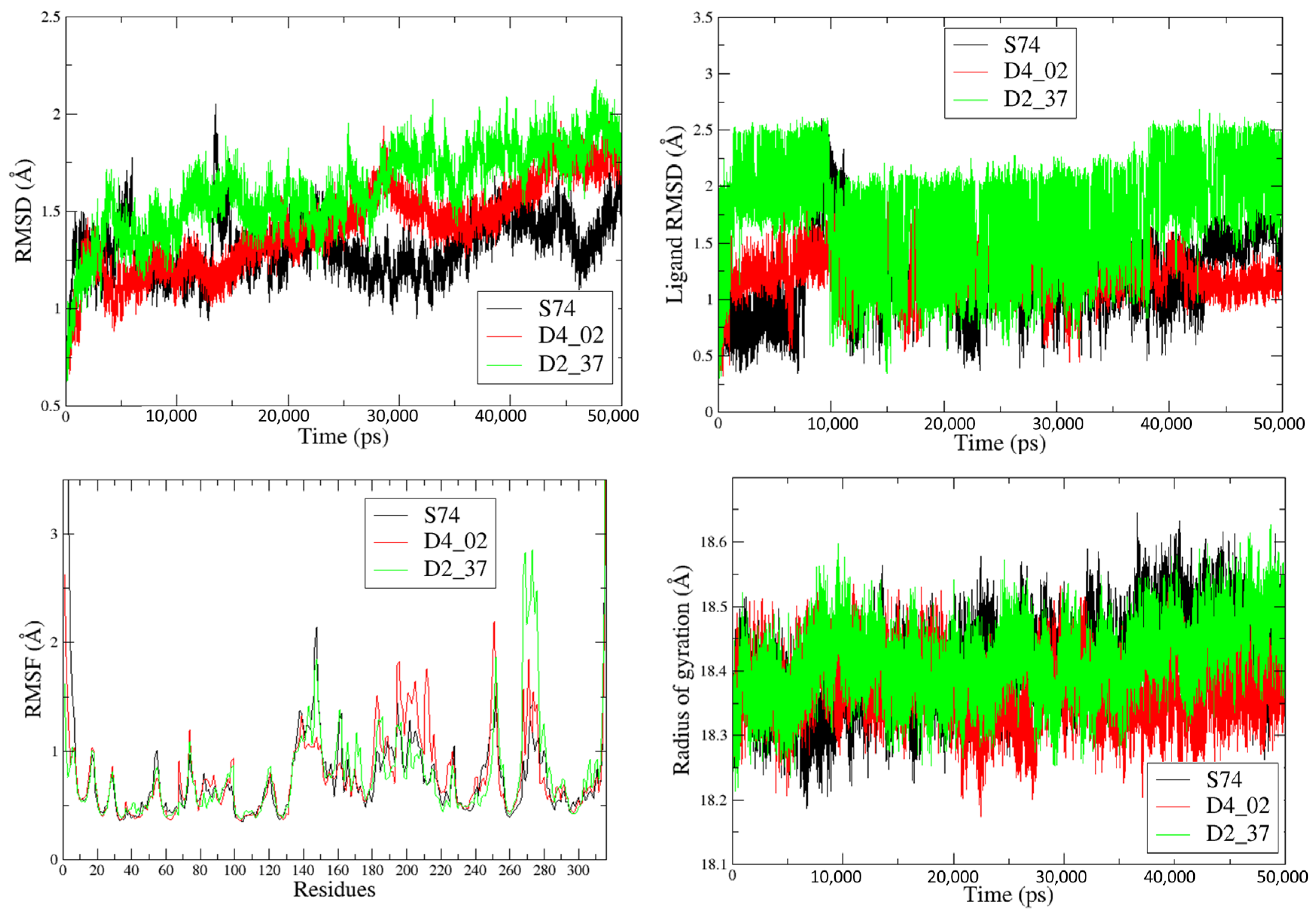
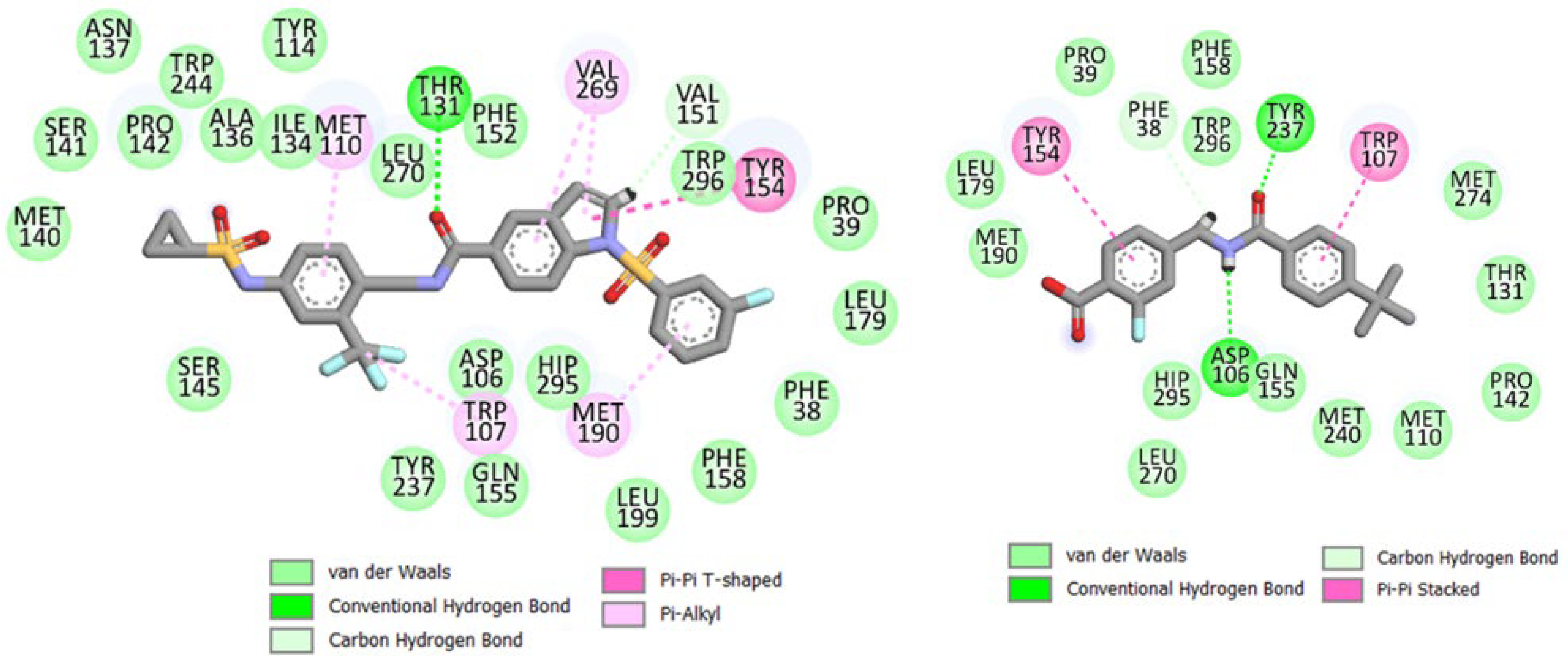
2.4. Molecular Dynamics Simulations
3. Materials and Methods
3.1. Conventional 2D-QSAR Modeling
3.1.1. Dataset Collection and Preparation
3.1.2. Calculation of Descriptors
3.1.3. Dataset Division and Feature Selection
3.1.4. Model Evaluation
3.1.5. Applicability Domain of the Models
3.1.6. Machine Learning Techniques and Partial Least Square (PLS)
3.1.7. Consensus Modeling
3.2. Transformer-CNN Based QSAR Modeling
3.3. 3D-QSAR Modeling
3.3.1. Alignment Techniques
3.3.2. Model Development
3.3.3. Molecular Docking and Molecular Dynamics Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Iyer, M.R.; Kundu, B.; Wood, C.M. Soluble epoxide hydrolase inhibitors: An overview and patent review from the last decade. Expert Opin. Ther. Pat. 2022, 32, 629–647. [Google Scholar] [CrossRef] [PubMed]
- Spector, A.A.; Fang, X.; Snyder, G.D. Weintraub NL. Epoxyeicosatrienoic acids (EETs): Metabolism and biochemical function. Prog. Lipid Res. 2004, 43, 55–90. [Google Scholar] [CrossRef] [PubMed]
- Nazari, M.; Rezaee, E.; Tabatabai, S.A. A Comprehensive Review of Soluble Epoxide Hyådrolase Inhibitors Evaluating their Structure-Activity Relationship. Mini Rev. Med. Chem. 2023, 23, 99–117. [Google Scholar] [CrossRef] [PubMed]
- Helmstädter, M.; Kaiser, A.; Brunst, S.; Schmidt, J.; Ronchetti, R.; Weizel, L.; Proschak, E.; Merk, D. Second-Generation Dual FXR/sEH Modulators with Optimized Pharmacokinetics. J. Med. Chem. 2021, 64, 9525–9536. [Google Scholar] [CrossRef]
- Shen, H.C. Soluble epoxide hydrolase inhibitors: A patent review. Expert Opin. Ther. Pat. 2010, 20, 941–956. [Google Scholar] [CrossRef]
- Fleming, I. The pharmacology of the cytochrome P450 epoxygenase/soluble epoxide hydrolase axis in the vasculature and cardiovascular disease. Pharmacol. Rev. 2014, 66, 1106–1140. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Y.; Schmelzer, K.; Lee, T.S.; Fang, X.; Zhu, Y.; Spector, A.A.; Gill, S.; Morisseau, C.; Hammock, B.D.; et al. The antiinflammatory effect of laminar flow: The role of PPARγ, epoxyeicosatrienoic acids, and soluble epoxide hydrolase. Proc. Natl. Acad. Sci. USA 2005, 46, 16747–16752. [Google Scholar] [CrossRef]
