Molecular Profiling, Characterization and Antimicrobial Efficacy of Silver Nanoparticles Synthesized from Calvatia gigantea and Mycena leaiana against Multidrug-Resistant Pathogens
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification
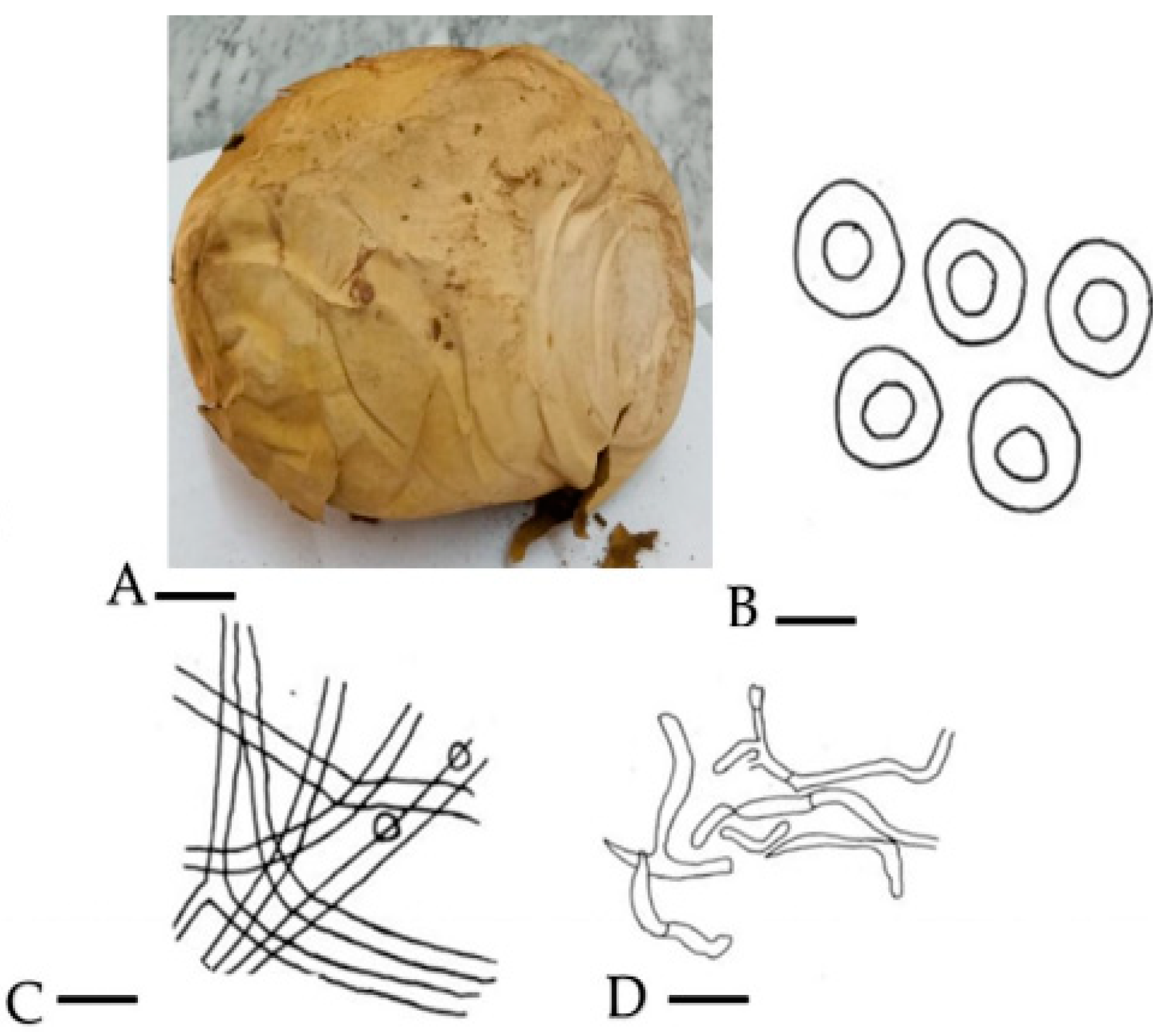
2.1.1. Morpho-Anatomical Study
2.1.2. Phylogenetic Analysis of C. gigantea
2.2. M. leaiana
Morpho-Anatomical Study
2.3. Phylogenetic Analysis of M. leaiana
2.4. Nanoparticle Characterization
2.4.1. XRD Analysis
2.4.2. UV-Vis Analysis
2.4.3. SEM Analysis
2.4.4. FT-IR Analysis
2.5. Antibacterial Potential of Organic Solvent
2.5.1. Methanol
2.5.2. Ethyl Acetate Extract
2.5.3. n-Hexane Extract
2.5.4. Aqueous Extract (AE)
2.5.5. Pure Water Extracts (PWE)
2.5.6. AgNPs’ Antibacterial Activity
2.6. Minimum Inhibitory Concentration
2.7. Antibiotic Sensitivity
3. Methodology
3.1. Collection of Mushroom
3.2. Identification
3.2.1. Morphological Study
3.2.2. Anatomical Study
3.3. Laboratory Procedure for Identification of Mushroom
3.3.1. DNA Isolation and Amplification
3.3.2. Sequencing and Blast Analysis
3.3.3. Phylogenetic Analysis
3.4. Extracts Preparation
3.5. Pure Water Extract Preparation
3.6. Synthesis of AgNPs
3.7. Characterization of AgNPs
3.8. Bacterial Strain
3.9. Antibacterial Test
3.10. Antibiotic Sensitivity
3.11. Minimum Inhibitory Concentration
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Bonner, J.T. Life Cycles: Reflections of an Evolutionary Biologist; Princeton University Press: Princeton, NJ, USA, 2015. [Google Scholar]
- Gholami-Shabani, M.; Shams-Ghahfarokhi, M.; Razzaghi-Abyaneh, M. Natural product synthesis by fungi: Recent trends and future prospects. In Recent Advancement in White Biotechnology through Fungi: Volume 2: Perspective for Value-Added Products and Environments; Springer: Cham, Switzerland, 2019; pp. 195–228. [Google Scholar]
- Yadav, A.N.; Singh, S.; Mishra, S.; Gupta, A. Recent Advancement in White Biotechnology through Fungi; Springer International Publishing: Cham, Switzerland, 2019; p. 528. [Google Scholar]
- Wang, N.; Zhao, Z.; Gao, J.; Tian, E.; Yu, W.; Li, H.; Zhang, J.; Xie, R.; Zhao, X.; Chen, A. Rapid and Visual Identification of Chlorophyllum Molybdites with Loop-Mediated Isothermal Amplification Method. Front. Microbiol. 2021, 12, 638315. [Google Scholar] [CrossRef]
- Romi, W.; Keisam, S.; Ahmed, G.; Jeyaram, K. Reliable Differentiation of Meyerozyma Guilliermondii from Meyerozyma Caribbica by Internal Transcribed Spacer Restriction Fingerprinting. BMC Microbiol. 2014, 14, 52. [Google Scholar] [CrossRef]
- Zimmerman, E.; St-Arnaud, M.; Hijri, M. Sustainable Agriculture and the Multigenomic Model: How Advances in the Genetics of Arbuscular Mycorrhizal Fungi Will Change Soil Management Practices. In Molecular Plant-Microbe Interactions; CABI: Wallingford, UK, 2009. [Google Scholar]
