The Impact of AAPH-Induced Oxidation on the Functional and Structural Properties, and Proteomics of Arachin
Abstract
:1. Introduction
2. Results and Discussion
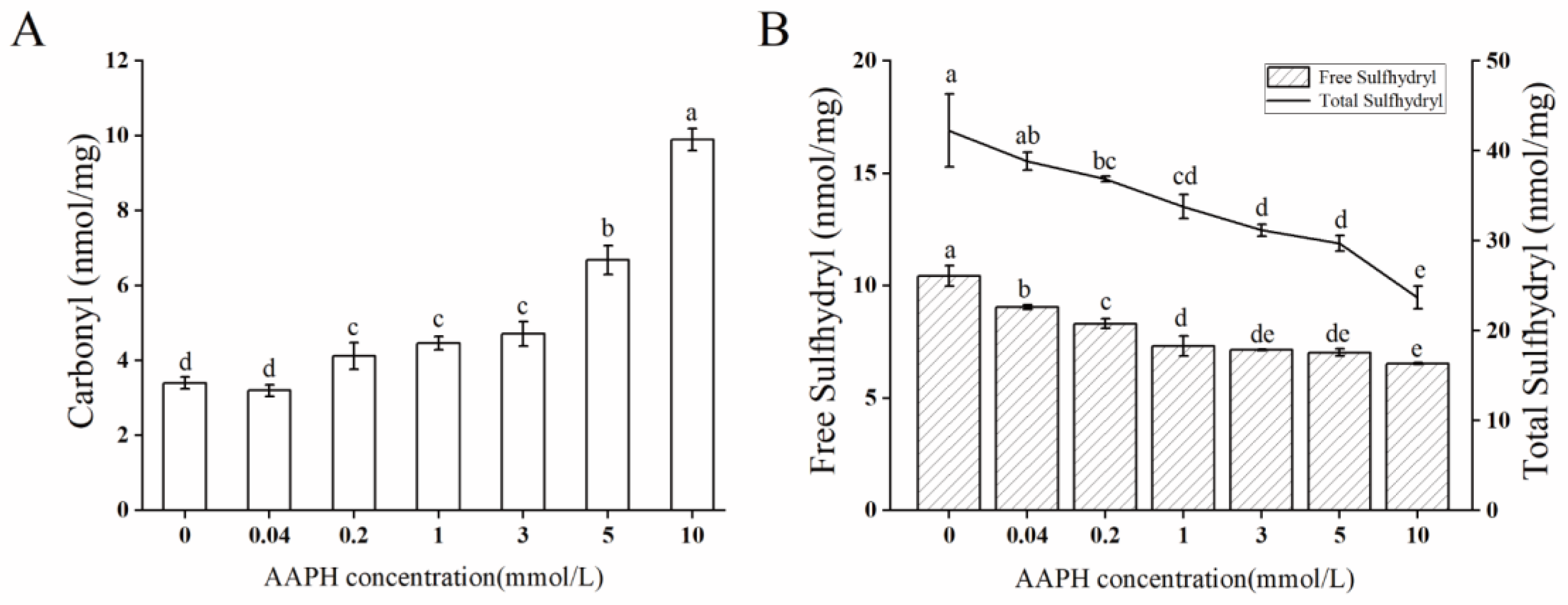
2.1. Oxidation Degree
2.2. Physicochemical Properties
2.3. Denaturation Temperature
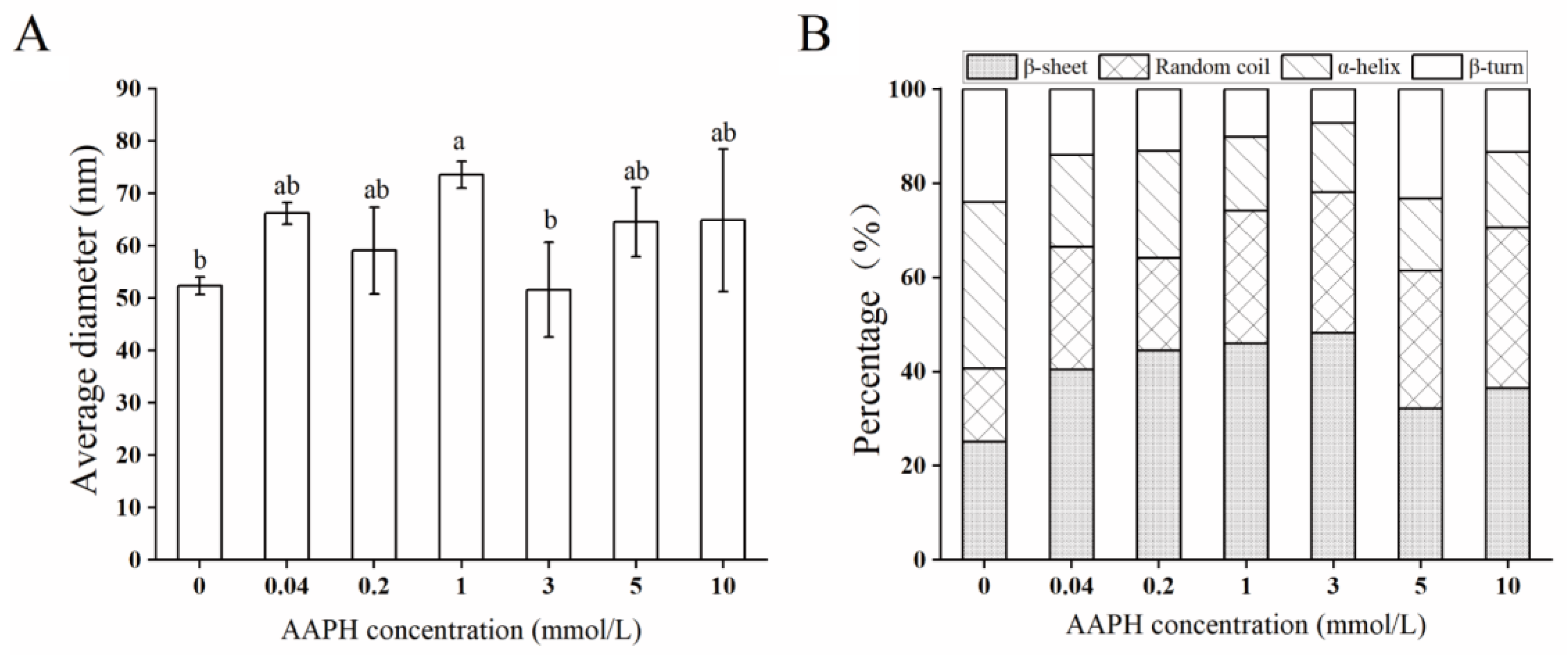
2.4. Structural Characteristics
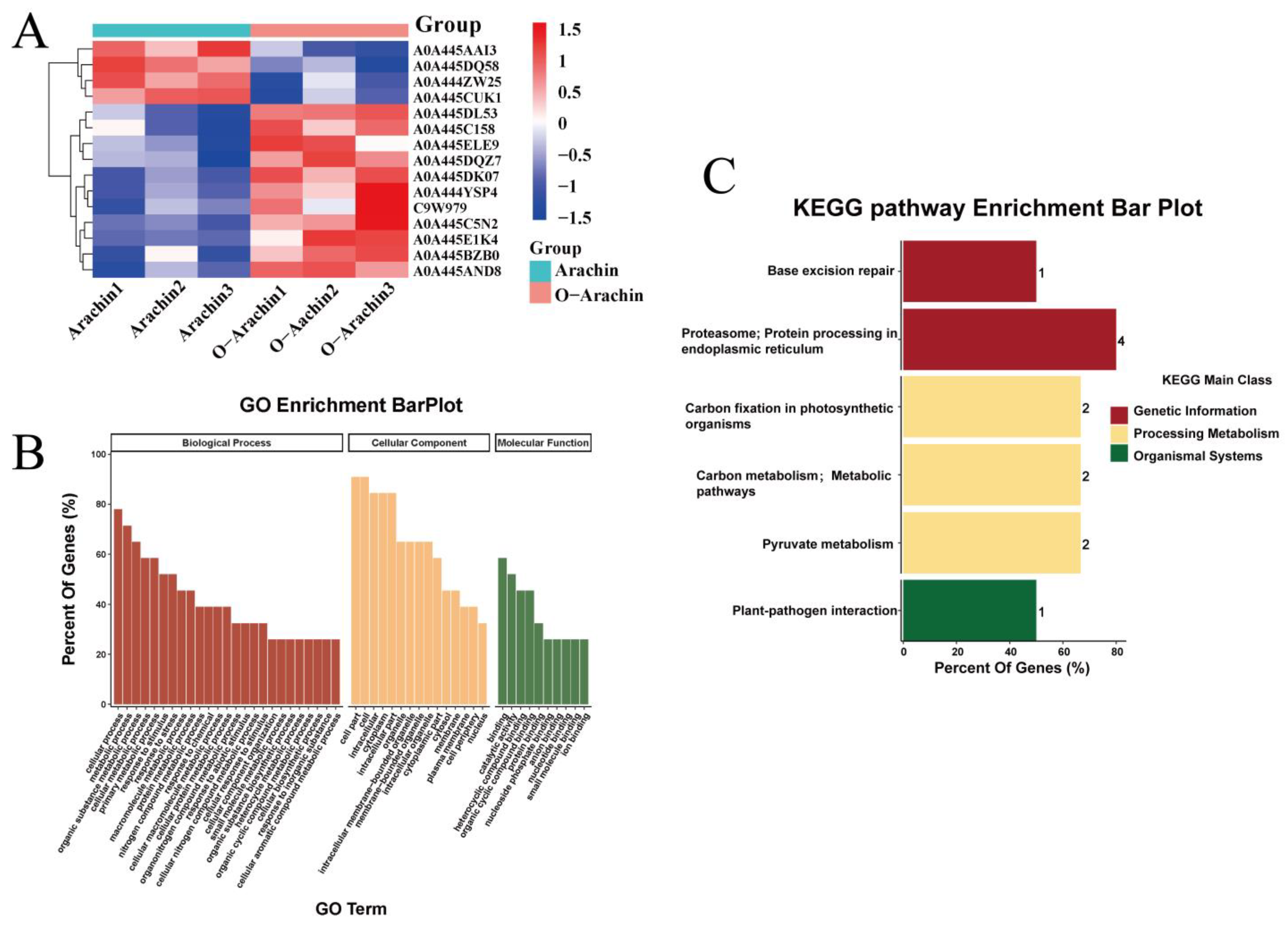
2.5. Proteomic Analysis
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Sample Preparation
4.2.1. Extraction of Arachin
4.2.2. Oxidation of Arachin
4.3. Determination of Carbonyl Content
4.4. Determination of Sulfhydryl Group Content
4.5. Solubility
4.6. Water/Oil Holding Capacity (WHC/OHC)
4.7. Emulsifying Activity Index (EAI) and Emulsifying Stability Index (ESI)
4.8. Particle Size Distributions
4.9. Fourier Transformed Infrared Spectroscopy (FT-IR)
4.10. Scanning Electron Microscope (SEM)
4.11. Determination of Differential Scanning Calorimetry (DSC)
4.12. LC-MS/MS
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Appendix A

References
- Li, S.; Li, Z.; Li, X.; Wang, P.; Yu, X.; Fu, Q.; Gao, S. Effect of AAPH oxidation on digestion characteristics of seed watermelon (Citrullus lanatus var) kernels protein isolates. Food Sci. Hum. Wellness 2020, 9, 402–410. [Google Scholar] [CrossRef]
- Ye, L.; Liao, Y.; Zhao, M.; Sun, W. Effect of Protein Oxidation on the Conformational Properties of Peanut Protein Isolate. J. Chem. 2013, 2013, 423254. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, J.; Zhao, J.; Wu, H.; Ditta, Y.A.; Zhang, J. The Interaction Between Lipoxygenase-Catalyzed Oxidation and Autoxidation in Dry-Cured Bacon and a Model System. J. Food Sci. 2015, 80, C2640–C2646. [Google Scholar] [CrossRef]
- Simion, A.M.C.; Aprodu, I.; Dumitrașcu, L.; Bahrim, G.E.; Alexe, P.; Stănciuc, N. Exploring the heat-induced structural changes of β-lactoglobulin -linoleic acid complex by fluorescence spectroscopy and molecular modeling techniques. J. Food Sci. Technol. 2015, 52, 8095–8103. [Google Scholar] [CrossRef] [PubMed]
