Marine Natural Compound (Neviotin A) Displays Anticancer Efficacy by Triggering Transcriptomic Alterations and Cell Death in MCF-7 Cells
Abstract
:1. Introduction
2. Results
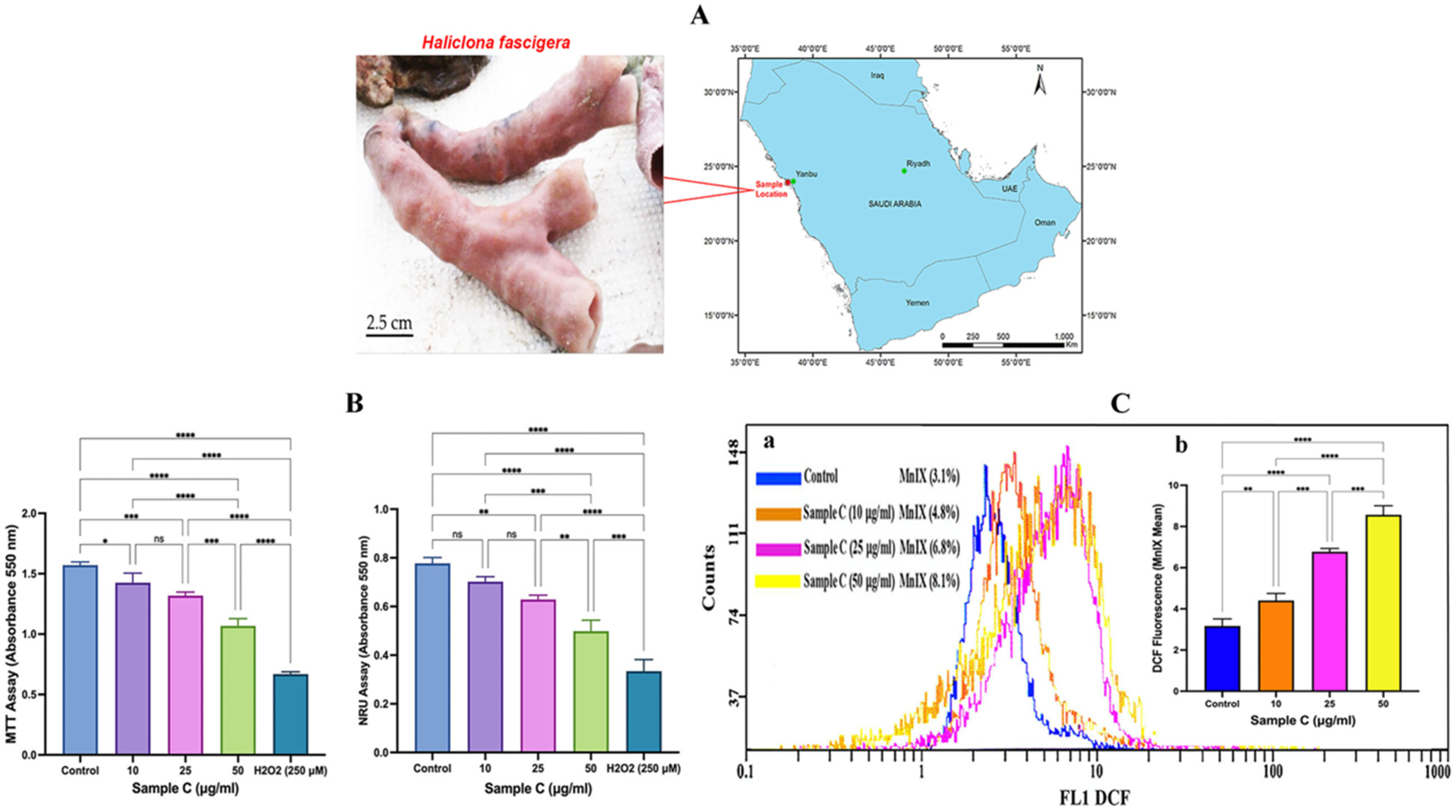
2.1. Cell Viability Analysis
2.2. Intracellular ROS generation
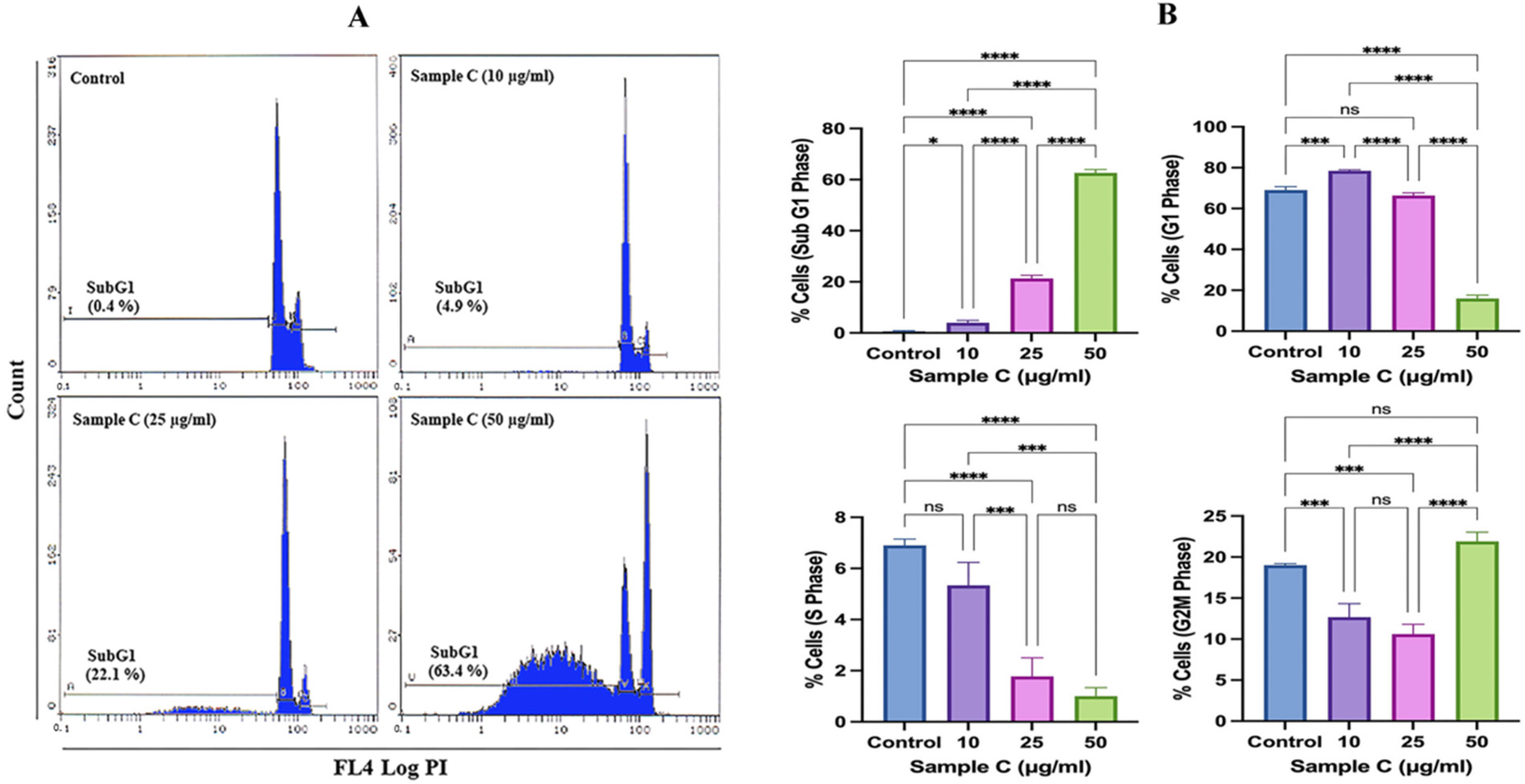
2.3. Cell Cycle Deregulation by Sample C
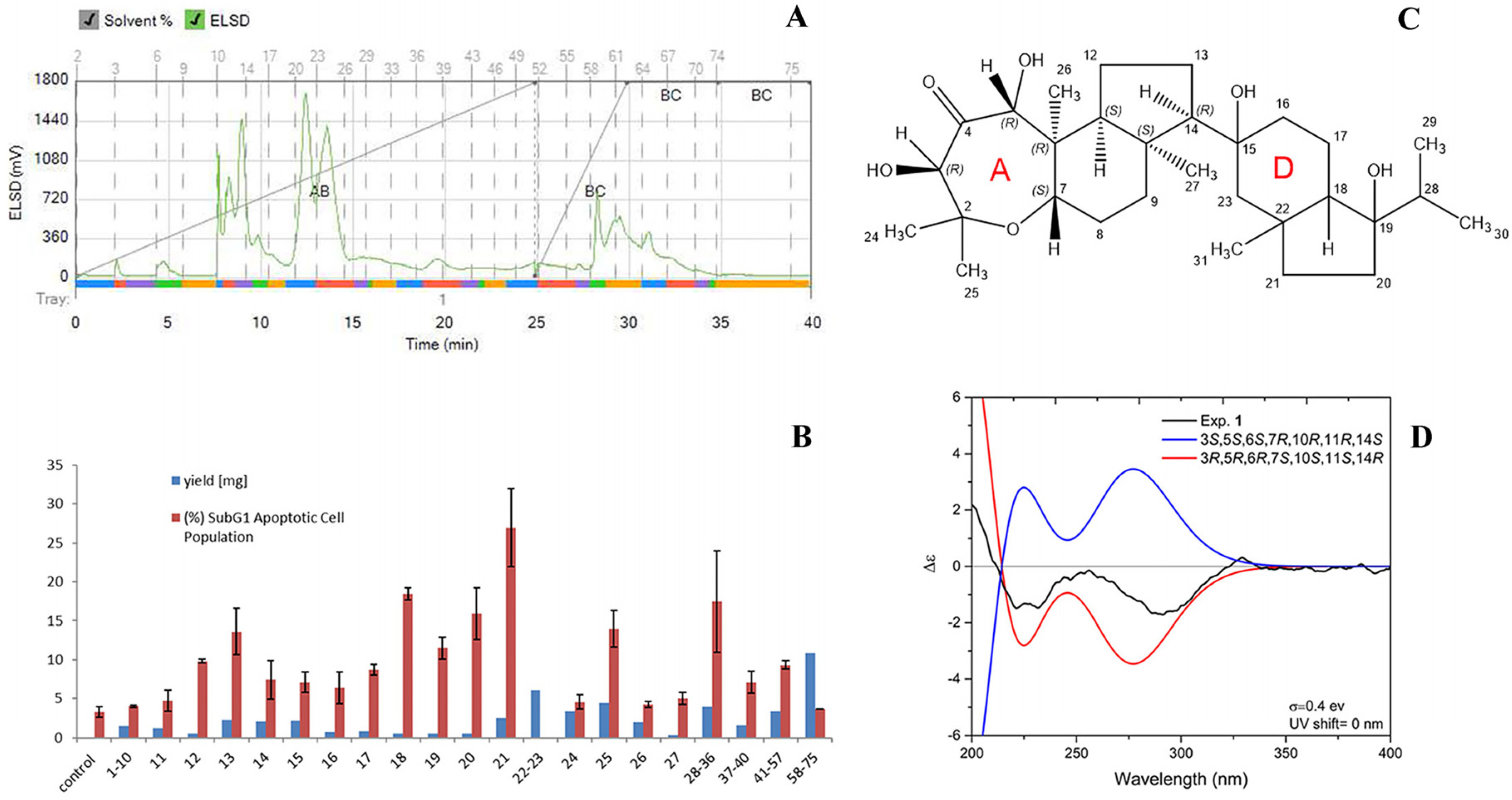
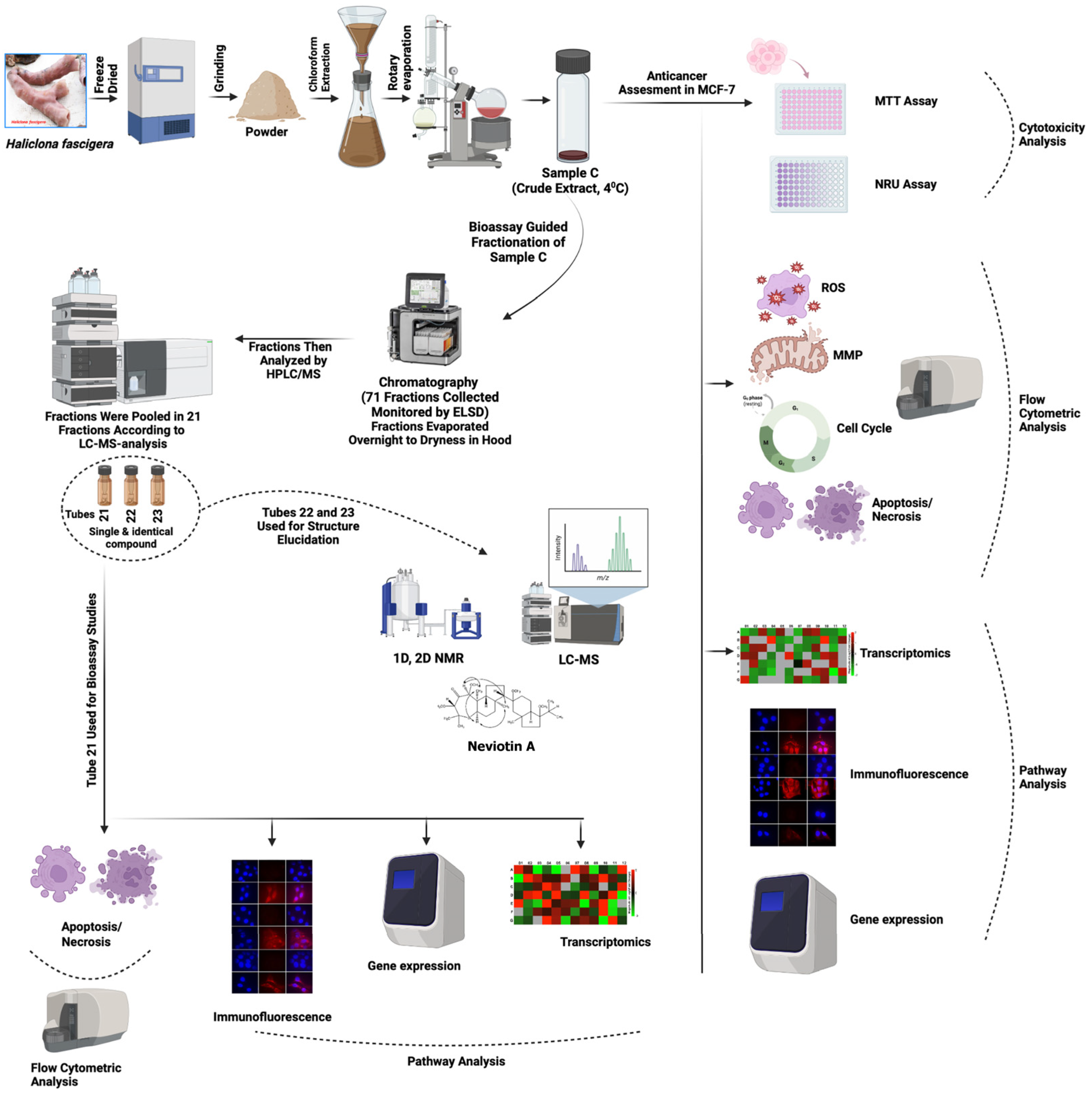
2.4. Bioassay-Guided Fractionation of Sample C
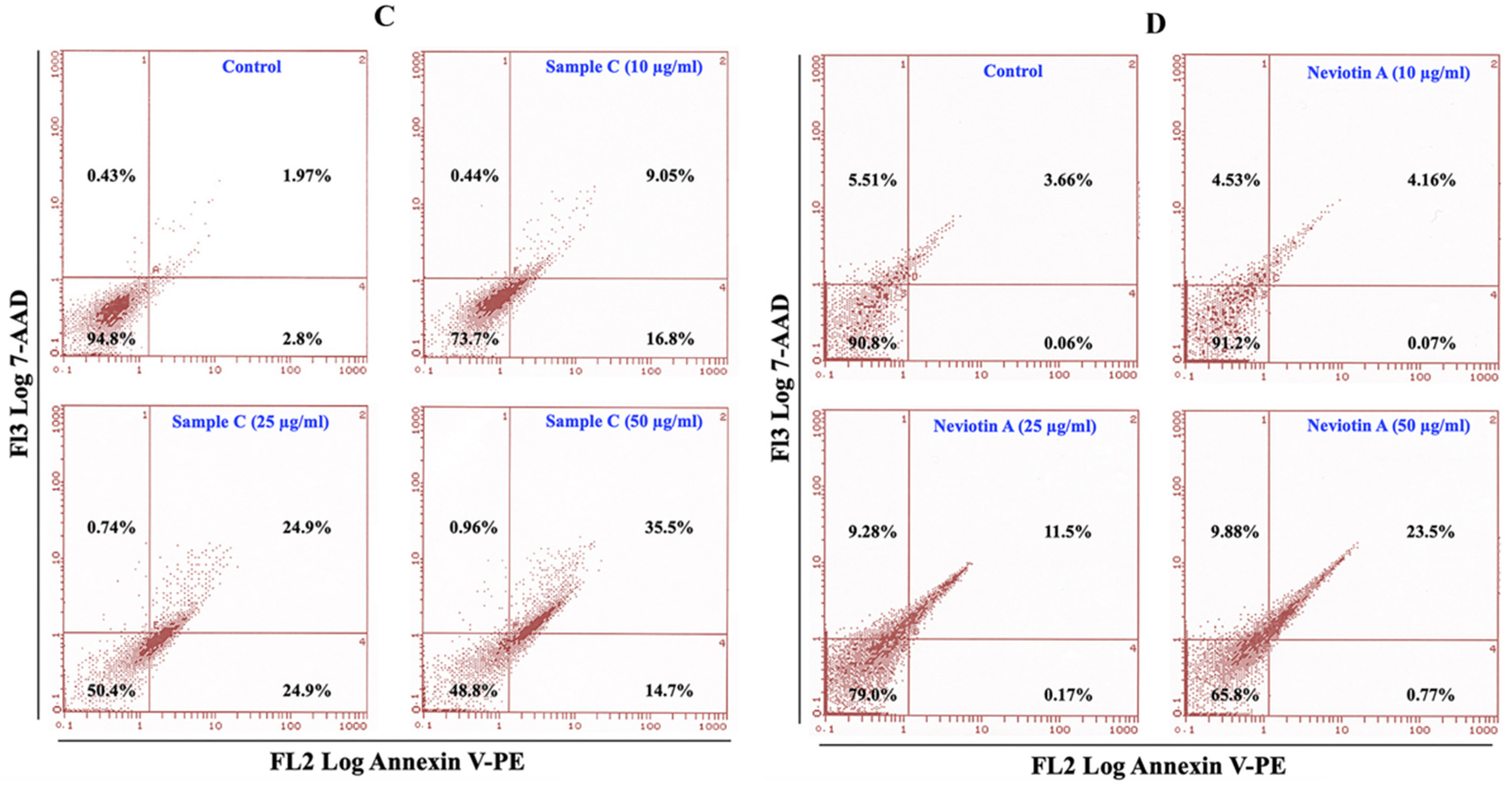
2.5. Effect of Sample C and Neviotin A on the Apoptotic Pathway
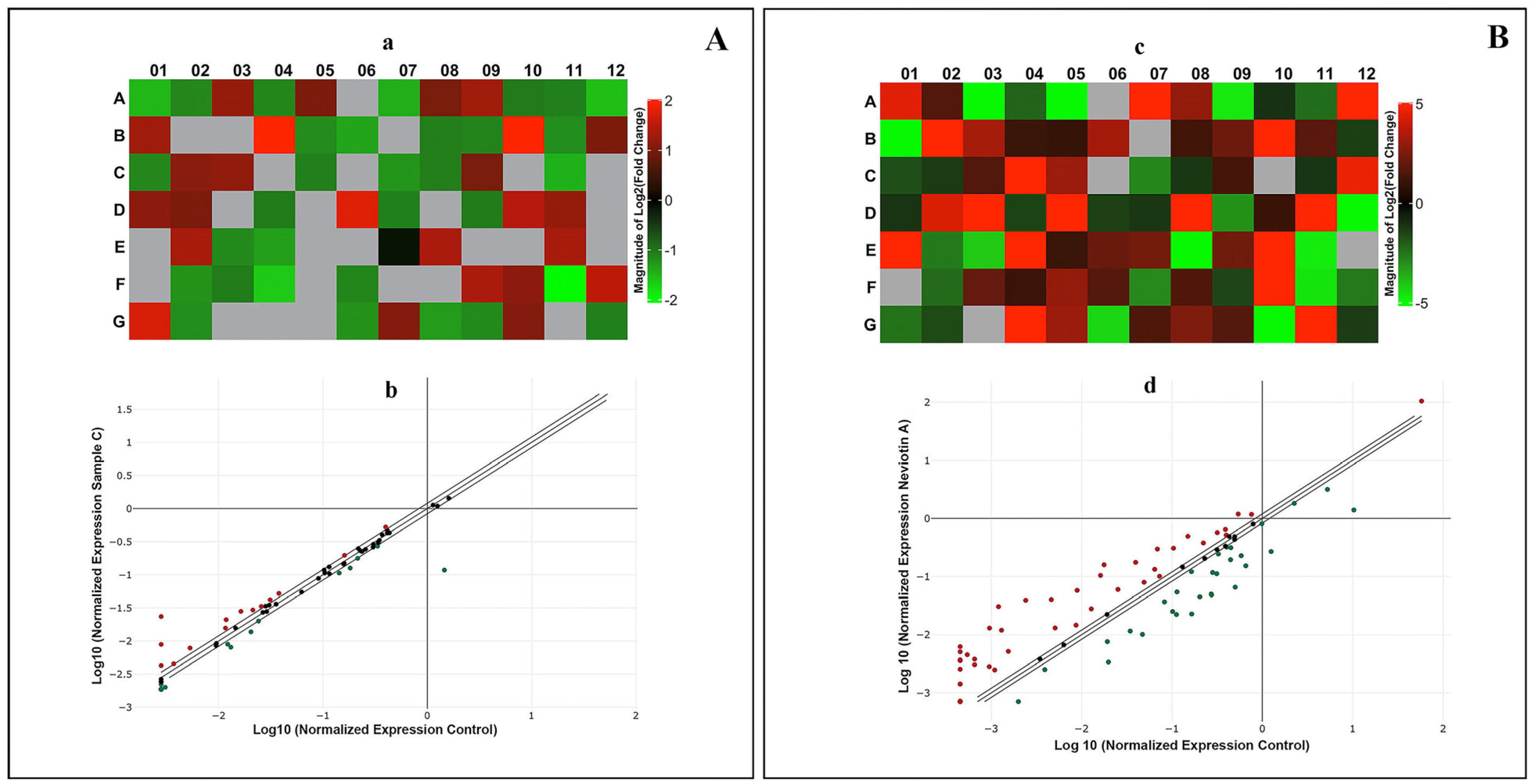
2.6. Translational and Transcriptional Activation of Apoptotic Proteins
3. Discussion
4. Materials and Methods
4.1. General
4.2. Collection of Sponge
4.3. Extract Preparation and Activity-Guided Isolation
4.4. Cell Culture and Cell Proliferation Assays
4.5. Flow Cytometric Quantitation of ROS
4.6. Cell Cycle Dysregulation
4.7. Apoptosis and Necrosis Analysis
4.8. Transcriptome Analysis by qPCR Array
4.9. Immunofluorescence Analysis
4.10. Transcriptional Upregulation of Apoptotic Genes
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Sarfaraj, H.M.; Sheeba, F.; Saba, A.; Khan, M. Marine natural products: A lead for Anti-cancer. Indian J. Geo-Mar. Sci. 2012, 41, 27–39. [Google Scholar]
- Van Minh, C.; Van Kiem, P.; Dang, N.H. Marine natural products and their potential application in the future. ASEAN J. Sci. Technol. Dev. 2005, 22, 297–311. [Google Scholar] [CrossRef]
- Burgess, J.G. New and emerging analytical techniques for marine biotechnology. Curr. Opin. Biotechnol. 2012, 23, 29–33. [Google Scholar] [CrossRef]
- Williams, D.H.; Stone, M.J.; Hauck, P.R.; Rahman, S.K. Why Are Secondary Metabolites (Natural Products) Biosynthesized? J. Nat. Prod. 1989, 52, 1189–1208. [Google Scholar] [CrossRef]
- Firn, R.D.; Jones, C.G. Natural products—A simple model to explain chemical diversity. Nat. Prod. Rep. 2003, 20, 382–391. [Google Scholar] [CrossRef]
- Hussain, A.; Bourguet-Kondracki, M.-L.; Majeed, M.; Ibrahim, M.; Imran, M.; Yang, X.-W.; Ahmed, I.; Altaf, A.A.; Khalil, A.A.; Rauf, A.; et al. Marine life as a source for breast cancer treatment: A comprehensive review. Biomed. Pharmacother. 2023, 159, 114165. [Google Scholar] [CrossRef]
- Lyu, C.; Chen, T.; Qiang, B.; Liu, N.; Wang, H.; Zhang, L.; Liu, Z. CMNPD: A comprehensive marine natural products database towards facilitating drug discovery from the ocean. Nucleic Acids Res. 2021, 49, D509–D515. [Google Scholar] [CrossRef]
- Ahmed, S.; Mirzaei, H.; Aschner, M.; Khan, A.; Al-Harrasi, A.; Khan, H. Marine peptides in breast cancer: Therapeutic and mechanistic understanding. Biomed. Pharmacother. 2021, 142, 112038. [Google Scholar] [CrossRef]
