Abstract
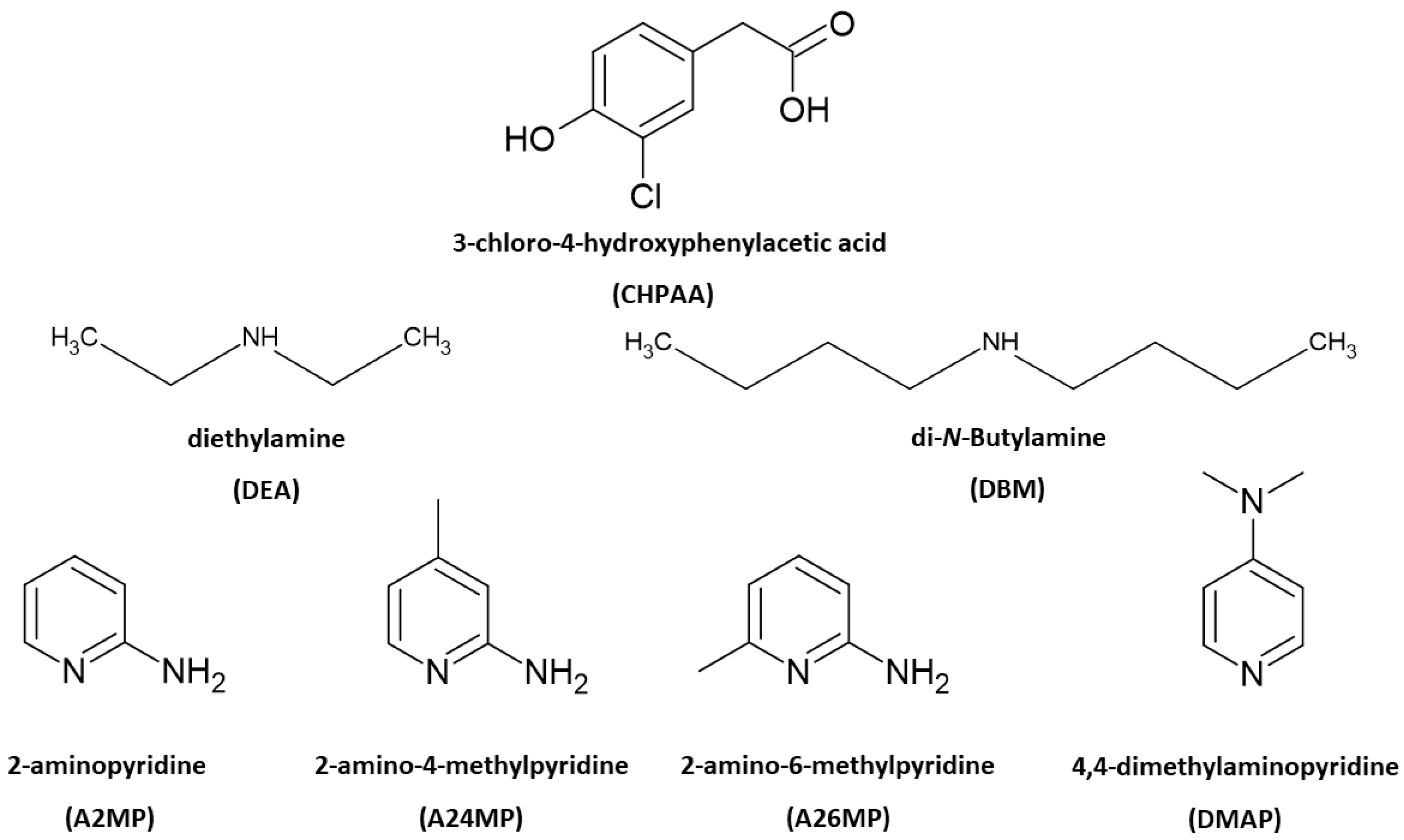
3-chloro-4-hydroxyphenylacetic acid (CHPAA) is a fungal metabolite. It is a small molecule that is useful in crystal engineering studies due to the functional groups present. Six amines were selected to form salts with CHPAA. Linear derivatives included diethylamine (DEA) and di-N-butylamine (DBM). The aromatic compounds chosen were 2-aminopyridine (A2MP), 2-amino-4-methylpyridine (A24MP), 2-amino-6-methylpyridine (A26MP) and 4-dimethylaminopyridine (DMAP). The salts were characterised using single-crystal X-ray diffraction, thermal analysis, FTIR spectroscopy and Hirshfeld surface analysis. For all the crystal structures, N-H···O and C-H···Cl contacts were present. O-H···O contacts were found in all the crystal structures except for (, which was also the only structure that displayed a Cl···Cl contact. Furthermore, C-H···O contacts were found in all the crystal structures except for The thermal stability trend showed that the DBM salt was more stable than the DEA salt. For the aromatic co-formers, the thermal stability trend showed the following: >.