- Gürbüz, G.; Heinonen, M. LC-MS investigations on interactions between isolated β-lactoglobulin peptides and lipid oxidation product malondialdehyde. Food Chem. 2015, 175, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Wu, X.J.; Hua, Y. Structural modification of soy protein by the lipid peroxidation product acrolein. LWT Food Sci. Technol. 2010, 43, 133–140. [Google Scholar] [CrossRef]
- Cucu, T.; Devreese, B.; Kerkaert, B.; Mestdagh, F.; Sučić, M.; Perre, I.V.D.; Meulenaer, B.D. A comparative study of lipid and hypochlorous acid induced oxidation of soybean proteins. LWT Food Sci. Technol. 2013, 50, 451–458. [Google Scholar] [CrossRef]
- Wu, W.; Lin, Q.; Hua, Y.; Wu, Y.; Liang, Y.; Fu, X.; Xiao, H. Study on Mechanism of Soy Protein Oxidation Induced by Lipid Peroxidation Products. Adv. J. Food Sci. Technol. 2013, 5, 46–53. [Google Scholar] [CrossRef]
- Hematyar, N.; Rustad, T.; Sampels, S.; Kastrup Dalsgaard, T. Relationship between lipid and protein oxidation in fish. Aquac. Res. 2019, 50, 1393–1403. [Google Scholar] [CrossRef]
- Cheng, Y.; Chi, Y.; Geng, X.; Chi, Y. Effect of 2,2′-azobis(2-amidinopropane) dihydrochloride (AAPH) induced oxidation on the physicochemical properties, in vitro digestibility, and nutritional value of egg white protein. LWT 2021, 143, 111103. [Google Scholar] [CrossRef]
- Gieseg, S.P.; Duggan, S.P.; Gebicki, J.M. Peroxidation of proteins before lipids in U937 cells exposed to peroxyl radicals. Biochem. J. 2000, 350 Pt 1, 215–218. [Google Scholar] [CrossRef]
- Keppler, J.K.; Heyn, T.R.; Meissner, P.M.; Schrader, K.; Schwarz, K. Protein oxidation during temperature-induced amyloid aggregation of beta-lactoglobulin. Food Chem. 2019, 289, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, W.; Zhou, M.; Yi, S.; Ye, B.; Mi, H.; Li, J.; Wang, J.; Li, X. Effect of oxidation modification induced by peroxyl radicals on the physicochemical and gel characteristics of grass carp myofibrillar protein. J. Food Meas. Charact. 2021, 15, 5572–5583. [Google Scholar] [CrossRef]
- Zhao, J.; Su, G.; Chen, C.; Sun, X.; Sun, W.; Zhao, M. Physicochemical and Structural Characteristics of Soybean Protein Isolates Induced by Lipoxygenase-Catalyzed Linoleic Acid Oxidation during In Vitro Gastric Digestion. J. Agric. Food Chem. 2020, 68, 12384–12392. [Google Scholar] [CrossRef]