- Zhang, Q.-T.; Liu, Z.-D.; Wang, Z.; Wang, T.; Wang, N.; Wang, N.; Zhang, B.; Zhao, Y.-F. Recent Advances in Small Peptides of Marine Origin in Cancer Therapy. Mar. Drugs 2021, 19, 115. [Google Scholar] [CrossRef]
- Su, B.-C.; Hung, G.-Y.; Tu, Y.-C.; Yeh, W.-C.; Lin, M.-C.; Chen, J.-Y. Marine Antimicrobial Peptide TP4 Exerts Anticancer Effects on Human Synovial Sarcoma Cells via Calcium Overload, Reactive Oxygen Species Production and Mitochondrial Hyperpolarization. Mar. Drugs 2021, 19, 93. [Google Scholar] [CrossRef]
- Hsu, H.-C.; Chen, M.-H.; Yeh, M.-L.; Chen, W.-J. Antibacterial and Anticancer Activities of Pleurocidin-Amide, a Potent Marine Antimicrobial Peptide Derived from Winter Flounder, Pleuronectes americanus. Mar. Drugs 2022, 20, 519. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, B.; Sachdeva, P.; Negi, A.; Ghosh, S.; Han, S.; Dewanjee, S.; Jha, S.K.; Bhaskar, R.; Sinha, J.K.; Paiva-Santos, A.C.; et al. Chitosan Nanoparticles-Based Cancer Drug Delivery: Application and Challenges. Mar. Drugs 2023, 21, 211. [Google Scholar] [CrossRef] [PubMed]
- González-Ballesteros, N.; Maietta, I.; Rey-Méndez, R.; Rodríguez-Argüelles, M.C.; Lastra-Valdor, M.; Cavazza, A.; Grimaldi, M.; Bigi, F.; Simón-Vázquez, R. Gold Nanoparticles Synthesized by an Aqueous Extract of Codium tomentosum as Potential Antitumoral Enhancers of Gemcitabine. Mar. Drugs 2023, 21, 20. [Google Scholar] [CrossRef] [PubMed]
- Kruk, K.; Winnicka, K. Alginates Combined with Natural Polymers as Valuable Drug Delivery Platforms. Mar. Drugs 2023, 21, 11. [Google Scholar] [CrossRef]
- American Cancer Society. Cancer Facts & Figures 2023. American Cancer Society, 2023. Available online: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2023/2023-cancer-facts-and-figures.pdf (accessed on 22 August 2023).
- Ghareeb, M.A.; Tammam, M.A.; El-Demerdash, A.; Atanasov, A.G. Insights about clinically approved and Preclinically investigated marine natural products. Curr. Res. Biotechnol. 2020, 2, 88–102. [Google Scholar] [CrossRef]
- Wang, E.; Sorolla, M.A.; Gopal Krishnan, P.D.; Sorolla, A. From Seabed to Bedside: A Review on Promising Marine Anticancer Compounds. Biomolecules 2020, 10, 248. [Google Scholar] [CrossRef]
- Faulkner, D.J. Marine natural products. Nat. Prod. Rep. 2002, 19, 1–49. [Google Scholar] [CrossRef]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2017, 34, 235–294. [Google Scholar] [CrossRef]
- Sipkema, D.; Franssen, M.C.R.; Osinga, R.; Tramper, J.; Wijffels, R.H. Marine sponges as pharmacy. Mar. Biotechnol. 2005, 7, 142–162. [Google Scholar] [CrossRef]
- Chianese, G.; Yu, H.-B.; Yang, F.; Sirignano, C.; Luciano, P.; Han, B.-N.; Khan, S.; Lin, H.-W.; Taglialatela-Scafati, O. PPAR Modulating Polyketides from a Chinese Plakortis simplex and Clues on the Origin of Their Chemodiversity. J. Org. Chem. 2016, 81, 5135–5143. [Google Scholar] [CrossRef]