- Xu, D.Y.; Davis, B.B.; Wang, Z.H.; Zhao, S.P.; Wasti, B.; Liu, Z.L.; Li, N.; Morisseau, C.; Chiamvimonvat, N.; Hammock, B.D. A potent soluble epoxide hydrolase inhibitor, t-AUCB, acts through PPARγ to modulate the function of endothelial progenitor cells from patients with acute myocardial infarction. Int. J. Cardiol. 2013, 4, 1298–1304. [Google Scholar] [CrossRef]
- Wagner, K.M.; McReynolds, C.B.; Schmidt, W.K.; Hammock, B.D. Soluble epoxide hydrolase as a therapeutic target for pain, inflammatory and neurodegenerative diseases. Pharmacol. Ther. 2017, 180, 62–76. [Google Scholar] [CrossRef]
- Blöcher, R.; Lamers, C.; Wittmann, S.K.; Merk, D.; Hartmann, M.; Weizel, L.; Diehl, O.; Brüggerhoff, A.; Boß, M.; Kaiser, A.; et al. N-Benzylbenzamides: A Novel Merged Scaffold for Orally Available Dual Soluble Epoxide Hydrolase/Peroxisome Proliferator-Activated Receptor γ Modulators. J. Med. Chem. 2016, 59, 61–81. [Google Scholar] [CrossRef]
- Morisseau, C.; Schebb, N.H.; Dong, H.; Ulu, A.; Aronov, P.A.; Hammock, B.D. Role of soluble epoxide hydrolase phosphatase activity in the metabolism of lysophosphatidic acids. Biochem. Biophys. Res. Commun. 2012, 419, 796–800. [Google Scholar] [CrossRef] [PubMed]
- Oguro, A.; Imaoka, S. Lysophosphatidic acids are new substrates for the phosphatase domain of soluble epoxide hydrolase. J. Lipid Res. 2012, 53, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.H.; Liao, Y.J.; Hsiao, S.H.; Shyue, S.K.; Lee, T.S. Role of phosphatase activity of soluble epoxide hydrolase in regulating simvastatin-activated endothelial nitric oxide synthase. Sci. Rep. 2015, 5, 13524. [Google Scholar] [CrossRef]
- Kramer, J.S.; Woltersdorf, S.; Duflot, T.; Hiesinger, K.; Lillich, F.F.; Knöll, F.; Wittmann, S.K.; Klingler, F.M.; Brunst, S.; Chaikuad, A.; et al. Discovery of the First in Vivo Active Inhibitors of the Soluble Epoxide Hydrolase Phosphatase Domain. J. Med. Chem. 2019, 62, 8443–8460. [Google Scholar] [CrossRef]
- Liu, Y.; Dang, H.; Li, D.; Pang, W.; Hammock, B.D.; Zhu, Y. Inhibition of soluble epoxide hydrolase attenuates high-fat-diet-induced hepatic steatosis by reduced systemic inflammatory status in mice. PLoS ONE 2012, 7, e39165. [Google Scholar] [CrossRef]
- Schmidt, J.; Rotter, M.; Weiser, T.; Wittmann, S.; Weizel, L.; Kaiser, A.; Heering, J.; Goebel, T.; Angioni, C.; Wurglics, M.; et al. A Dual Modulator of Farnesoid X Receptor and Soluble Epoxide Hydrolase to Counter Nonalcoholic Steatohepatitis. J. Med. Chem. 2017, 60, 7703–7724. [Google Scholar] [CrossRef] [PubMed]