- Jani, B.L. Peanut Protein: Rich Source As Vegan Protein. Food Sci. Nutr. 2020, 6, 59. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Liu, Y.; Chen, F. Effect of storage temperature on lipid oxidation and changes in nutrient contents in peanuts. Food Sci. Nutr. 2019, 7, 2280–2290. [Google Scholar] [CrossRef]
- Li, W.; Zhang, C.; Xu, N.; Hu, Y.; Wang, C.; Li, D.; Li, W. Effect of lipoxygenase-induced oxidation on molecular structure and digestive properties of arachin and conarachin. J. Food Process. Preserv. 2021, 46, e15874. [Google Scholar] [CrossRef]
- Tian, Y.; Rao, H.; Zhang, K.; Tao, S.; Xue, W. Effects of different thermal processing methods on the structure and allergenicity of peanut allergen Ara h 1. Food Sci. Nutr. 2018, 6, 1706–1714. [Google Scholar] [CrossRef]
- Khorasani, S.; Boroumand, N.; Pasavei, A.G.; Sahebari, M.; Hashemy, S.I. A Study on Association Between Protein Carbonyl and Anti-cyclic Citrullinated Peptide Antibody in Rheumatoid Arthritis: Introducing a New Supplementary Biomarker. Indian J. Clin. Biochem. 2019, 35, 347–352. [Google Scholar] [CrossRef]
- Zhang, X.C.; Yang, X.; LI, Y.Q.; Wang, Z.X.; He, X.M.; Sun, J. Effect of Peroxyl Radical-Induced Oxidation on Functional and Structural Characteristics of Walnut Protein Isolates Revealed by High-Resolution Mass Spectrometry. Foods 2022, 11, 385. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, C.; Kong, X.; Hua, Y. Oxidative modification of soy protein by peroxyl radicals. Food Chem. 2009, 116, 295–301. [Google Scholar] [CrossRef]
- Yu, L.; Yang, W.; Sun, J.; Zhang, C.; Bi, J.; Yang, Q. Preparation, characterisation and physicochemical properties of the phosphate modified peanut protein obtained from Arachin conarachin L. Food Chem. 2015, 170, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Li, H.; Wang, Z.; Emara, A.M.; Hu, Y.; He, Z. Effects of in vitro oxidation on myofibrillar protein charge, aggregation, and structural characteristics. Food Chem. 2020, 332, 127396. [Google Scholar] [CrossRef]