- Lai, K.-H.; Liu, Y.-C.; Su, J.-H.; El-Shazly, M.; Wu, C.-F.; Du, Y.-C.; Hsu, Y.-M.; Yang, J.-C.; Weng, M.-K.; Chou, C.-H.; et al. Antileukemic Scalarane Sesterterpenoids and Meroditerpenoid from Carteriospongia (Phyllospongia) sp., Induce Apoptosis via Dual Inhibitory Effects on Topoisomerase II and Hsp90. Sci. Rep. 2016, 6, 36170. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Tietjen, I.; Chen, M.; Williams, D.E.; Daoust, J.; Brockman, M.A.; Andersen, R.J. Sesterterpenoids Isolated from the Sponge Phorbas sp. Activate Latent HIV-1 Provirus Expression. J. Org. Chem. 2016, 81, 11324–11334. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, M.; Konno, H. Cytotoxic evaluation of natural and synthetic callipeltins: A revision of cytotoxicity of callipeltin B. Biosci. Biotechnol. Biochem. 2016, 80, 1066–1069. [Google Scholar] [CrossRef] [PubMed]
- Goey, A.K.L.; Chau, C.H.; Sissung, T.M.; Cook, K.M.; Venzon, D.J.; Castro, A.; Ransom, T.R.; Henrich, C.J.; McKee, T.C.; McMahon, J.B.; et al. Screening and Biological Effects of Marine Pyrroloiminoquinone Alkaloids: Potential Inhibitors of the HIF-1α/p300 Interaction. J. Nat. Prod. 2016, 79, 1267–1275. [Google Scholar] [CrossRef] [PubMed]
- Alonso, E.; Alvariño, R.; Leirós, M.; Tabudravu, J.N.; Feussner, K.; Dam, M.A.; Rateb, M.E.; Jaspars, M.; Botana, L.M. Evaluation of the Antioxidant Activity of the Marine Pyrroloiminoquinone Makaluvamines. Mar. Drugs 2016, 14, 197. [Google Scholar] [CrossRef]
- Florean, C.; Schnekenburger, M.; Lee, J.-Y.; Kim, K.R.; Mazumder, A.; Song, S.; Kim, J.-M.; Grandjenette, C.; Kim, J.-G.; Yoon, A.-Y.; et al. Discovery and characterization of Isofistularin-3, a marine brominated alkaloid, as a new DNA demethylating agent inducing cell cycle arrest and sensitization to TRAIL in cancer cells. Oncotarget 2016, 7, 24027–24049. [Google Scholar] [CrossRef]
- Bae, W.; Lim, H.K.; Kim, K.M.; Cho, H.; Lee, S.Y.; Jeong, C.-S.; Lee, H.-S.; Jung, J. Apoptosis-Inducing Activity of Marine Sponge Haliclona sp. Extracts Collected from Kosrae in Nonsmall Cell Lung Cancer A549 Cells. Evid.-Based Complement. Alternat. Med. 2015, 2015, 717959. [Google Scholar] [CrossRef]
- Alarif, W.M.; Abdel-Lateff, A.; Al-Lihaibi, S.S.; Ayyad, S.-E.N.; Badria, F.A. A New Cytotoxic Brominated Acetylenic Hydrocarbon from the Marine Sponge Haliclona sp. with a Selective Effect against Human Breast Cancer. Z. Naturforsch. C Biosci. 2013, 68, 70–75. [Google Scholar] [CrossRef]
- Choi, K.; Lim, H.K.; Oh, S.R.; Chung, W.-H.; Jung, J. Anticancer Effects of the Marine Sponge Lipastrotethya sp. Extract on Wild-Type and p53 Knockout HCT116 Cells. Evid.-Based Complement. Alternat. Med. 2017, 2017, 7174858. [Google Scholar] [CrossRef]
- Carmely, S.; Kashman, Y. Neviotine-A, a new triterpene from the Red Sea sponge Siphonochalina siphonella. J. Org. Chem. 1986, 51, 784–788. [Google Scholar] [CrossRef]