- Munir, N.; Xiang, T.C.; Bhuyar, P.; Ramli, A.N.M. Effective microbes (EM) and their potential on mushroom commercialization in Malaysia. Maejo Int. J. Energy Environ. Commun. 2021, 3, 45–55. [Google Scholar] [CrossRef]
- Anand, U.; Carpena, M.; Kowalska-Góralska, M.; Garcia-Perez, P.; Sunita, K.; Bontempi, E.; Dey, A.; Prieto, M.A.; Proćków, J.; Simal-Gandara, J. Safer Plant-Based Nanoparticles for Combating Antibiotic Resistance in Bacteria: A Comprehensive Review on Its Potential Applications, Recent Advances, and Future Perspective. Sci. Total Environ. 2022, 821, 153472. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Gulati, S.S.; Sharma, N.; Chaudhary, A. Sustainable Synthesis of Silver Nanoparticles Using Various Biological Sources and Waste Materials: A Review. Emergent Mater. 2022, 5, 1649–1678. [Google Scholar] [CrossRef]
- Iqtedar, M.; Aslam, M.; Akhyar, M.; Shehzaad, A.; Abdullah, R.; Kaleem, A. Extracellular Biosynthesis, Characterization, Optimization of Silver Nanoparticles (AgNPs) Using Bacillus Mojavensis BTCB15 and Its Antimicrobial Activity against Multidrug Resistant Pathogens. Prep. Biochem. Biotechnol. 2019, 49, 136–142. [Google Scholar] [CrossRef]
- Pantidos, N. Biological Synthesis of Metallic Nanoparticles by Bacteria, Fungi and Plants. J. Nanomed. Nanotechnol. 2014, 05, 1. [Google Scholar] [CrossRef]
- Pandey, S.; Mishra, A.; Giri, V.P.; Kumari, M.; Soni, S. A Green Nano-Synthesis to Explore the Plant Microbe Interactions. In New and Future Developments in Microbial Biotechnology and Bioengineering: Microbial Biotechnology in Agro-Environmental Sustainability; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Ullah, T.S.; Firdous, S.S.; Mehmood, A.; Shaheen, H.; Dar, M. Ethnomycological and nutritional analyses of some wild edible mushrooms from Western Himalayas, Azad Jammu and Kashmir (Pakistan). Int. J. Med. Mushrooms 2017, 19, 949–955. [Google Scholar] [CrossRef]
- Wang, S.; Liu, Y.; Xu, J. Comparison of Different Drying Methods for recovery of mushroom DNA. Sci. Rep. 2017, 7, 3008. [Google Scholar] [CrossRef]
- Bates, S.T.; Roberson, R.W.; Desjardin, D.E. Arizona Gasteroid Fungi I: Lycoperdaceae (Agaricales, Basidiomycota). Fungal Divers. 2009, 37, 153. [Google Scholar]
- Das, K.; Akhtar, N.; Aminuzzaman, F.M. Diversity of fleshy macro fungi in mangrove forest regions of Bangladesh. J. Biol. Nat. 2016, 6, 218–241. [Google Scholar]
- Damm, U.; Verkley, G.J.M.; Crous, P.W.; Fourie, P.H.; Haegi, A.; Riccioni, L. Novel Paraconiothyrium Species on Stone Fruit Trees and Other Woody Hosts. Persoonia Mol. Phylogeny Evol. Fungi 2008, 20, 9–17. [Google Scholar] [CrossRef]