- Neves, B.J.; Braga, R.C.; Melo-Filho, C.C.; Moreira-Filho, J.T.; Muratov, E.N.; Andrade, C.H. QSAR-Based Virtual Screening: Advances and Applications in Drug Discovery. Front. Pharmacol. 2018, 9, 1275. [Google Scholar] [CrossRef]
- Halder, A.K.; Moura, A.S.; Cordeiro, M.N.D.S. QSAR modelling: A therapeutic patent review 2010-present. Expert Opin. Ther. Pat. 2018, 28, 467–476. [Google Scholar] [CrossRef]
- Halder, A.K.; Cordeiro, M.N.D.S. Development of Multi-Target Chemometric Models for the Inhibition of Class I PI3K Enzyme Isoforms: A Case Study Using QSAR-Co Tool. Int. J. Mol. Sci. 2019, 20, 4191. [Google Scholar] [CrossRef]
- Halder, A.K.; Cordeiro, M.N.D.S. QSAR-Co-X: An open source toolkit for multitarget QSAR modelling. J. Cheminform. 2021, 13, 29. [Google Scholar] [CrossRef]
- Muratov, E.N.; Bajorath, J.; Sheridan, R.P.; Tetko, I.V.; Filimonov, D.; Poroikov, V.; Oprea, T.I.; Baskin, I.I.; Varnek, A.; Roitberg, A.; et al. QSAR without borders. Chem. Soc. Rev. 2020, 49, 3525–3564. [Google Scholar] [CrossRef] [PubMed]
- Roy, K. Advances in QSAR Modeling, Applications in Pharmaceutical, Chemical, Food, Agricultural and Environmental Sciences; Challenges and Advances in Computational Chemistry and Physics; Springer: Cham, Switzerland, 2017; Volume 24. [Google Scholar] [CrossRef]
- Tetko, I.V.; Tanchuk, V.Y.; Villa, A.E. Prediction of n-octanol/water partition coefficients from PHYSPROP database using ar-tificial neural networks and E-state indices. J. Chem. Inf. Comput. Sci. 2001, 41, 1407–1421. [Google Scholar] [CrossRef] [PubMed]
- Golbraikh, A.; Tropsha, A. Beware of q2! J. Mol. Graph. Model. 2002, 20, 269–276. [Google Scholar] [CrossRef]
- Gramatica, P.; Sangion, A. A Historical Excursus on the Statistical Validation Parameters for QSAR Models: A Clarification Concerning Metrics and Terminology. J. Chem. Inf. Model. 2016, 56, 1127–1131. [Google Scholar] [CrossRef]
- Todeschini, R.; Consonni, V. Handbook of Molecular Descriptors; Wiley-VCH Verlag GmbH: Weinheim, Germany, 2000. [Google Scholar]
- Sliwoski, G.; Mendenhall, J.; Meiler, J. Autocorrelation descriptor improvements for QSAR: 2DA_Sign and 3DA_Sign. J. Comput. Aided Mol. Des. 2016, 30, 209–217. [Google Scholar] [CrossRef]