- Angawi, R.F.; Saqer, E.; Abdel-Lateff, A.; Badria, F.A.; Ayyad, S.-E.N. Cytotoxic neviotane triterpene-type from the red sea sponge Siphonochalina siphonella. Pharmacogn. Mag. 2014, 10, S334–S341. [Google Scholar] [CrossRef] [PubMed]
- Beesoo, R.; Neergheen-Bhujun, V.; Bhagooli, R.; Bahorun, T. Apoptosis inducing lead compounds isolated from marine organisms of potential relevance in cancer treatment. Mutat. Res.-Fundam. Mol. Mech. Mutagen. 2014, 768, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, M.; Kelkel, M.; Dicato, M.; Diederich, M. Gold from the sea: Marine compounds as inhibitors of the hallmarks of cancer. Biotechnol. Adv. 2011, 29, 531–547. [Google Scholar] [CrossRef]
- Gerwick, W.H.; Moore, B.S. Lessons from the Past and Charting the Future of Marine Natural Products Drug Discovery and Chemical Biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.M.; Glaser, K.B.; Cuevas, C.; Jacobs, R.S.; Kem, W.; Little, R.D.; McIntosh, J.M.; Newman, D.J.; Potts, B.C.; Shuster, D.E. The odyssey of marine pharmaceuticals: A current pipeline perspective. Trends Pharmacol. Sci. 2010, 31, 255–265. [Google Scholar] [CrossRef]
- Hoppers, A.; Stoudenmire, J.; Wu, S.; Lopanik, N.B. Antibiotic activity and microbial community of the temperate sponge, Haliclona sp. J. Appl. Microbiol. 2015, 118, 419–430. [Google Scholar] [CrossRef]
- Nazemi, M.; Alidoust Salimi, M.; Alidoust Salimi, P.; Motallebi, A.; Tamadoni Jahromi, S.; Ahmadzadeh, O. Antifungal and antibacterial activity of Haliclona sp. from the Persian Gulf, Iran. J. Mycol. Med. 2014, 24, 220–224. [Google Scholar] [CrossRef]
- Yamazaki, H.; Wewengkang, D.S.; Kanno, S.; Ishikawa, M.; Rotinsulu, H.; Mangindaan, R.E.; Namikoshi, M. Papuamine and haliclonadiamine, obtained from an Indonesian sponge Haliclona sp., inhibited cell proliferation of human cancer cell lines. Nat. Prod. Res. 2013, 27, 1012–1015. [Google Scholar] [CrossRef]
- Shirazi, F.H.; Zarghi, A.; Ashtarinezhad, A.; Kobarfard, F.; Nakhjavani, M.; Anjidani, N.; Zendehdel, R.; Arfaiee, S.; Shoeibi, S.; Mohebi, S. Remarks in Successful Cellular Investigations for Fighting Breast Cancer Using Novel Synthetic Compounds; INTECH Open Access Publisher: Rijeka, Croatia, 2011. [Google Scholar]
- Holliday, D.L.; Speirs, V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011, 13, 215. [Google Scholar] [CrossRef]
- Gest, C.; Joimel, U.; Huang, L.; Pritchard, L.-L.; Petit, A.; Dulong, C.; Buquet, C.; Hu, C.-Q.; Mirshahi, P.; Laurent, M.; et al. Rac3 induces a molecular pathway triggering breast cancer cell aggressiveness: Differences in MDA-MB-231 and MCF-7 breast cancer cell lines. BMC Cancer 2013, 13, 63. [Google Scholar] [CrossRef]
- ŞErban, C.; Anca Maria, C.; Marius, R. The Story of MCF-7 Breast Cancer Cell Line: 40 years of Experience in Research. Anticancer Res. 2015, 35, 3147–3154. [Google Scholar]