- Bindhani, B.K.; Panigrahi, A.K. Biosynthesis and Characterization of Silver Nanoparticles (Snps) by Using Leaf Extracts of Ocimum Sanctum L (Tulsi) and Study of Its Antibacterial Activities. J. Nanomed. Nanotechnol. 2015, s6, 2157–7439. [Google Scholar] [CrossRef]
- Philip, D.; Unni, C. Extracellular Biosynthesis of Gold and Silver Nanoparticles Using Krishna Tulsi (Ocimum sanctum) Leaf. Phys. E Low-Dimens. Syst. Nanostruct. 2011, 43, 1318–1322. [Google Scholar] [CrossRef]
- Senapati, U.S.; Sarkar, D. Characterization of Biosynthesized Zinc Sulphide Nanoparticles Using Edible Mushroom Pleurotuss Ostreatu. Indian J. Phys. 2014, 88, 557–562. [Google Scholar] [CrossRef]
- Philip, D. Biosynthesis of Au, Ag and Au-Ag Nanoparticles Using Edible Mushroom Extract. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 2009, 73, 374–381. [Google Scholar] [CrossRef]
- Bhardwaj, A.K.; Shukla, A.; Maurya, S.; Singh, S.C.; Uttam, K.N.; Sundaram, S.; Singh, M.P.; Gopal, R. Direct sunlight enabled photo-biochemical synthesis of silver nanoparticles and their bactericidal efficacy: Photon energy as key for size and distribution control. J. Photochem. Photobiol. B Biol. 2018, 188, 42–49. [Google Scholar] [CrossRef]
- Bhuvaneswari, T.; Thiyagarajan, M.; Geetha, N.; Venkatachalam, P. Bioactive Compound Loaded Stable Silver Nanoparticle Synthesis from Microwave Irradiated Aqueous Extracellular Leaf Extracts of Naringi Crenulata and Its Wound Healing Activity in Experimental Rat Model. Acta Trop. 2014, 135, 55–61. [Google Scholar] [CrossRef]
- Siddiqi, K.S.; Husen, A.; Rao, R.A.K. A Review on Biosynthesis of Silver Nanoparticles and Their Biocidal Properties. J. Nanobiotechnol. 2018, 16, 14. [Google Scholar] [CrossRef]
- Prasannaraj, G.; Sahi, S.V.; Ravikumar, S.; Venkatachalam, P. Enhanced Cytotoxicity of Biomolecules Loaded Metallic Silver Nanoparticles against Human Liver (HepG2) and Prostate (PC3) Cancer Cell Lines. J. Nanosci. Nanotechnol. 2016, 16, 4948–4959. [Google Scholar] [CrossRef]
- Maraki, S.; Mantadakis, E.; Mavromanolaki, V.E.; Kofteridis, D.P.; Samonis, G. A 5-Year Surveillance Study on Antimicrobial Resistance of Acinetobacter baumannii Clinical Isolates from a Tertiary Greek Hospital. Infect. Chemother. 2016, 48, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Quereshi, S.; Pandey, A.K.; Sandhu, S.S. Evaluation of Antibacterial Activity of Different Ganoderma Lucidum Extracts. People’s J. Sci. Res. 2010, 3, 9–13. [Google Scholar]