1. Introduction
Multicomponent crystals are important in the pharmaceutical industry as the formation of new solid forms can improve the physicochemical properties of active pharmaceutical ingredients [1,2]. The choice of co-crystal formers can affect properties such as the solubility, hygroscopicity and compaction of the resultant pharmaceutical co-crystals and salts. Natural products, including small molecules derived from plants and fungi, also contribute to drug design. These metabolites are of interest due to their potential biological activity [3,4]. Crystal engineering principles of exploiting intermolecular interactions are utilised to obtain multicomponent crystals with suitable properties. These concepts are also relevant in the agrochemical industry [5]. One example is the improved solubility of the herbicide atrazine through co-crystallisation with fumaric acid [6]. Another herbicide 2,4-dichlorophenoxyacetic acid, formed salts with imidazole, 2-aminopyridine and 3-aminopyridine and co-crystals with isonicotinamide and pyrazinamide. All five multicomponent crystals showed enhanced solubility compared to the 2,4-dichlorophenoxyacetic acid alone [7]. Intermolecular interactions such as hydrogen bonding, π–π stacking and halogen bonding are typically employed in the formation of new solid forms. Strong hydrogen bonds include O-H···O and N-H···O. Examples of weak hydrogen bonds are C-H···O, C-H···π, O-H···π and C-H···Cl [8]. Crystal engineering employs supramolecular synthons whereby functional groups present in molecules form complementary units [9]. Etter’s hydrogen bond rules provide insights into the organisation of crystal structures with a focus on the best proton donor pairing with the best acceptor group [10]. The interaction of identical functional groups gives rise to homosynthons, whereas heterosynthons result when different functional groups interact. Notable heterosynthons are the robust carboxylic acid pyridine and carboxylic acid 2-aminopyridinium building blocks [11,12]. Amines and carboxylic acids also form the useful N-H···O heterosynthon [13]. In this study, we continue our focus on the crystal landscape of the fungal metabolite 3-chloro-4-hydroxyphenylacetic acid (CHPAA). CHPAA has previously been isolated from natural sources [14] and has also been used in the development of a screening library for applications in medicinal chemistry [15]. A library of compounds based on the skeletal framework of 3-chloro-4-hydroxyphenylacetamide and CHPAA has also been developed for potential use in pharmaceutical and agrochemical screening [16]. CHPAA is useful in crystal engineering studies as it possesses carboxylic acid and hydroxyl functional groups. Furthermore, CHPAA also contains the chlorine atom in the meta position, which can be involved in hydrogen and/or halogen bonds. We have previously reported co-crystals of CHPAA with nicotinamide, isonicotinamide, phenazine and 4,4′-bipyridine [17]. These are the only crystal structures of CHPAA in the CSD version 5.44, June 2023 update [18]. In the current study, a series of linear amines and aminopyridine derivatives were selected to co-crystallise with CHPAA. These included diethylamine (DEA), di-N-butylamine (DBM), 2-aminopyridine (A2MP), 2-amino-4-methylpyridine (A24MP), 2-amino-6-methylpyridine (A26MP) and 4,4-dimethylaminopyridine (DMAP). These are depicted in Scheme 1. The pKa [19] of CHPAA is 3.06 and the pKa values of the amines are DEA (10.58), DBM (10.75), A2MP (6.84), A24MP (7.62), A26MP (7.60) DMAP (8.78). Thus, the ∆pKa values ranged from 3.78 to 7.69. Salt formation is expected when ∆pKa > 4. If <−1 ∆pKa < 4, then either salt or co-crystal can form [20]. In this study, proton transfer occurred between the CHPAA and the respective bases forming salts. This work aimed to prepare multicomponent crystals of CHPAA with the abovementioned amines and to study their crystal structures and thermal stability. Fourier transform infrared (FTIR) spectroscopy was used to confirm the formation of the new solid forms. Furthermore, alternate methods of preparation of the salts were also explored, for example, liquid-assisted grinding and slurry conversion, and the products were analysed using powder X-ray diffraction. Hirshfeld surface analysis was used to compare the intermolecular interactions in the salts, and the resulting fingerprint plots were interpreted.
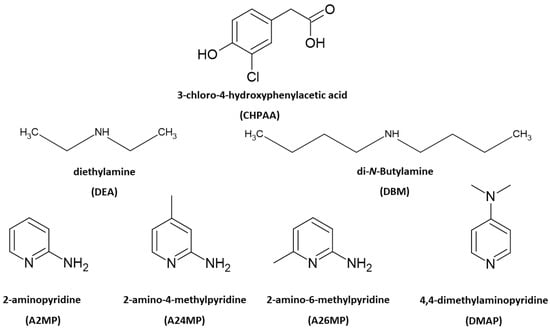
Scheme 1.
Chemical structures of CHPAA and the amines.