- Zhang, H.J.; Zhang, H.; Wang, L.; Guo, X.N. Preparation and functional properties of rice bran proteins from heat-stabilized defatted rice bran. Food Res. Int. 2012, 47, 359–363. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, X.; Zhong, J.; Tan, L.-h.; Liu, C. Effect of pH on emulsification performance of a new functional protein from jackfruit seeds. Food Hydrocoll. 2019, 93, 325–334. [Google Scholar] [CrossRef]
- Srinivasan, S.; Hultin, H.O. Chemical, Physical, and Functional Properties of Cod Proteins Modified by a Nonenzymic Free-Radical-Generating System. J. Agric. Food Chem. 1997, 45, 310–320. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, G.; Zhao, M.; Ren, J.; Yang, B. Improvement of functional properties of peanut protein isolate by conjugation with dextran through Maillard reaction. Food Chem. 2012, 131, 901–906. [Google Scholar] [CrossRef]
- Colombo, A.; Ribotta, P.D.; Leon, A.E. Differential scanning calorimetry (DSC) studies on the thermal properties of peanut proteins. J. Agric. Food Chem. 2010, 58, 4434–4439. [Google Scholar] [CrossRef]
- Tang, C.-H.; Sun, X.; Foegeding, E.A. Modulation of physicochemical and conformational properties of kidney bean vicilin (phaseolin) by glycation with glucose: Implications for structure-function relationships of legume vicilins. J. Agric. Food Chem. 2011, 59, 10114–10123. [Google Scholar] [CrossRef]
- Shen, H.R.; Stephen Elmore, J.; Zhao, M.; Sun, W. Effect of oxidation on the gel properties of porcine myofibrillar proteins and their binding abilities with selected flavour compounds. Food Chem. 2020, 329, 127032. [Google Scholar] [CrossRef]
- Li, X.; Liu, C.; Wang, J.; Li, W.; Lin, B.; Zhu, W.; Xu, Y.; Yi, S.; Mi, H.; Li, J. Tea Polyphenols Affect Oxidative Modification and Solution Stability of Myofibrillar Protein from Grass Carp (Ctenopharyngodon idellus). Food Biophys. 2020, 15, 397–408. [Google Scholar] [CrossRef]
- Huang, Y.; Hua, Y.; Qiu, A. Soybean protein aggregation induced by lipoxygenase catalyzed linoleic acid oxidation. Food Res. Int. 2006, 39, 240–249. [Google Scholar] [CrossRef]
- Wu, W.; Li, F.; Wu, X.J. Effects of rice bran rancidity on oxidation, structural characteristics and interfacial properties of rice bran globulin. Food Hydrocoll. 2021, 110, 106123. [Google Scholar] [CrossRef]