- Agarwal, H.; Menon, S.; Venkat Kumar, S.; Rajeshkumar, S. Mechanistic Study on Antibacterial Action of Zinc Oxide Nanoparticles Synthesized Using Green Route. Chem. Biol. Interact. 2018, 286, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.N.; Azad, A.K.; Sultana, F.; Anisuzzaman, A.S.M. In-Vitro Antimicrobial Activity of Ethyl Acetate Extract of Two Common Edible Mushrooms. J. Phytopharm. 2016, 5, 79–82. [Google Scholar] [CrossRef]
- Muzammal, M.; Khan, M.A.; Al Mohaini, M.; Alsalman, A.J.; Al Hawaj, M.A.; Farid, A. In Silico Analysis of Honeybee Venom Protein Interaction with Wild Type and Mutant (A82V + P375S) Ebola Virus Spike Protein. Biologics 2022, 2, 45–55. [Google Scholar] [CrossRef]
- Dixit, S.; Tripathi, A.; Kumar, P. Medicinal Properties of Moringa Oleifera: A Review. Int. J. Educ. Sci. Res. Rev. 2016, 3, 173–185. [Google Scholar]
- Garcia, J.; Rodrigues, F.; Castro, F.; Aires, A.; Marques, G.; Saavedra, M.J. Antimicrobial, antibiofilm, and antioxidant properties of Boletus edulis and Neoboletus luridiformis against multidrug-resistant ESKAPE pathogens. Front. Nutr. 2022, 8, 773346. [Google Scholar] [CrossRef]
- Mariadoss, A.V.A.; Ramachandran, V.; Shalini, V.; Agilan, B.; Franklin, J.H.; Sanjay, K.; Alaa, Y.G.; Tawfiq, M.A.A.; Ernest, D. Green synthesis, characterization and antibacterial activity of silver nanoparticles by Malus domestica and its cytotoxic effect on (MCF-7) cell line. Microb. Pathog. 2019, 135, 103609. [Google Scholar] [CrossRef]
- Joshi, A.S.; Singh, P.; Mijakovic, I. Interactions of Gold and Silver Nanoparticles with Bacterial Biofilms: Molecular Interactions behind Inhibition and Resistance. Int. J. Mol. Sci. 2020, 21, 7658. [Google Scholar] [CrossRef]
- Singh, M.; Singh, S.; Prasad, S.; Gambhir, I.S. Nanotechnology in Medicine and Antibacterial Effect of Silver Nanoparticles. Dig. J. Nanomater. Biostruct. 2008, 3, 115–122. [Google Scholar]
- Dibrov, P.; Dzioba, J.; Gosink, K.K.; Häse, C.C. Chemiosmotic Mechanism of Antimicrobial Activity of Ag+ in Vibrio Cholerae. Antimicrob. Agents Chemother. 2002, 46, 2668–2670. [Google Scholar] [CrossRef] [PubMed]
- Sondi, I.; Salopek-Sondi, B. Silver Nanoparticles as Antimicrobial Agent: A Case Study on E. coli as a Model for Gram-Negative Bacteria. J. Colloid Interface Sci. 2004, 275, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Leont’ev, V.K.; Pogorel’skii, I.P.; Frolov, G.A.; Karasenkov, Y.N.; Gusev, A.A.; Latuta, N.V.; Borozdkin, L.L.; Stefantsova, D.S. Antibacterial Properties of Aqueous Colloid Solutions of Metal and Metal Oxide Nanoparticles against Dental Plaque Bacteria. Nanotechnol. Russ. 2018, 13, 195–198. [Google Scholar] [CrossRef]
- Rabe, T.; Van Staden, J. Antibacterial Activity of South African Plants Used for Medicinal Purposes. J. Ethnopharmacol. 1997, 56, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Parekh, J.; Chanda, S. In-Vitro Antimicrobial Activities of Extracts of Launaea Procumbens Roxb. (Labiateae), Vitis vinifera L. (Vitaceae) and Cyperus rotundus L. (Cyperaceae). Afr. J. Biomed. Res. 2009, 9, 89–93. [Google Scholar] [CrossRef]