2. Results and Discussion
2.1. Crystal Structures
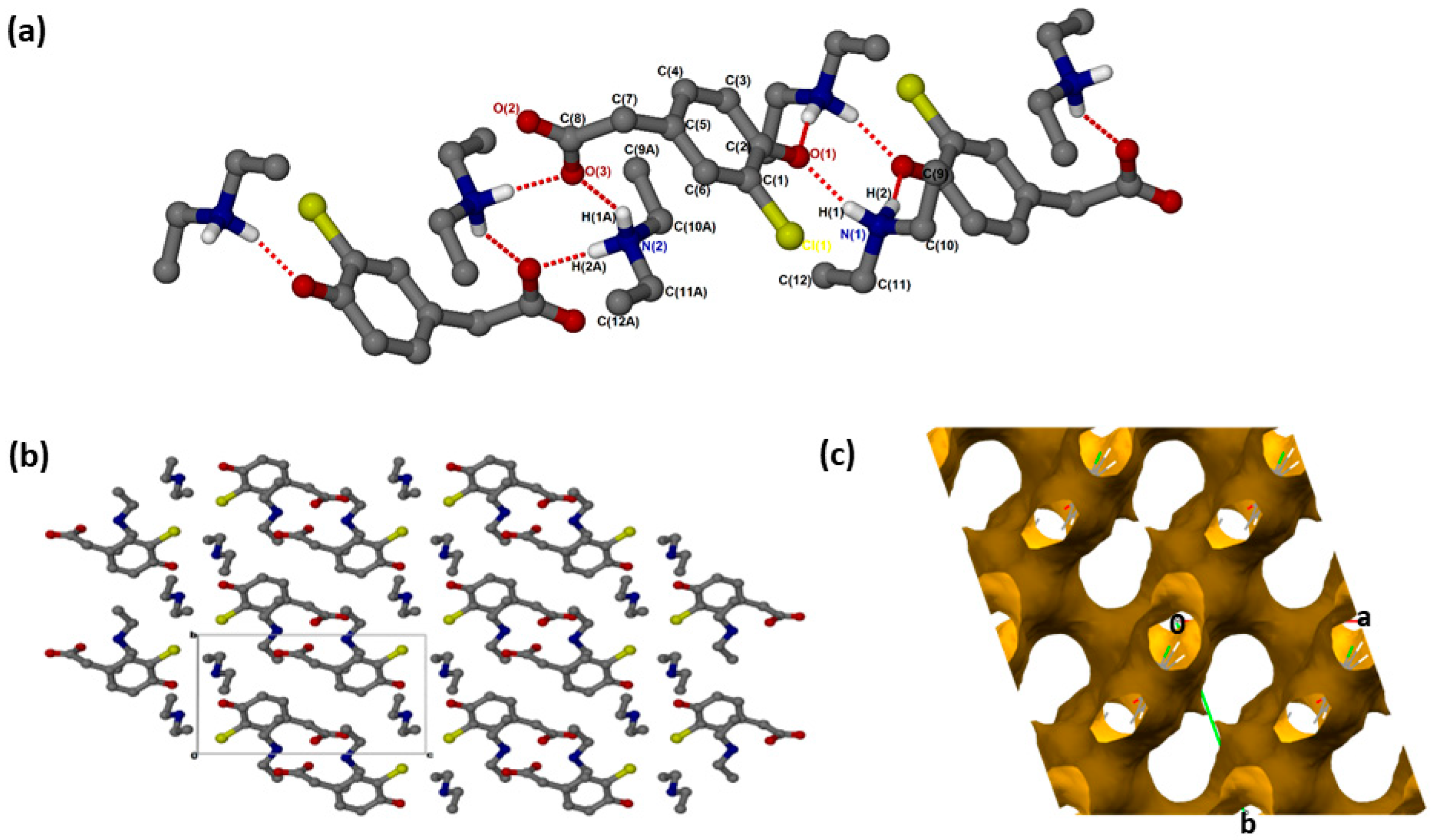
(crystallised in the triclinic space group PĪ, Z = 2 (Figure 1a). The crystal data is summarised in Supplementary Materials: Table S1. In addition to proton transfer from the carboxylic acid to the nitrogen of one of the DEA bases, the hydroxyl hydrogen of the CHPAA molecule was transferred to the nitrogen of the second DEA base. ( presents two major N-H···O intermolecular interactions, N1-H1···O1 and N2-H1A···O3, generating two rings which can be described in graph set notation as rings [21]. There are C-H···O contacts with d(C11···O2) of 3.4706(18) Å as well as contacts to the chlorine atom; C-H···Cl (d(C11···Cl1) = 3.821(2) Å) and N-H···Cl (d(N1···Cl1) = 3.4818(16) Å). The hydrogen bond metrics are summarised in Table S2. The chlorine atoms of adjacent CHPAA2− anions point directly towards each other, forming a type I halogen bond with d(Cl···Cl) = 3.517(2) Å and an angle of 174° (Figure 1b). The Cl···Cl distance in ( is close to twice the value of the van der Waals radius of Cl (1.76 Å) [22]. Two models have been proposed for Cl···Cl interactions: the Nyburg model [23,24] and William’s model [25]. Subsequently, the nature of Cl···Cl interactions has been the subject of much debate [26,27,28]. The Cl···Cl distance in ( is also similar to those found in substituted 2-chloroquinoline derivatives [29]. The DEA+ cations occupy channels (Figure 1c) within the crystal structure; the voids were calculated using Mercury, with a probe radius of 1.2 Å, and found to constitute 62.6% of the unit cell.
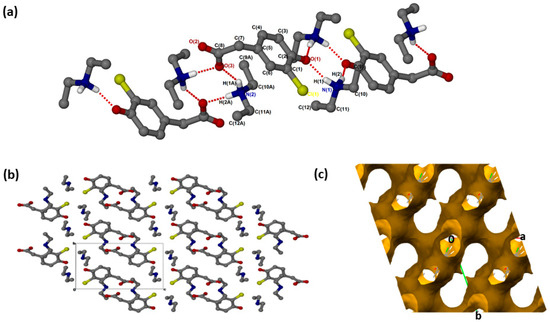
Figure 1.
(a) Hydrogen bonding in (, (b) Packing diagram along [100] and (c) removed to show voids along [001].