- Reutlinger, M.; Koch, C.P.; Reker, D.; Todoroff, N.; Schneider, P.; Rodrigues, T.; Schneider, G. Chemically Advanced Template Search (CATS) for Scaffold-Hopping and Prospective Target Prediction for ‘Orphan’ Molecules. Mol. Inform. 2013, 32, 133–138. [Google Scholar] [CrossRef]
- Lillich, F.F.; Willems, S.; Ni, X.; Kilu, W.; Borkowsky, C.; Brodsky, M.; Kramer, J.S.; Brunst, S.; Hernandez-Olmos, V.; Heering, J.; et al. Structure-Based Design of Dual Partial Peroxisome Proliferator-Activated Receptor γ Agonists/Soluble Epoxide Hydrolase Inhibitors. J. Med. Chem. 2021, 64, 17259–17276. [Google Scholar] [CrossRef] [PubMed]
- ACD/ChemSketch, version 2021.1.2; Advanced Chemistry Development, Inc. (ACD/Labs): Toronto, ON, Canada; Available online: www.acdlabs.com (accessed on 25 April 2023).
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Sushko, I.; Novotarskyi, S.; Körner, R.; Pandey, A.K.; Rupp, M.; Teetz, W.; Brandmaier, S.; Abdelaziz, A.; Prokopenko, V.V.; Tanchuk, V.Y.; et al. Online chemical modeling environment (OCHEM): Web platform for data storage, model development and publishing of chemical information. J. Comput. Aided Mol. Des. 2011, 25, 533–554. [Google Scholar] [CrossRef]
- Sadowski, J.; Gasteiger, J.; Klebe, G. Comparison of Automatic Three-Dimensional Model Builders Using 639 X-ray Structures. J. Chem. Inf. Model. 2002, 4, 1000–1008. [Google Scholar] [CrossRef]
- Durant, J.L.; Leland, B.A.; Henry, D.R.; Nourse, J.G. Reoptimization of MDL keys for use in drug discovery. J. Chem. Inf. Comp. Sci. 2002, 42, 1273–1280. [Google Scholar] [CrossRef]
- Van der Maaten, L.J.P.; Hinton, G.E. Visualizing High-Dimensional Data Using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Mauri, A. alvaDesc: A Tool to Calculate and Analyze Molecular Descriptors and Fingerprints. In Ecotoxicological QSARs. Methods in Pharmacology and Toxicology; Humana: New York, NY, USA, 2020; pp. 801–820. [Google Scholar]
- De Sousa, J.M.A. Descriptors Generation Using the CDK Toolkit and Web Services. In Tutorials in Chemoinformatics; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017; pp. 127–134. [Google Scholar]
- Varnek, A.; Fourches, D.; Hoonakker, F.; Solov’ev, V.P. Substructural fragments: An universal language to encode reactions, molecular and supramolecular structures. J. Comput. Aided Mol. Des. 2005, 19, 693–703. [Google Scholar] [CrossRef]