- Sweeney, E.E.; McDaniel, R.E.; Maximov, P.Y.; Fan, P.; Jordan, V.C. Models and mechanisms of acquired antihormone resistance in breast cancer: Significant clinical progress despite limitations. Horm. Mol. Biol. Clin. Investig. 2012, 9, 143–163. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Tang, S.; Zheng, X.; Zhu, Y.; Ma, Z.; Liu, C.; Hecker, M.; Saunders, D.M.; Giesy, J.P.; Zhang, X.; et al. Bioaccumulation, biotransformation, and toxicity of BDE-47, 6-OH-BDE-47, and 6-MeO-BDE-47 in early life-stages of zebrafish (Danio rerio). Environ. Sci. Technol. 2015, 49, 1823–1833. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef]
- Zhao, M.; Antunes, F.; Eaton, J.W.; Brunk, U.T. Lysosomal enzymes promote mitochondrial oxidant production, cytochrome c release and apoptosis. Eur. J. Biochem. 2003, 270, 3778–3786. [Google Scholar] [CrossRef] [PubMed]
- Todkar, K.; Ilamathi, H.S.; Germain, M. Mitochondria and Lysosomes: Discovering Bonds. Front. Cell Dev. Biol. 2017, 5, 106. [Google Scholar] [CrossRef]
- Demers-Lamarche, J.; Guillebaud, G.; Tlili, M.; Todkar, K.; Bélanger, N.; Grondin, M.; Nguyen, A.P.; Michel, J.; Germain, M. Loss of Mitochondrial Function Impairs Lysosomes. J. Biol. Chem. 2016, 291, 10263–10276. [Google Scholar] [CrossRef]
- Conte da Frota, M.L.; Braganhol, E.; Delgado Canedo, A.; Klamt, F.; Apel, M.A.; Mothes, B.; Lerner, C.; Oliveira Battastini, A.M.; Henriques, A.T.; Fonseca Moreira, J.C. Extracts of marine sponge Polymastia janeirensis induce oxidative cell death through a caspase-9 apoptotic pathway in human U138MG glioma cell line. Investig. New Drugs 2009, 27, 440–446. [Google Scholar] [CrossRef]
- Azad, N.; Iyer, A.K.V. Reactive Oxygen Species and Apoptosis. In Systems Biology of Free Radicals and Antioxidants; Laher, I., Ed.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 113–135. [Google Scholar]
- Neumann, C.A.; Fang, Q. Are peroxiredoxins tumor suppressors? Curr. Opin. Pharmacol. 2007, 7, 375–380. [Google Scholar] [CrossRef]
- Nicoletti, I.; Migliorati, G.; Pagliacci, M.C.; Grignani, F.; Riccardi, C. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J. Immunol. Methods 1991, 139, 271–279. [Google Scholar] [CrossRef]
- Ravi, S.; Chiruvella, K.K.; Rajesh, K.; Prabhu, V.; Raghavan, S.C. 5-Isopropylidene-3-ethyl rhodanine induce growth inhibition followed by apoptosis in leukemia cells. Eur. J. Med. Chem. 2010, 45, 2748–2752. [Google Scholar] [CrossRef] [PubMed]
- Leist, M.; Jäättelä, M. Four deaths and a funeral: From caspases to alternative mechanisms. Nat. Rev. Mol. Cell Biol. 2001, 2, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Kim, D.K.; Seo, Y.R.; Woo, K.M.; Kim, C.S.; Cho, M.H. Nickel (II)-induced apoptosis and G2/M enrichment. Exp. Mol. Med. 1998, 30, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Saquib, Q.; Al-Khedhairy, A.A.; Siddiqui, M.A.; Abou-Tarboush, F.M.; Azam, A.; Musarrat, J. Titanium dioxide nanoparticles induced cytotoxicity, oxidative stress and DNA damage in human amnion epithelial (WISH) cells. Toxicol. Vitr. 2012, 26, 351–361. [Google Scholar] [CrossRef]