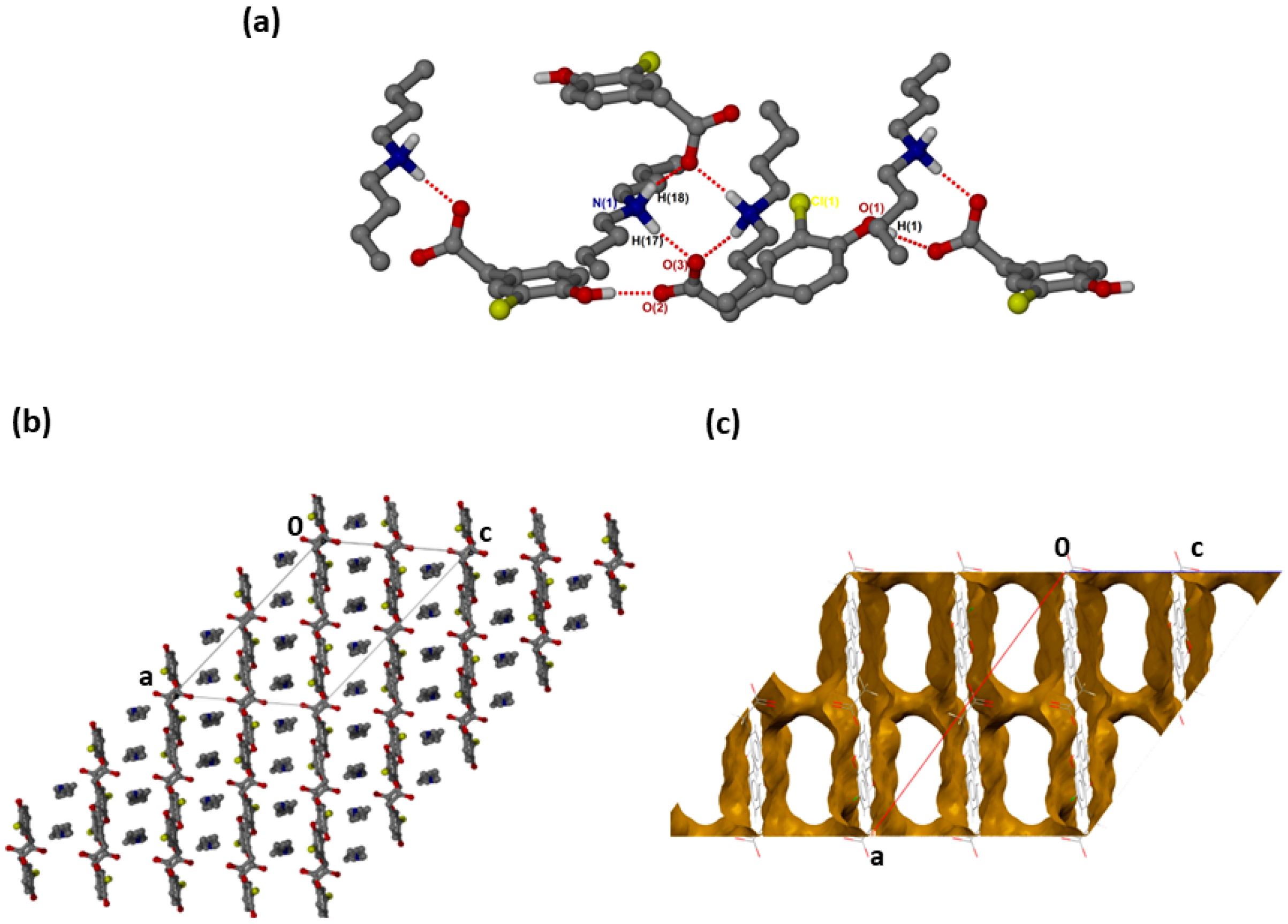
was successfully solved in the monoclinic space group C2/c with Z = 8 (Figure 2a). Both N-H groups are involved in hydrogen bonding to a carboxylate forming rings. The CHPAA− hydroxyl group also forms O-H···O hydrogen bonds to neighbouring carboxylate oxygens, generating a chain with a graph set notation of. The closest contacts to the chlorine atom are from the alkyl chain of with d(C10···Cl) of 3.7662(15) Å and d(C16···Cl) of 3.6809(17) Å. CHPAA− is oriented to form chains, allowing the DBM+ cations to accommodate in channels (Figure 2b). The void space occupied by DBM+ cations is 59.7% of the unit cell (Mercury, probe radius 1.2 Å).
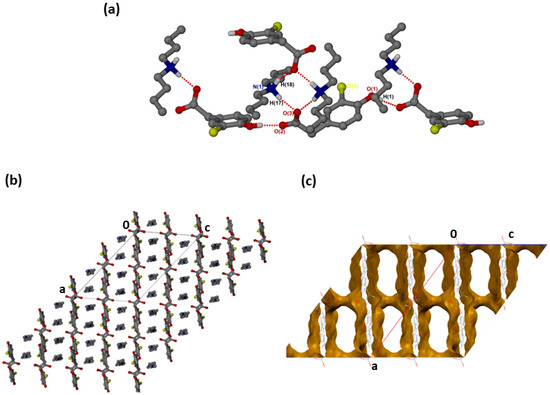
Figure 2.
(a) Hydrogen bonding in (CHPAA−)(DBM+), (b) Packing diagram along [010] and (c) DBM+ removed to show voids along [010].