- El-Mahmood, A.M.; Ogbonna, O.B.; Raji, M. The Antibacterial Activity of Azadarichta Indica (Neem) Seeds Extracts against Bacterial Pathogens Associated with Eye and Ear Infections. J. Med. Plants Res. 2010, 4, 1414–1421. [Google Scholar]
- Berrio, I.; Maldonado, N.; De Bedout, C.; Arango, K.; Cano, L.E.; Valencia, Y.; Jiménez-Ortigosa, C.; Perlin, D.S.; Gómez, B.L.; Robledo, C.; et al. Comparative Study of Candida spp. Isolates: Identification and Echinocandin Susceptibility in Isolates Obtained from Blood Cultures in 15 Hospitals in Medellín, Colombia. J. Glob. Antimicrob. Resist. 2018, 13, 254–260. [Google Scholar] [CrossRef]
- McMahon, S. Earth’s Earliest and Deepest Purported Fossils May Be Iron-Mineralized Chemical Gardens. Proc. R. Soc. B Biol. Sci. 2019, 286, 20192410. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, X.; Wang, J.; Cheng, M. Synthesis and characterization of konjac glucomannan/carrageenan/nano-silica films for the preservation of postharvest white mushrooms. Polymers 2018, 11, 6. [Google Scholar] [CrossRef]
- Li, J.; Mahoney, B.D.; Jacob, M.S.; Caron, S.J.C. Visual Input into the Drosophila Melanogaster Mushroom Body. Cell Rep. 2020, 32, 108138. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Zhang, S.; Liu, X.Z.; Wen, H.A.; Wang, M. A Simple Method of Genomic DNA Extraction Suitable for Analysis of Bulk Fungal Strains. Lett. Appl. Microbiol. 2010, 51, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Amey, R.C.; Athey-Pollard, A.; Mills, P.R.; Foster, G.D.; Bailey, A. Investigations into the Taxonomy of the Mushroom Pathogen Verticillium Fungicola and Its Relatives Based on Sequence Analysis of Nitrate Reductase and ITS Regions. Microbiology 2007, 76, 757–768. [Google Scholar] [CrossRef]
- Ao, T.; Deb, C.R.; Rao, S.R. Molecular Strategies for Identification and Characterization of Some Wild Edible Mushrooms of Nagaland, India. Mol. Biol. Rep. 2020, 47, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Papoutsis, K.; Zhang, J.; Bowyer, M.C.; Brunton, N.; Gibney, E.R.; Lyng, J. Fruit, Vegetables, and Mushrooms for the Preparation of Extracts with α-Amylase and α-Glucosidase Inhibition Properties: A Review. Food Chem. 2021, 338, 128119. [Google Scholar] [CrossRef] [PubMed]