- Moriwaki, H.; Tian, Y.S.; Kawashita, N.; Takagi, T. Mordred: A molecular descriptor calculator. J. Cheminform. 2018, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Filimonov, D.; Poroikov, V.; Borodina, Y.; Gloriozova, T. Chemical Similarity Assessment through Multilevel Neighborhoods of Atoms: Definition and Comparison with the Other Descriptors. J. Chem. Inf. Model. 1999, 4, 666–670. [Google Scholar] [CrossRef]
- Potemkin, V.A.; Grishina, M.A.; Bartashevich, E.V. Modeling of drug molecule orientation within a receptor cavity in the BiS algorithm framework. J. Struct. Chem. 2007, 48, 155–160. [Google Scholar] [CrossRef][Green Version]
- Masand, V.H.; Rastija, V. PyDescriptor: A new PyMOL plugin for calculating thousands of easily understandable molecular descriptors. Chemom. Intell. Lab. Syst. 2017, 169, 12–18. [Google Scholar] [CrossRef]
- Halder, A.K.; Delgado, A.H.S.; Cordeiro, M.N.D.S. First multi-target QSAR model for predicting the cytotoxicity of acrylic acid-based dental monomers. Dent. Mater. 2022, 38, 333–346. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Müller, A.; Nothman, J.; Louppe, G.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Ambure, P.; Aher, R.B.; Gajewicz, A.; Puzyn, T.; Roy, K. “NanoBRIDGES” software: Open access tools to perform QSAR and nano-QSAR modeling. Chemom. Intell. Lab. Syst. 2015, 147, 1–13. [Google Scholar] [CrossRef]
- Gramatica, P. Principles of QSAR models validation: Internal and external. QSAR Comb. Sci. 2007, 26, 694–701. [Google Scholar] [CrossRef]
- Roy, P.P.; Paul, S.; Mitra, I.; Roy, K. On two novel parameters for validation of predictive QSAR models. Molecules 2009, 14, 1660–1701. [Google Scholar] [CrossRef]
- Yoo, W.; Mayberry, R.; Bae, S.; Singh, K.; He, Q.P.; Lillard, J.W., Jr. A Study of Effects of MultiCollinearity in the Multivariable Analysis. Int. J. Appl. Sci. Technol. 2014, 4, 9–19. [Google Scholar]
- Gramatica, P.; Chirico, N.; Papa, E.; Cassani, S.; Kovarich, S. QSARINS: A new software for the development, analysis, and validation of QSAR MLR models. J. Comput. Chem. 2013, 34, 2121–2132. [Google Scholar] [CrossRef]
- Todeschini, R.; Consonni, V.; Maiocchi, A. The K correlation index: Theory development and its application in chemometrics. Chem. Intell. Lab. Sys. 1999, 46, 13–29. [Google Scholar] [CrossRef]
- Ojha, P.K.; Roy, K. Comparative QSARs for antimalarial endochins: Importance of descriptor-thinning and noise reduction prior to feature selection. Chem. Intell. Lab. Sys. 2011, 109, 146–161. [Google Scholar] [CrossRef]