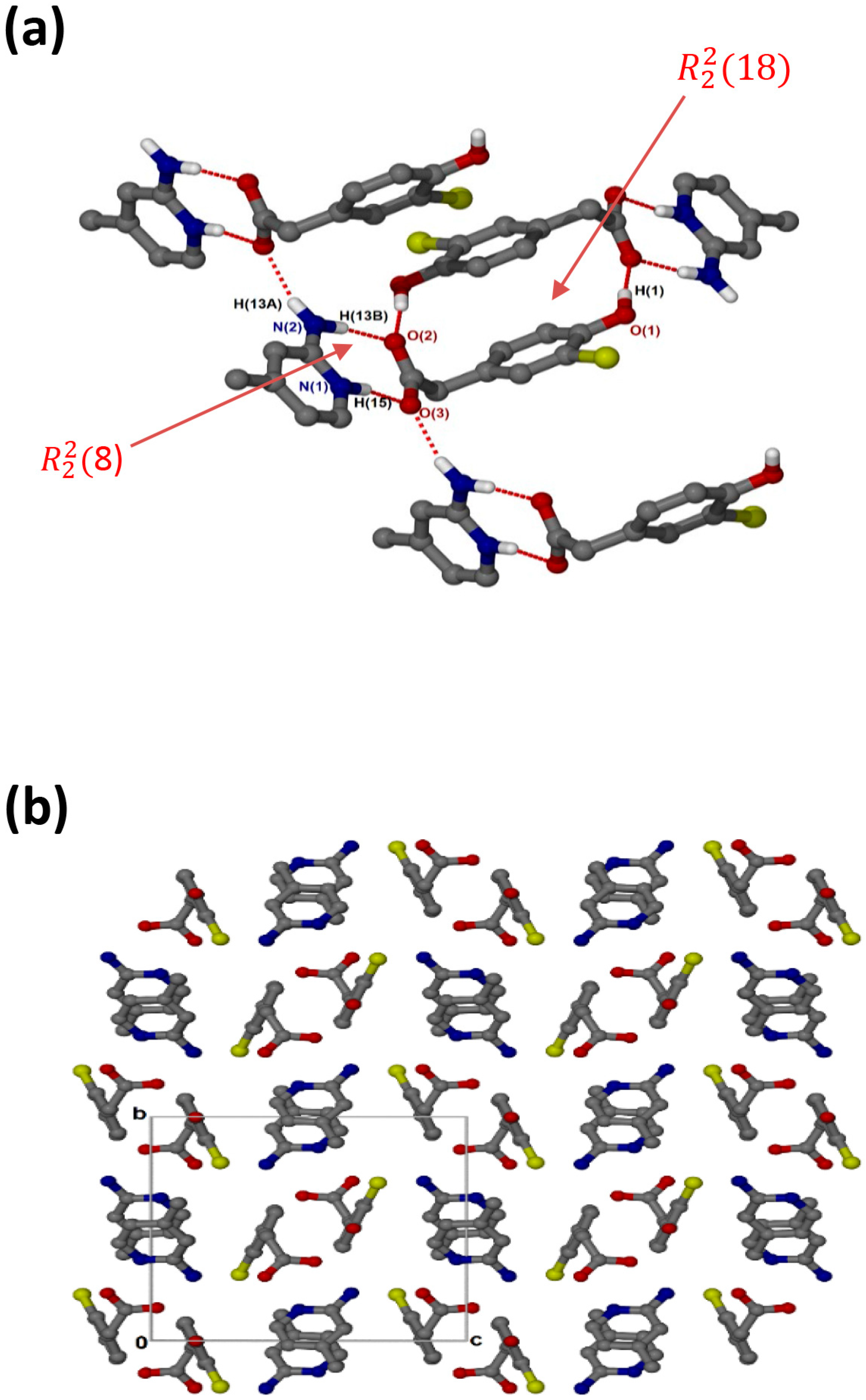
crystallised in the triclinic space group PĪ, Z = 2 (Figure 3a). There are N-H···O hydrogen bonds, as well as O-H···O linkages between the hydroxyl group and neighbouring carboxylate oxygens. The cations and the carboxylate oxygens form aminopyridinium carboxylate heterosynthon motifs with the hydrogen bonding extended to include the pyridinium nitrogen, which forms additional motifs. There are weak contacts C12-H12···π (3.452(1) Å), C6-H6···π (3.857(1) Å) and C10-H10···Cl (3.7854(17) Å). As seen for (, C-H···O contacts (d(C10···O1) = 3.367(2) Å and (d(C9···O3) = 3.461(2) Å) are also present. In the packing diagram below (Figure 3b), the cations and anions are arranged in alternate columns with the packing stabilised by π–π stacking between CHPAA− anions (3.639(1) Å) and between 2AMP+ cations (3.835(1) Å).
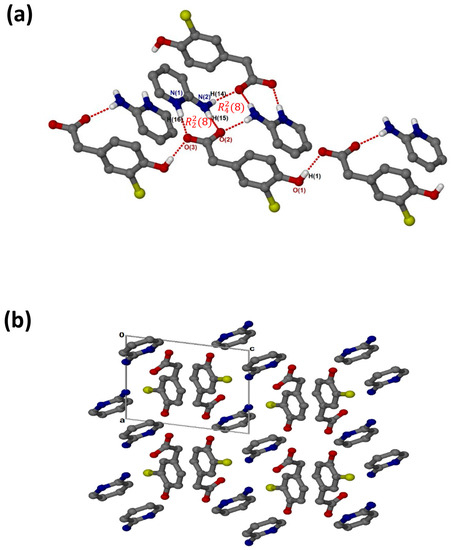
Figure 3.
(a) Hydrogen bonding in and (b) Packing diagram along [010].