- Nagajyothi, P.C.; Sreekanth, T.V.M.; Lee, J.I.; Lee, K.D. Mycosynthesis: Antibacterial, Antioxidant and Antiproliferative Activities of Silver Nanoparticles Synthesized from Inonotus obliquus (Chaga Mushroom) Extract. J. Photochem. Photobiol. B Biol. 2014, 130, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Bhuyar, P.; Rahim, M.H.A.; Sundararaju, S.; Ramaraj, R.; Maniam, G.P.; Govindan, N. Synthesis of silver nanoparticles using marine macroalgae Padina sp. and its antibacterial activity towards pathogenic bacteria. Beni-Suef Univ. J. Basic Appl. Sci. 2020, 9, 3. [Google Scholar] [CrossRef]
- Narasimha, G.; Praveen, B.; Mallikarjuna, K.; Deva, P.R.B. Mushrooms (Agaricus bisporus) mediated biosynthesis of sliver nanoparticles, characterization and their antimicrobial activity. Int. J. Nano Dimens. 2011, 2, 29–36. [Google Scholar]
- Liu, K.; Wang, J.; Zhao, L.; Wang, Q. Anticancer, Antioxidant and Antibiotic Activities of Mushroom Ramaria Flava. Food Chem. Toxicol. 2013, 58, 375–380. [Google Scholar] [CrossRef]
- Zeynali Aghdam, S.; Minaeian, S.; Sadeghpour Karimi, M.; Tabatabaee Bafroee, A.S. The Antibacterial Effects of the Mixture of Silver Nanoparticles with the Shallot and Nettle Alcoholic Extracts. J. Appl. Biotechnol. Rep. 2019, 6, 158–164. [Google Scholar] [CrossRef]














| Mushroom Composites (100 μL) | E. coli | K. pneumoniae | P. aeruginosa | E. cloacae | A. baumannii | S. aureus |
|---|---|---|---|---|---|---|
| Methanolic | 22 ± 2.64 | 23 ± 1.73 | 12 ± 2.62 | 11 ± 1.62 | 18 ± 3.51 | 21 ± 2.64 |
| Ethyl acetate | 23 ± 1 | R | R | R | 16 ± 0.57 | R |
| n-hexane | 20 ± 1.72 | 19 ± 1.52 | 22 ± 3.51 | 18 ± 1.43 | 17 ± 2.53 | 16 ± 0.67 |
| Aqueous extract | 24 ± 3.05 | 16 ± 1.52 | 16 ± 2.08 | R | 15 ± 1.16 | 16 ± 2.51 |
| Pure water extract | 20 ± 3 | 20 ± 1.15 | 19 ± 2.28 | 17 ± 2.06 | 18 ± 1.76 | 20 ± 2.03 |
| AgNPs | 19 ± 2 | 18 ± 0.75 | 25 ± 1.52 | 13 ± 1.25 | 13 ± 0.56 | 18 ± 1.73 |
| Mushroom Composites (100 μL) | E. coli | K. pneumoniae | P. aeruginosa | E. cloacae | A. baumannii | S. aureus |
|---|---|---|---|---|---|---|
| Methanolic | 20 ± 2.57 | 18 ± 1.16 | 17 ± 3.05 | 20 ± 1.25 | 15 ± 0.46 | 15 ± 0.57 |
| Ethyl acetate | 10 ± 1.87 | 4 ± 3 | 9 ± 0.62 | 8 ± 0.73 | R | R |
| n-hexane | 19 ± 2.62 | 10 ± 0.72 | 14 ± 0.57 | 17 ± 3.51 | 13 ± 1.35 | 16 ± 1.43 |
| Aqueous extract | 8 ± 2.08 | 5 ± 1.52 | 6 ± 1.15 | 14 ± 0.57 | 10 ± 0.53 | R |
| Pure water extract | 19 ± 2.57 | 16 ± 2.08 | 20 ± 0.17 | 19 ± 0.52 | 18 ± 0.53 | 22 ± 1.64 |
| AgNPs | 15 ± 0.57 | 16 ± 1.52 | 10 ± 1.43 | 9 ± 0.62 | 10 ± 1.36 | 14 ± 1.45 |
| AgNPs synergy | 22 ± 2.64 | 24 ± 2.08 | 20 ± 3.05 | 21 ± 3 | 23 ± 2.08 | 29 ± 2 |
| Mushroom Composites Dilutions (100 μL) | MIC Values (mg mL−1) | |||||
|---|---|---|---|---|---|---|
| E. coli | K. pneumoniae | P. aeruginosa | E. cloacae | A. baumannii | S. aureus | |
| Methanolic | 20 ± 0.179 | 10 ± 0.117 | 40 ± 0.127 | 40 ± 0.227 | 30 ± 0.139 | 20 ± 0.103 |
| Ethyl acetate | 30 ± 0.217 | --- | --- | --- | 40 ± 1.13 | --- |