- Sun, W.; Zhou, F.; Sun, D.W.; Zhao, M. Effect of Oxidation on the Emulsifying Properties of Myofibrillar Proteins. Food Bioprocess Technol. 2012, 6, 1703–1712. [Google Scholar] [CrossRef]
- Shi, Y.; Carroll, K.S. Activity-Based Sensing for Site-Specific Proteomic Analysis of Cysteine Oxidation. Acc. Chem. Res. 2020, 53, 20–31. [Google Scholar] [CrossRef]
- Mergner, J.; Frejno, M.; List, M.; Papacek, M.; Chen, X.; Chaudhary, A.; Samaras, P.; Richter, S.; Shikata, H.; Messerer, M.; et al. Mass-spectrometry-based draft of the Arabidopsis proteome. Nature 2020, 579, 409–414. [Google Scholar] [CrossRef]
- Wang, H.; Wang, S.; Lu, Y.; Alvarez, S.; Hicks, L.M.; Ge, X.; Xia, Y. Proteomic analysis of early-responsive redox-sensitive proteins in Arabidopsis. J. Proteome. Res. 2012, 11, 412–424. [Google Scholar] [CrossRef]
- Duque-Estrada, P.; Berton-Carabin, C.C.; Nieuwkoop, M.; Dekkers, B.L.; Janssen, A.E.M.; van der Goot, A.J. Protein Oxidation and In Vitro Gastric Digestion of Processed Soy-Based Matrices. J. Agric. Food Chem. 2019, 67, 9591–9600. [Google Scholar] [CrossRef]
- Zhang, Z.; Xiong, Z.; Lu, S.; Walayat, N.; Hu, C.; Xiong, H. Effects of oxidative modification on the functional, conformational and gelling properties of myofibrillar proteins from Culter alburnus. Int. J. Biol. Macromol. 2020, 162, 1442–1452. [Google Scholar] [CrossRef]
- Lv, L.; Lin, H.; Li, Z.; Wang, J.; Ahmed, I.; Chen, H. Changes of structure and IgE binding capacity of shrimp (Metapenaeus ensis) tropomyosin followed by acrolein treatment. Food Funct. 2017, 8, 1028–1036. [Google Scholar] [CrossRef]
- Xu, J.; Chen, Z.; Han, D.-Y.; Li, Y.; Sun, X.; Wang, Z.; Jin, H. Structural and Functional Properties Changes of β-Conglycinin Exposed to Hydroxyl Radical-Generating Systems. Mol. J. Synth. Chem. Nat. Prod. Chem. 2017, 22, 1893. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhou, F.; Peng, W.; Zheng, J.; Dziugan, P.; Zhang, B. The effects of carrageenan on stability of arachin and the interactions between them. Food Hydrocoll. 2015, 43, 763–768. [Google Scholar] [CrossRef]