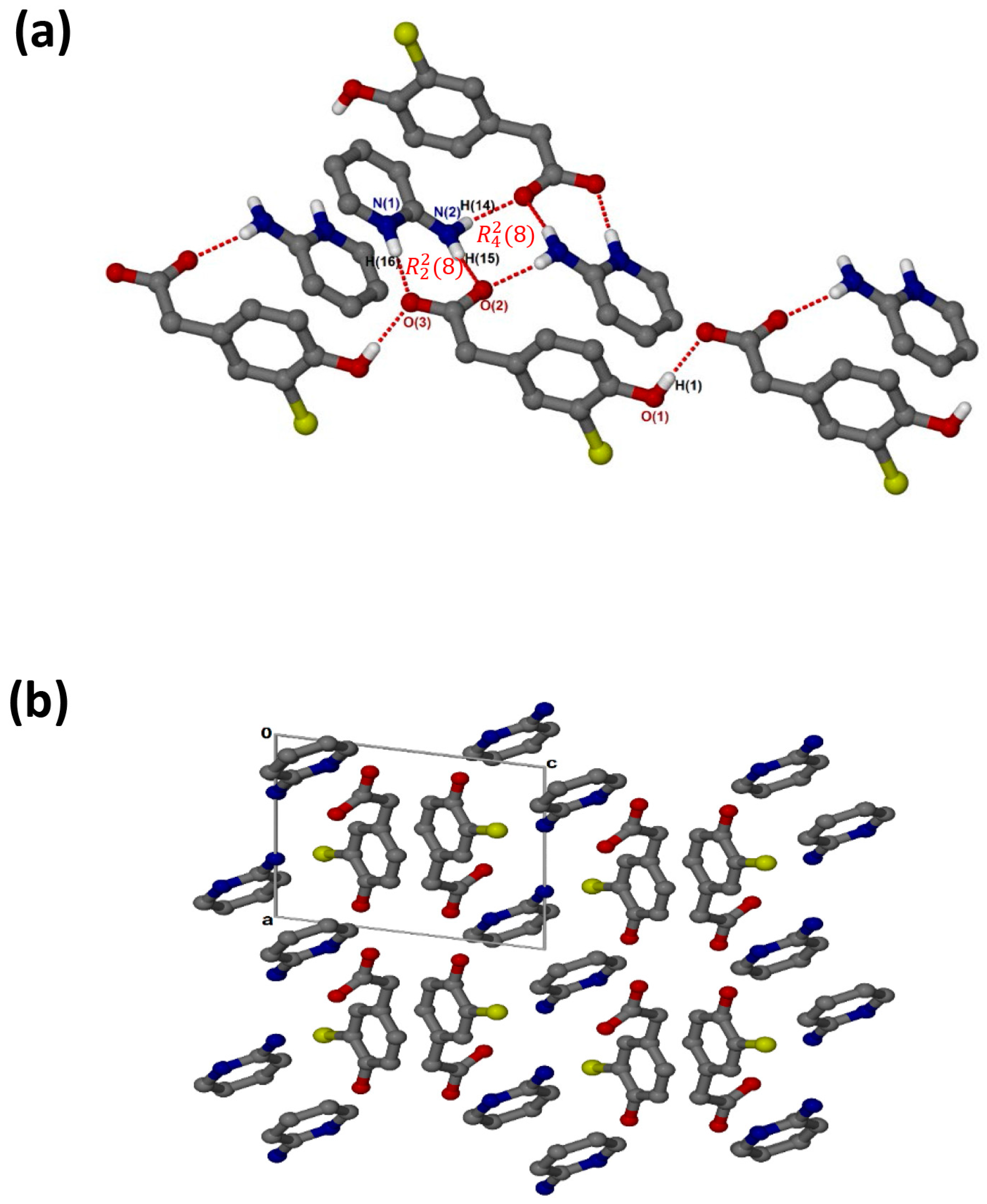
crystallises in the monoclinic space group P21/c and its asymmetric unit contains one anion and one cation (Z = 4). The 1:1 salt of (, shows that the carboxylate and aminopyridinium interactions also form the supramolecular heterosynthon. However, in this structure (Figure 4a), the acid anions dimerise to form the homosynthon involving the hydroxyl group and carboxylate oxygen (O-H···O interaction). Furthermore, the hydrogen anti-oriented (H13A) on the primary amine forms an additional hydrogen bond, allowing the structure to generate an extended network system described as chains of and motifs via N-H···O interactions. The chlorine atom also interacts with one of the carbons of the cation, forming a weak C3-H3···Cl1 interaction. This generated the closest contact of 3.5088(16) Å for d(C3···Cl1) with the angle of 99.9°. There are also C-H···O contacts with C14-H14···O1 (3.405(2) Å) and C7-H7A···O2 (3.527(2) Å). Columns of cations and anions (Figure 4b) are arranged in an alternating fashion along [100]. The packing is further stabilised by π–π stacking, including acid–acid (3.602(1) Å) and cation–cation (3.746(1) Å).
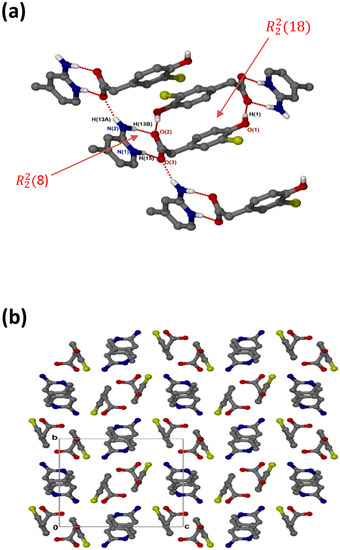
Figure 4.