| n-hexane | 30 ± 0.132 | 40 ± 1.46 | 20 ± 1.53 | 20 ± 0.971 | 40 ± 0.083 | 30 ± 0.471 |
| Aqueous extract | 10 ± 0.181 | 40 ± 0.134 | 40 ± 0.72 | --- | 40 ± 0.107 | 40 ± 1.12 |
| Pure water extract | 20 ± 0.161 | 20 ± 0.503 | 30 ± 0.136 | 40 ± 0.127 | 30 ± 0.941 | 20 ± 1.08 |
| AgNPs | 20 ± 0.113 | 20 ± 0.145 | 10 ± 0.181 | 30 ± 0.929 | 30 ± 0.169 | 20 ± 0.209 |
| Mushroom Composites Dilutions (100 μL) | MIC Values (mg mL−1) | |||||
|---|---|---|---|---|---|---|
| E. coli | K. pneumoniae | P. aeruginosa | E. cloacae | A. baumannii | S. aureus | |
| Methanolic | 20 ± 0.119 | 30 ± 0.459 | 30 ± 0.138 | 20 ± 1.06 | 40 ± 0.171 | 40 ± 0.117 |
| Ethyl acetate | 40 ± 0.221 | --- | --- | --- | --- | --- |
| n-hexane | 20 ± 0.147 | 40 ± 0.173 | 40 ± 0.093 | 20 ± 0.197 | 30 ± 0.203 | 40 ± 0.119 |
| Aqueous extract | --- | --- | --- | 20 ± 0.186 | 40 ± 0.145 | --- |
| Pure water extract | 20 ± 0.121 | 40 ± 0.136 | 20 ± 0.187 | 20 ± 0.127 | 30 ± 0.139 | 10 ± 0.163 |
| AgNPs | 10 ± 0.831 | 10 ± 0.651 | 30 ± 0.712 | 40 ± 0.143 | 40 ± 0.163 | 30 ± 0.209 |
| Antibiotics | E. coli | Staph | K. p | E. b | A. b | P. a | P. m |
|---|---|---|---|---|---|---|---|
| Cefpirome (CPR) | R | R | R | S | S | I | I |
| Tazobactam (TZP) | R | S | R | S | S | I | S |
| Gentamicin (GEN) | S | I | R | S | S | I | R |
| Ceftazidime (CAZ) | R | R | R | S | R | I | R |
| Doxycycline (DO) | R | R | R | R | R | R | R |
| Cefepime (FEP) | R | R | R | R | R | R | R |
| Ceftxime (CFM) | R | R | R | R | R | R | R |
| Chloramphinicole (C) | R | I | S | R | I | R | S |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, S.; Fiaz, M.; Yasmin, H.; Ahmad, J.; Ullah, A.; Niaz, Z.; Hayat, S.; Ahmad, A.; Kaushik, P.; Farid, A. Molecular Profiling, Characterization and Antimicrobial Efficacy of Silver Nanoparticles Synthesized from Calvatia gigantea and Mycena leaiana against Multidrug-Resistant Pathogens. Molecules 2023, 28, 6291. https://doi.org/10.3390/molecules28176291
Khan S, Fiaz M, Yasmin H, Ahmad J, Ullah A, Niaz Z, Hayat S, Ahmad A, Kaushik P, Farid A. Molecular Profiling, Characterization and Antimicrobial Efficacy of Silver Nanoparticles Synthesized from Calvatia gigantea and Mycena leaiana against Multidrug-Resistant Pathogens. Molecules. 2023; 28(17):6291. https://doi.org/10.3390/molecules28176291
Chicago/Turabian StyleKhan, Sayab, Muhammad Fiaz, Humaira Yasmin, Junaid Ahmad, Amin Ullah, Zeeshan Niaz, Shubana Hayat, Ajaz Ahmad, Prashant Kaushik, and Arshad Farid. 2023. "Molecular Profiling, Characterization and Antimicrobial Efficacy of Silver Nanoparticles Synthesized from Calvatia gigantea and Mycena leaiana against Multidrug-Resistant Pathogens" Molecules 28, no. 17: 6291. https://doi.org/10.3390/molecules28176291