- Zhang, J.-J.; Tu, Z.C.; Wang, H.; Hu, Y.M.; Du, P.-C.; Yang, Y. Mechanism of the effect of 2, 2'-azobis (2-amidinopropane) dihydrochloride simulated lipid oxidation on the IgG/IgE binding ability of ovalbumin. Food Chem. 2020, 327, 127037. [Google Scholar] [CrossRef]
- Abbou, A.; Kadri, N.; Dahmoune, F.; Chergui, A.; Remini, H.; Berkani, F.; Adel, K.; Boukhalfa, F.; Madani, K. Optimising functional properties and chemical composition of Pinus halepensis Mill. Seeds protein concentrates. Food Hydrocoll. 2020, 100, 105416. [Google Scholar] [CrossRef]
- Yan, C.; Zhou, Z. Solubility and emulsifying properties of phosphorylated walnut protein isolate extracted by sodium trimetaphosphate. LWT Food Sci. Technol. 2021, 143, 111117. [Google Scholar] [CrossRef]
- Duan, X.; Li, M.; Shao, J.; Chen, H.; Xu, X.; Jin, Z.; Liu, X. Effect of oxidative modification on structural and foaming properties of egg white protein. Food Hydrocoll. 2018, 75, 223–228. [Google Scholar] [CrossRef]
- Liu, R.; Shi, C.; Song, Y.; Wu, T.; Zhang, M. Impact of oligomeric procyanidins on wheat gluten microstructure and physicochemical properties. Food Chem. 2018, 260, 37–43. [Google Scholar] [CrossRef]
- Zahari, I.; Ferawati, F.; Helstad, A.; Ahlström, C.; Östbring, K.; Rayner, M.; Purhagen, J.K. Development of High-Moisture Meat Analogues with Hemp and Soy Protein Using Extrusion Cooking. Foods 2020, 9, 772. [Google Scholar] [CrossRef]
- Xu, M.; Luo, J.; Li, Y.; Shen, L.-L.; Zhang, X.; Yu, J.; Guo, Z.; Wu, J.; Chi, Y.; Yang, J. First comprehensive proteomics analysis of lysine crotonylation in leaves of peanut (Arachis hypogaea L.). Proteomics 2020, 21, e2000156. [Google Scholar] [CrossRef]


| Properties | AAPH (mmol/L) | |||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 0.04 | 0.2 | 1 | 3 | 5 | 10 | ||
| Functional | Solubility (%) | 94 ± 4 a | 89 ± 7 ab | 84 ± 7 b | 71 ± 2 c | 0.68 ± 0.05 c | 0.66 ± 0.02 c | 0.65 ± 0.01 c |
| WHC (g/g) | 1.73 ± 0.08 d | 1.67 ± 0.11 d | 1.79 ± 0.07 d | 2.39 ± 0.02 c | 2.98 ± 0.16 a | 2.47 ± 0.19 bc | 2.81 ± 0.31 ab | |
| OHC (g/g) | 7.81 ± 0.21 a | 8.09 ± 0.99 a | 7.98 ± 1.03 a | 7.24 ± 0.36 a | 8.25 ± 0.53 a | 6.82 ± 0.15 ab | 5.50 ± 0.48 b | |