- Ghosh, A.; Panda, P.; Halder, A.K.; Cordeiro, M.N.D.S. In silico characterization of aryl benzoyl hydrazide derivatives as potential inhibitors of RdRp enzyme of H5N1 influenza virus. Front. Pharmacol. 2022, 13, 1004255. [Google Scholar] [CrossRef]
- Serra, A.; Önlü, S.; Festa, P.; Fortino, V.; Greco, D. MaNGA: A novel multi-niche multi-objective genetic algorithm for QSAR modelling. Bioinformatics 2020, 36, 145–153. [Google Scholar] [CrossRef]
- Gajewicz-Skretna, A.; Wyrzykowska, E.; Gromelski, M. Quantitative multi-species toxicity modeling: Does a multi-species, machine learning model provide better performance than a single-species model for the evaluation of acute aquatic toxicity by organic pollutants? Sci. Total Environ. 2023, 861, 160590. [Google Scholar] [CrossRef]
- Huang, G.B.; Babri, H.A. Upper bounds on the number of hidden neurons in feedforward networks with arbitrary bounded nonlinear activation functions. IEEE Trans. Neural Netw. 1998, 9, 224–229. [Google Scholar]
- Boser, B.E.; Guyon, I.M.; Vapnik, V.N. A training algorithm for optimal margin classifers. In Proceedings of the Fifth Annual Workshop on Computational Learning Theory, Pittsburgh, PA, USA, 27–29 July 1992; ACM: Rochester, NY, USA, 1992; pp. 144–152. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2000, 45, 5–32. [Google Scholar] [CrossRef]
- Urias, R.W.P.; Barigye, S.J.; Marrero-Ponce, Y.; García-Jacas, C.R.; Valdes-Martiní, J.R.; Perez-Gimenez, F. IMMAN: Free software for information theory-based chemometric analysis. Mol. Divers. 2015, 19, 305–319. [Google Scholar] [CrossRef]
- Khan, K.; Kumar, V.; Colombo, E.; Lombardo, A.; Benfenati, E.; Roy, K. Intelligent consensus predictions of bioconcentration factor of pharmaceuticals using 2D and fragment-based descriptors. Environ. Int. 2022, 170, 107625. [Google Scholar] [CrossRef]
- Halder, A.K.; Haghbakhsh, R.; Voroshylova, I.V.; Duarte, A.R.C.; Cordeiro, M.N.D.S. Density of Deep Eutectic Solvents: The Path Forward Cheminformatics-Driven Reliable Predictions for Mixtures. Molecules 2021, 26, 5779. [Google Scholar] [CrossRef]
- Karpov, P.; Godin, G.; Tetko, I.V. Transformer-CNN: Swiss knife for QSAR modeling and interpretation. J. Cheminform. 2020, 12, 17. [Google Scholar] [CrossRef] [PubMed]