(a) Hydrogen bonding in and (b) Packing diagram along [100].
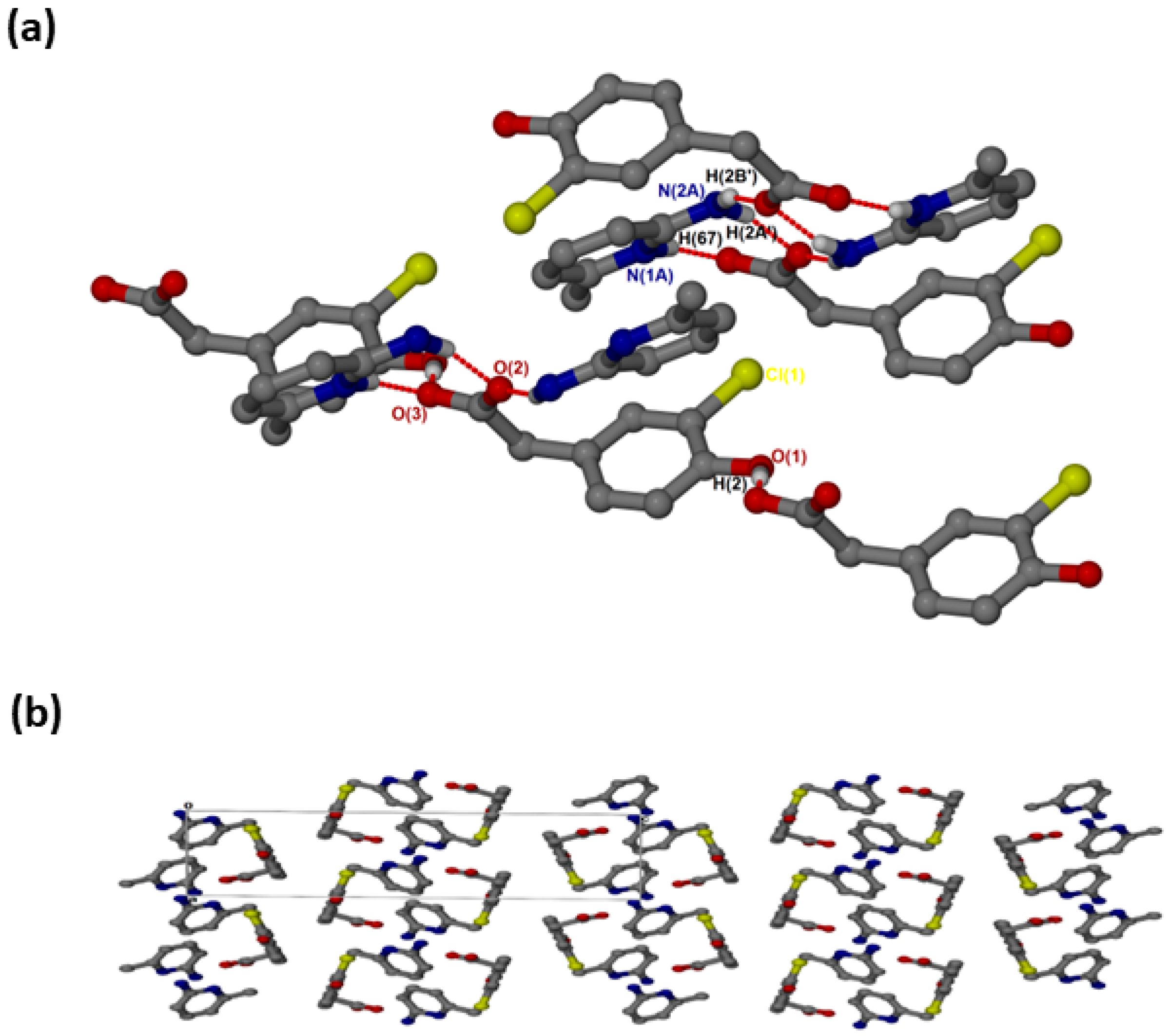
crystallised in the triclinic space group PĪ and its asymmetric unit contains two anions and two cations (Z = 4). In the 2:2 salt of , the anions interact with via supramolecular heterosynthons (Figure 5a) consisting of and rings. The hydroxyl group is also involved in O-H···O interactions with the carboxylate oxygen of neighbouring anions forming chains. A weak C9A-H9A’···Cl intermolecular interaction (3.823(4) Å) was also observed. The packing diagram (Figure 5b) shows columns of cations located between two columns of anions parallel to the a-axis. The π-π stacking interactions between cations (4.433(1) Å) are much weaker in this structure compared to and . C-H···π contacts between cations and the aromatic ring of the anion are also present with C13-H13···π (3.420(1) Å, 145°) and C13A-H13A···π (3.440(1) Å, 145°). There are also C-H···O contacts: C9A-H9A’···O1 (3.507(4)) Å and C9-H9A···O1A (3.523(5) Å).