| EAI (m2/g) | 0.13 ± 0.01 d | 0.14 ± 0.01 d | 0.18 ± 0.00 cd | 0.26 ± 0.02 abc | 0.25 ± 0.09 bc | 0.28 ± 0.02 ab | 0.35 ± 0.00 a | |
| ESI (min) | 12.411 ± 0.533 c | 12.631 ± 0.800 c | 12.994 ± 1.004 bc | 15.823 ± 0.133 abc | 20.093 ± 3.989 a | 17.365 ± 2.683 ab | 15.077 ± 0.601 bc | |
| Thermographic | Onset denaturation temperature (To/°C) | 39.05 ± 0.212 | 39.45 ± 1.485 | 41.40 ± 0.707 | 43.53 ± 1.127 | 37.97 ± 0.321 | 38.15 ± 1.909 | 35.25 ± 0.778 |
| Denaturation temperature (Td/°C) | 64.37 ± 3.000 | 76.70 ± 4.952 | 76.60 ± 1.300 | 73.70 ± 7.862 | 60.9 ± 3.818 | 62.83 ± 4.761 | 68.90 ± 3.111 | |
| Maximum denaturation temperature (Tm/°C) | 102.80 ± 2.179 | 114.07 ± 3.166 | 105.65 ± 4.596 | 109.13 ± 5.498 | 100.25 ± 1.061 | 101.85 ± 5.445 | 105.10 ± 1.838 | |
| Denaturation enthalpy (∆H∙J−1∙g−1) | 101.21 ± 11.158 | 105.57 ± 8.393 | 91.17 ± 5.616 | 102.16 ± 23.967 | 81.14 ± 1.308 | 94.43 ± 11.413 | 95.62 ± 3.500 | |
| Amino Acid | Sequence | Modification type | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbamidomethyl | Oxidation | Deoxidation | Dehydroalanine | Dehydration | Trioxidation | HNE | Kynurenin | Hydroxykynurenine | GGS | Carboxyethylation | Carbonylation | Acetyl | AAS | AAA | MDA | ||
| Cysteine | NAVMAPHYNLNCHAVIYGTEGR | √ | |||||||||||||||
| Cysteine | ANLRPCEEHIRQRVEQEQEQEQDEYPYIQR | √ | |||||||||||||||
| Cysteine | NSDCQPCCEGFFCPPGLTCMIPCPLGAYCPR | √ | |||||||||||||||
| Cysteine | EEGALLCLHCCHLFAFCCRR | √ | |||||||||||||||
| Cysteine | QCPNELRVSNR | √ | |||||||||||||||
| Methionine | GAPIIAEYLGGAVNCDAYHMTDPR | √ | |||||||||||||||
| Methionine | EDECDICLEPCTKMVLPNCCHAMCIR | √ | |||||||||||||||
| Proline | CCVSFSAFFNESVVPCQTCACGCSAKPER | √ | |||||||||||||||
| Proline | IVPIAER | √ | |||||||||||||||
| Phenylalanine | CCVSFSAFFNESVVPCQTCACGCSAKPER | √ | |||||||||||||||
| Phenylalanine | CMCEALQQIMENQCDRLQDRQMVQQFK | √ | |||||||||||||||
| Phenylalanine | EEGALLCLHCCHLFAFCCRR | √ | |||||||||||||||
| Histidine | CNAPCLRCNGHDQRCMAPCLR | √ | |||||||||||||||
| Histidine | MCAFCISLCSAEFCSTPPCSSAPLCSSPR | √ | |||||||||||||||
| Histidine | CFFLSFVGCHLFYFCCHAIICPSGLAAFESCPITR | √ | |||||||||||||||
| Histidine | CQGMTLENVSINFSGSFGVGMVYTALSR | √ | |||||||||||||||