- Saquib, Q.; Musarrat, J.; Siddiqui, M.A.; Dutta, S.; Dasgupta, S.; Giesy, J.P.; Al-Khedhairy, A.A. Cytotoxic and necrotic responses in human amniotic epithelial (WISH) cells exposed to organophosphate insecticide phorate. Mutat. Res./Genet. Toxicol. Environ. Mutagen. 2012, 744, 125–134. [Google Scholar] [CrossRef]
- Sheikh, M.S.; Fornace, A.J., Jr. Role of p53 family members in apoptosis. J. Cell. Physiol. 2000, 182, 171–181. [Google Scholar] [CrossRef]
- Porter, A.G.; Jänicke, R.U. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999, 6, 99–104. [Google Scholar] [CrossRef]
- Hong, S.J.; Dawson, T.M.; Dawson, V.L. PARP and the Release of Apoptosis-Inducing Factor from Mitochondria. In Poly(ADP-Ribosyl)ation; Bürkle, A., Ed.; Springer: Boston, MA, USA, 2006; pp. 103–117. [Google Scholar]
- Kruidering, M.; Evan, G.I. Caspase-8 in Apoptosis: The Beginning of “The End”? IUBMB Life 2000, 50, 85–90. [Google Scholar] [CrossRef]
- Jänicke, R.U.; Sprengart, M.L.; Wati, M.R.; Porter, A.G. Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J. Biol. Chem. 1998, 273, 9357–9360. [Google Scholar] [CrossRef]
- Green, D.R.; Amarante-Mendes, G.P. The Point of No Return: Mitochondria, Caspases, and the Commitment to Cell Death. In Apoptosis: Mechanisms and Role in Disease; Kumar, S., Ed.; Springer: Berlin/Heidelberg, Germany, 1998; pp. 45–61. [Google Scholar]
- Letai, A.G. Diagnosing and exploiting cancer’s addiction to blocks in apoptosis. Nat. Rev. Cancer. 2008, 8, 121–132. [Google Scholar] [CrossRef]
- Locksley, R.M.; Killeen, N.; Lenardo, M.J. The TNF and TNF receptor superfamilies: Integrating mammalian biology. Cell 2001, 104, 487–501. [Google Scholar] [CrossRef] [PubMed]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef] [PubMed]
- Basaiyye, S.S.; Naoghare, P.K.; Kanojiya, S.; Bafana, A.; Arrigo, P.; Krishnamurthi, K.; Sivanesan, S. Molecular mechanism of apoptosis induction in Jurkat E6-1 cells by Tribulus terrestris alkaloids extract. J. Tradit. Complement. Med. 2018, 8, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Packham, G.; Stevenson, F.K. Bodyguards and assassins: Bcl-2 family proteins and apoptosis control in chronic lymphocytic leukaemia. Immunology 2005, 114, 441–449. [Google Scholar] [CrossRef]
- Genini, D.; Sheeter, D.; Rought, S.; Zaunders, J.J.; Susin, S.A.; Kroemer, G.; Richman, D.D.; Carson, D.A.; Corbeil, J.; Leoni, L.M. HIV induces lymphocyte apoptosis by a p53-initiated, mitochondrial-mediated mechanism. FASEB J. 2001, 15, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Corbeil, J.; Sheeter, D.; Genini, D.; Rought, S.; Leoni, L.; Du, P.; Ferguson, M.; Masys, D.R.; Welsh, J.B.; Fink, J.L.; et al. Temporal gene regulation during HIV-1 infection of human CD4+ T cells. Genome Res. 2001, 11, 1198–1204. [Google Scholar] [CrossRef]