- Tosco, P.; Balle, T. Open3DQSAR: A new open-source software aimed at high-throughput chemometric analysis of molecular interaction fields. J. Mol. Model. 2011, 17, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Tosco, P.; Balle, T.; Shiri, F. Open3DALIGN: An open-source software aimed at unsupervised ligand alignment. J. Comput. Aided Mol. Des. 2011, 25, 777–783. [Google Scholar] [CrossRef]
- Takai, K.; Chiyo, N.; Nakajima, T.; Nariai, T.; Ishikawa, C.; Nakatani, S.; Ikeno, A.; Yamamoto, S.; Sone, T. Three-dimensional rational approach to the discovery of potent substituted cyclopropyl urea soluble epoxide hydrolase inhibitors. Bioorg Med. Chem. Lett. 2015, 25, 1705–1708. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Halder, A.K.; Cordeiro, M.N.D.S. Multi-Target In Silico Prediction of Inhibitors for Mitogen-Activated Protein Kinase-Interacting Kinases. Biomolecules 2021, 11, 1670. [Google Scholar] [CrossRef]
- Halder, A.K.; Honarparvar, B. Molecular alteration in drug susceptibility against subtype B and C-SA HIV-1 proteases: MD study. Struct. Chem. 2019, 30, 1715–1727. [Google Scholar] [CrossRef]
- Cheatham, T.E., 3rd; Srinivasan, J.; Case, D.A.; Kollman, P.A. Molecular dynamics and continuum solvent studies of the stability of polyG-polyC and polyA-polyT DNA duplexes in solution. J. Biomol. Struct. Dyn. 1998, 16, 265–280. [Google Scholar] [CrossRef] [PubMed]

| Name | Definition | Class |
|---|---|---|
| ATS6m | Broto-Moreau autocorrelation of lag 6 (log function) weighted by mass | 2D Autocorrelation |
| J_Dz(p) | Balaban-like index from Barysz matrix weighted by polarizability | 2D Matrix-based |
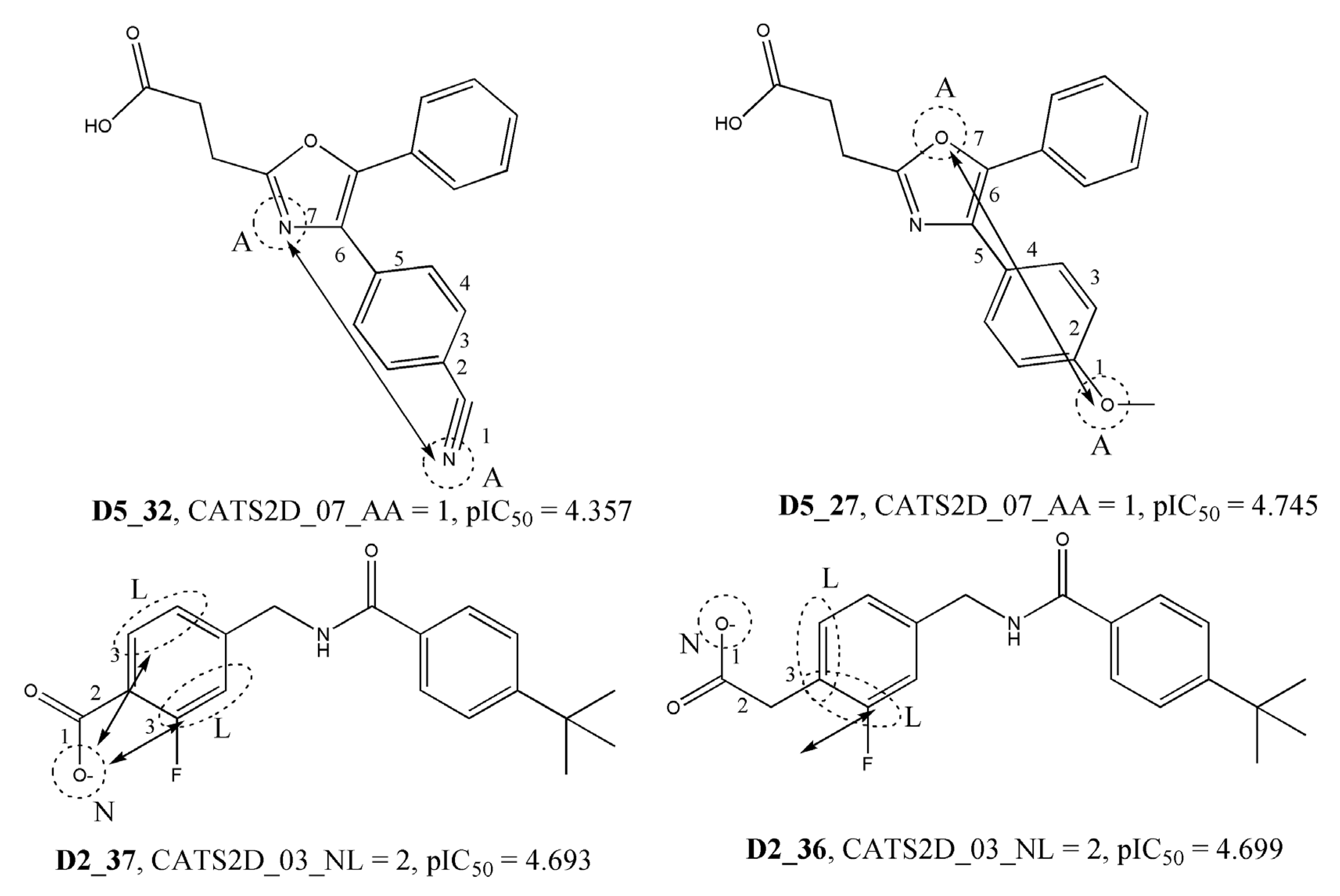
| CATS2D_07_AA | CATS2D Acceptor-Acceptor at lag 07 | 2D Pharmacophore |
| CATS2D_03_NL | CATS2D Negative-Lipophilic at lag 03 | 2D Pharmacophore |
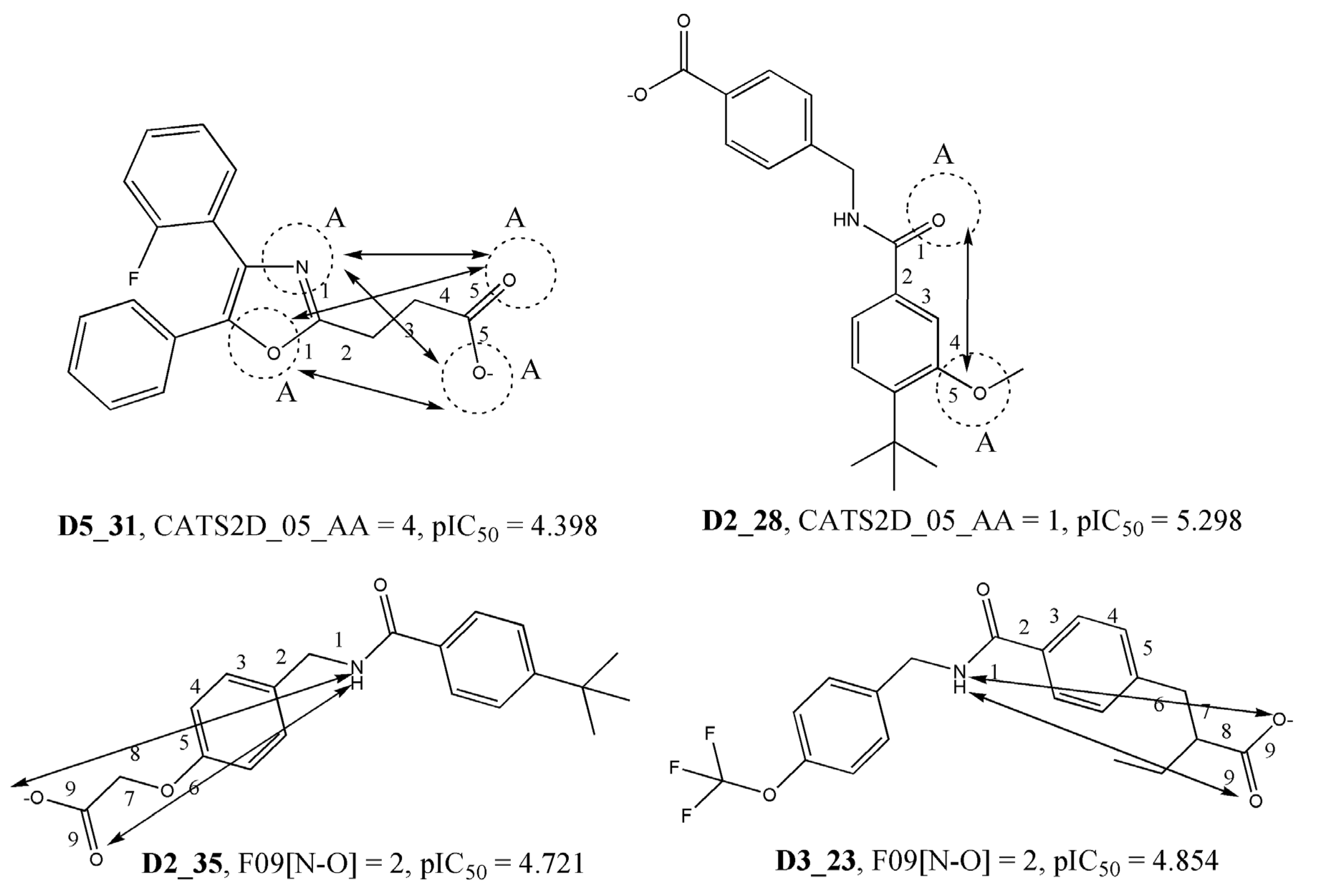
| CATS2D_05_AA | CATS2D Acceptor-Acceptor at lag 05 | 2D Pharmacophore |
| SM14_AEA(dm) | Spectral moment of order 14 from augmented edge adjacency matrix weighted by the dipole moment | Edge adjacency indices |
| F09[N-O] | Frequency of N–O at topological distance 9 | 2D atom-pairs |
| RDF140v | Radial Distribution Function at a distance of 14.0 Å weighted by van der Waals volume | 3D (RDF) |
| Descriptors | ML | Q2LOO (5-fold) | R2Pred | Average | Selected Parameters * |
|---|---|---|---|---|---|