| Lysine | CKCRSIGGSCGPSCGCK | √ | |||||||||||||||
| Lysine | AYEELHQEDLIK | √ | |||||||||||||||
| Lysine | CVKELFHFEEDSGGIIK | √ | |||||||||||||||
| Lysine | CCAKCLCVPPGYYGNK | √ | |||||||||||||||
| Lysine | ALCKSNETIDVALQIFREMPNR | √ | |||||||||||||||
| Lysine | CACCGATSTKSSVSK | √ | |||||||||||||||
| Lysine | KGSEEGDITNPINLR | √ | |||||||||||||||
| Tyrosine | DDVELVAVNDPFITTDYMTYMFK | √ | |||||||||||||||
| Tyrosine | NENQNLVLAYLFIPLTTPGDYK | √ | |||||||||||||||
| Tyrosine | YSLKPLVPRLTELLGVDVK | √ | |||||||||||||||
| Tyrosine | YFLMLAILNIVFLLGFLVYYYK | √ | |||||||||||||||
| Tryptophan | WDAPSRGDDQCQR | √ | |||||||||||||||
| Tryptophan | EWLDELK | √ | |||||||||||||||
| Tryptophan | EREEDWRQPR | √ | |||||||||||||||
| Tryptophan | IKYIETWNPNNQEFQCAGVALSR | √ | |||||||||||||||
| Tryptophan | MCCCSWCTCISNWIKMEGSSGSAWQK | √ | |||||||||||||||
| Arginine | RRPTEEIEER | √ | |||||||||||||||
| Arginine | SLPYSPYSPHSRPR | √ | |||||||||||||||
| Arginine | RCNLSPETICHCLFFCQK | √ | |||||||||||||||
| Arginine | REEEEEEEEER | √ | |||||||||||||||
| Serine | SSSSTTFTCCFQR | √ | |||||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, M.; Yang, X.; Wang, Z.; Sha, X.; Zhang, X.; Sun, J. The Impact of AAPH-Induced Oxidation on the Functional and Structural Properties, and Proteomics of Arachin. Molecules 2023, 28, 6277. https://doi.org/10.3390/molecules28176277
Shen M, Yang X, Wang Z, Sha X, Zhang X, Sun J. The Impact of AAPH-Induced Oxidation on the Functional and Structural Properties, and Proteomics of Arachin. Molecules. 2023; 28(17):6277. https://doi.org/10.3390/molecules28176277
Chicago/Turabian StyleShen, Mingjuan, Xi Yang, Zhenxing Wang, Xiaomei Sha, Xuechun Zhang, and Jian Sun. 2023. "The Impact of AAPH-Induced Oxidation on the Functional and Structural Properties, and Proteomics of Arachin" Molecules 28, no. 17: 6277. https://doi.org/10.3390/molecules28176277
APA StyleShen, M., Yang, X., Wang, Z., Sha, X., Zhang, X., & Sun, J. (2023). The Impact of AAPH-Induced Oxidation on the Functional and Structural Properties, and Proteomics of Arachin. Molecules, 28(17), 6277. https://doi.org/10.3390/molecules28176277