- Chinnaiyan, A.M.; O’Rourke, K.; Tewari, M.; Dixit, V.M. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell 1995, 81, 505–512. [Google Scholar] [CrossRef]
- Boldin, M.P.; Varfolomeev, E.E.; Pancer, Z.; Mett, I.L.; Camonis, J.H.; Wallach, D. A novel protein that interacts with the death domain of Fas/APO1 contains a sequence motif related to the death domain. J. Biol. Chem. 1995, 270, 7795–7798. [Google Scholar] [CrossRef]
- Hsu, H.; Huang, J.; Shu, H.-B.; Baichwal, V.; Goeddel, D.V. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity 1996, 4, 387–396. [Google Scholar] [CrossRef]
- Hsu, H.; Xiong, J.; Goeddel, D.V. The TNF receptor 1-associated protein TRADD signals cell death and NF-κB activation. Cell 1995, 81, 495–504. [Google Scholar] [CrossRef]
- Hsu, H.; Shu, H.-B.; Pan, M.-G.; Goeddel, D.V. TRADD–TRAF2 and TRADD–FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell 1996, 84, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Stanger, B.Z.; Leder, P.; Lee, T.-H.; Kim, E.; Seed, B. RIP: A novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell 1995, 81, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Itoh, N.; Nagata, S. A novel protein domain required for apoptosis. Mutational analysis of human Fas antigen. J. Biol. Chem. 1993, 268, 10932–10937. [Google Scholar] [CrossRef] [PubMed]
- Tartaglia, L.A.; Ayres, T.M.; Wong, G.H.; Goeddel, D.V. A novel domain within the 55 kd TNF receptor signals cell death. Cell 1993, 74, 845–853. [Google Scholar] [CrossRef] [PubMed]
- Rothe, M.; Sarma, V.; Dixit, V.M.; Goeddel, D.V. TRAF2-mediated activation of NF-kappa B by TNF receptor 2 and CD40. Science 1995, 269, 1424–1427. [Google Scholar] [CrossRef]
- Cheng, G.; Baltimore, D. TANK, a co-inducer with TRAF2 of TNF-and CD 40L-mediated NF-kappaB activation. Genes Dev. 1996, 10, 963–973. [Google Scholar] [CrossRef]
- Lee, S.Y.; Lee, S.Y.; Kandala, G.; Liou, M.L.; Liou, H.C.; Choi, Y. CD30/TNF receptor-associated factor interaction: NF-kappa B activation and binding specificity. Proc. Natl. Acad. Sci. USA 1996, 93, 9699–9703. [Google Scholar] [CrossRef]
- Chawla-Sarkar, M.; Bae, S.I.; Reu, F.J.; Jacobs, B.S.; Lindner, D.J.; Borden, E.C. Downregulation of Bcl-2, FLIP or IAPs (XIAP and survivin) by siRNAs sensitizes resistant melanoma cells to Apo2L/TRAIL-induced apoptosis. Cell Death Differ. 2004, 11, 915–923. [Google Scholar] [CrossRef]
- Nomura, T.; Yamasaki, M.; Nomura, Y.; Mimata, H. Expression of the inhibitors of apoptosis proteins in cisplatin-resistant prostate cancer cells. Oncol. Rep. 2005, 14, 993–997. [Google Scholar] [CrossRef]
- Li, F.; Yang, J.; Ramnath, N.; Javle, M.M.; Tan, D. Nuclear or cytoplasmic expression of survivin: What is the significance? Int. J. Cancer 2005, 114, 509–512. [Google Scholar] [CrossRef]