| Linear model | MLP | 0.767 | 0.797 | 0.780 | activation = Identity, solver = Lbfgs, hidden layer Sizes = (5) |
| Linear model | RF | 0.673 | 0.741 | 0.707 | max_depth = 10, max features = Sqrt, min samples leaf = 2 |
| Linear model | SVM | 0.757 | 0.805 | 0.781 | gamma = 1.0, kernel = Linear |
| dSe | MLP | 0.391 | 0.632 | 0.442 | activation = Identity, solver = lbfgs, hidden layer Sizes = (5) |
| dSe | RF | 0.531 | 0.601 | 0.566 | criterion: MAE, maximum depth = 30, max_features = Sqrt, n_estimators = 200 |
| dSe | SVM | 0.405 | 0.626 | 0.516 | C = 100.0, gamma = 1.0, kernel = Linear |
| Parameter | FFD-SEL | UVE-PLS |
|---|---|---|
| Ntraining | 148 | 148 |
| R2 | 0.756 | 0.778 |
| F | 148.89 | 168.68 |
| Q2LOO | 0.615 | 0.643 |
| Q2LTO | 0.614 | 0.643 |
| Q2LMO | 0.603 | 0.631 |
| Ntest | 36 | 36 |
| R2Pred | 0.631 | 0.657 |
| Compound | ΔEvdW | ΔEelec | ΔGpolar | ΔGnonpolar | TΔS | ΔGbind(T) a |
|---|---|---|---|---|---|---|
| S74 | −65.26 | 21.92 | −0.43 | −8.33 | −28.43 | −23.67 |
| D4_02 | −64.85 | −27.18 | 71.00 | −8.25 | −12.43 | −16.85 |
| D2_37 | −42.49 | 53.17 | −32.44 | −5.07 | −27 | 0.17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sar, S.; Mitra, S.; Panda, P.; Mandal, S.C.; Ghosh, N.; Halder, A.K.; Cordeiro, M.N.D.S. In Silico Modeling and Structural Analysis of Soluble Epoxide Hydrolase Inhibitors for Enhanced Therapeutic Design. Molecules 2023, 28, 6379. https://doi.org/10.3390/molecules28176379
Sar S, Mitra S, Panda P, Mandal SC, Ghosh N, Halder AK, Cordeiro MNDS. In Silico Modeling and Structural Analysis of Soluble Epoxide Hydrolase Inhibitors for Enhanced Therapeutic Design. Molecules. 2023; 28(17):6379. https://doi.org/10.3390/molecules28176379
Chicago/Turabian StyleSar, Shuvam, Soumya Mitra, Parthasarathi Panda, Subhash C. Mandal, Nilanjan Ghosh, Amit Kumar Halder, and Maria Natalia D. S. Cordeiro. 2023. "In Silico Modeling and Structural Analysis of Soluble Epoxide Hydrolase Inhibitors for Enhanced Therapeutic Design" Molecules 28, no. 17: 6379. https://doi.org/10.3390/molecules28176379
APA StyleSar, S., Mitra, S., Panda, P., Mandal, S. C., Ghosh, N., Halder, A. K., & Cordeiro, M. N. D. S. (2023). In Silico Modeling and Structural Analysis of Soluble Epoxide Hydrolase Inhibitors for Enhanced Therapeutic Design. Molecules, 28(17), 6379. https://doi.org/10.3390/molecules28176379