- Hinnis, A.R.; Luckett, J.C.A.; Walker, R.A. Survivin is an independent predictor of short-term survival in poor prognostic breast cancer patients. Br. J. Cancer 2007, 96, 639–645. [Google Scholar] [CrossRef]
- Altieri, D.C. Validating survivin as a cancer therapeutic target. Nat. Rev. Cancer 2003, 3, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wu, W.; Li, L.; He, J.; Huang, S.; Chen, S.; Chen, J.; Long, M.; Yang, S.; Li, P. Analysis of the miRNA Expression Profiles in the Zearalenone-Exposed TM3 Leydig Cell Line. Int. J. Mol. Sci. 2019, 20, 635. [Google Scholar] [CrossRef] [PubMed]
- Saquib, Q.; Al-Khedhairy, A.A.; Ahmad, J.; Siddiqui, M.A.; Dwivedi, S.; Khan, S.T.; Musarrat, J. Zinc ferrite nanoparticles activate IL-1b, NFKB1, CCL21 and NOS2 signaling to induce mitochondrial dependent intrinsic apoptotic pathway in WISH cells. Toxicol. Appl. Pharmacol. 2013, 273, 289–297. [Google Scholar] [CrossRef]
- Gene Globe (Qiagen). Available online: https://geneglobe.qiagen.com/sa/ (accessed on 22 August 2023).
- Al-Salem, A.M.; Saquib, Q.; Siddiqui, M.A.; Ahmad, J.; Wahab, R.; Al-Khedhairy, A.A. Organophosphorus flame retardant (tricresyl phosphate) trigger apoptosis in HepG2 cells: Transcriptomic evidence on activation of human cancer pathways. Chemosphere 2019, 237, 124519. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saquib, Q.; Schwaiger, S.; Alilou, M.; Ahmed, S.; Siddiqui, M.A.; Ahmad, J.; Faisal, M.; Abdel-Salam, E.M.; Wahab, R.; Al-Rehaily, A.J.; et al. Marine Natural Compound (Neviotin A) Displays Anticancer Efficacy by Triggering Transcriptomic Alterations and Cell Death in MCF-7 Cells. Molecules 2023, 28, 6289. https://doi.org/10.3390/molecules28176289
Saquib Q, Schwaiger S, Alilou M, Ahmed S, Siddiqui MA, Ahmad J, Faisal M, Abdel-Salam EM, Wahab R, Al-Rehaily AJ, et al. Marine Natural Compound (Neviotin A) Displays Anticancer Efficacy by Triggering Transcriptomic Alterations and Cell Death in MCF-7 Cells. Molecules. 2023; 28(17):6289. https://doi.org/10.3390/molecules28176289
Chicago/Turabian StyleSaquib, Quaiser, Stefan Schwaiger, Mostafa Alilou, Sarfaraz Ahmed, Maqsood A. Siddiqui, Javed Ahmad, Mohammad Faisal, Eslam M. Abdel-Salam, Rizwan Wahab, Adnan J. Al-Rehaily, and et al. 2023. "Marine Natural Compound (Neviotin A) Displays Anticancer Efficacy by Triggering Transcriptomic Alterations and Cell Death in MCF-7 Cells" Molecules 28, no. 17: 6289. https://doi.org/10.3390/molecules28176289
APA StyleSaquib, Q., Schwaiger, S., Alilou, M., Ahmed, S., Siddiqui, M. A., Ahmad, J., Faisal, M., Abdel-Salam, E. M., Wahab, R., Al-Rehaily, A. J., Stuppner, H., & Al-Khedhairy, A. A. (2023). Marine Natural Compound (Neviotin A) Displays Anticancer Efficacy by Triggering Transcriptomic Alterations and Cell Death in MCF-7 Cells. Molecules, 28(17), 6289. https://doi.org/10.3390/molecules28176289