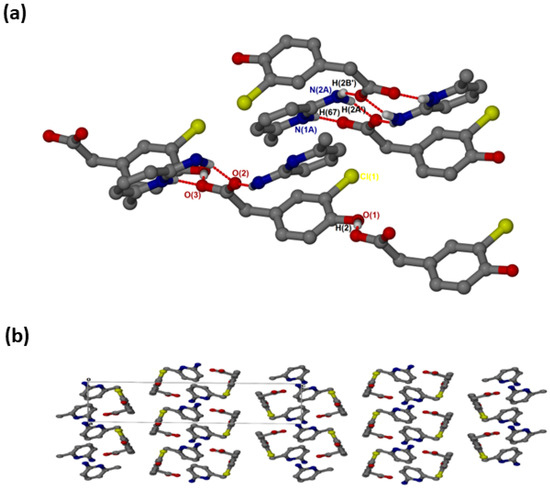
Figure 5.
(a) Hydrogen bonding in and (b) Packing diagram along [100].
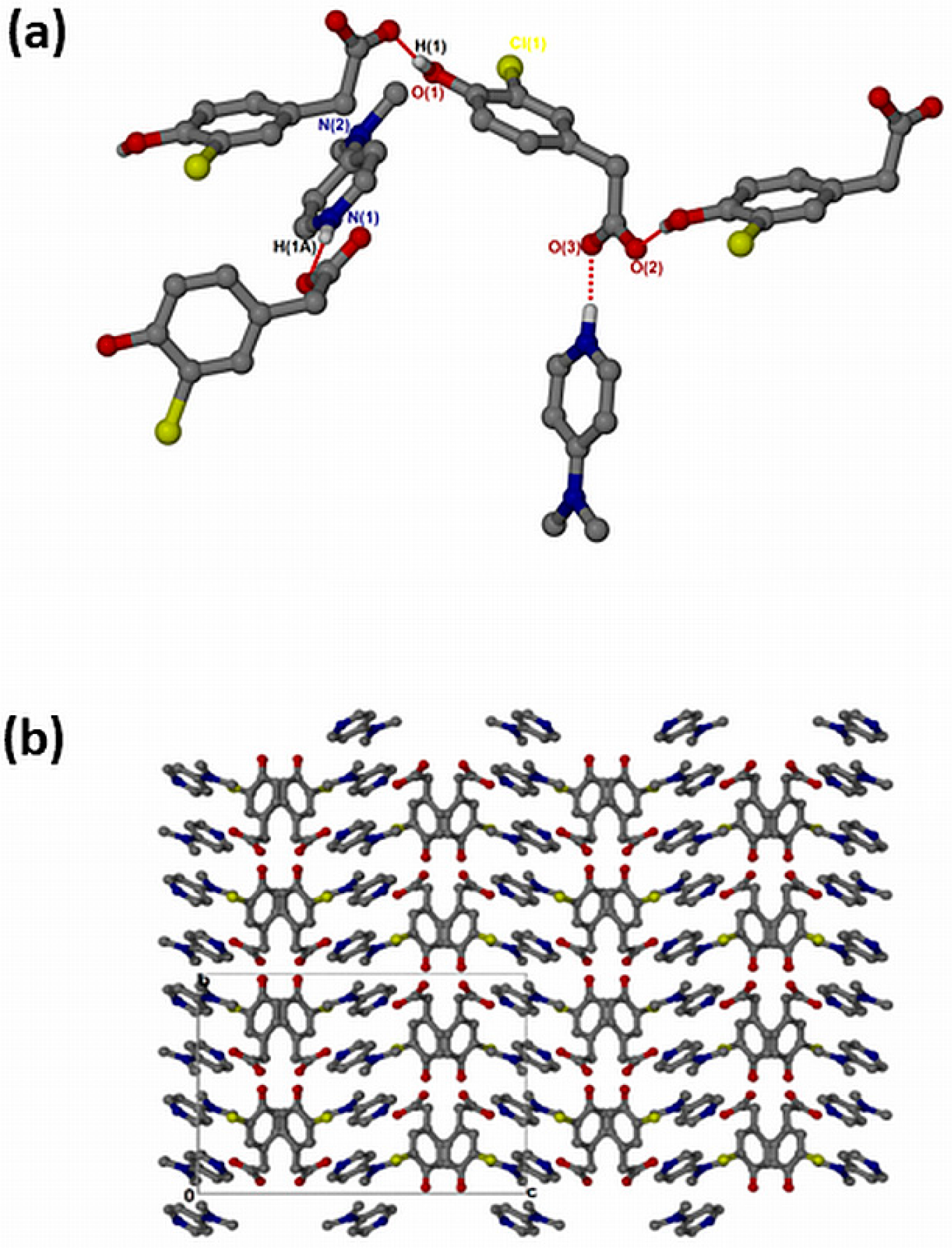
The salt of 4-dimethylaminopyridinium-3-chloro-4-hydroxyphenylacetate, , crystallised in the orthorhombic space group Pbca and its asymmetric unit contains one anion and one cation (Z = 8). The 1:1 salt of forms intermolecular interactions via N-H···O. The neighbouring hydroxyl group also interacts with the free O2 of another carboxylate forming extended columns with a motif via O-H···O interactions (Figure 6a). The structure does not contain any hydrogen-bonded ring motifs as was seen in the previous structures due to the absence of the NH2 group in DMAP. There are π–π stacking interactions between adjacent cations (3.807(1) Å). The structure also contains C-H···Cl contacts ranging from 3.474(3) Å for C13-H13···Cl1 to 3.846(4) for C15-H15B···Cl1 and C-H···O contacts range from 3.281(3) Å for C10-H10···O1 to 3.386(3) for C7-H7B···O1. The packing is shown in Figure 6b, highlighting a columnar arrangement parallel to the b-axis.
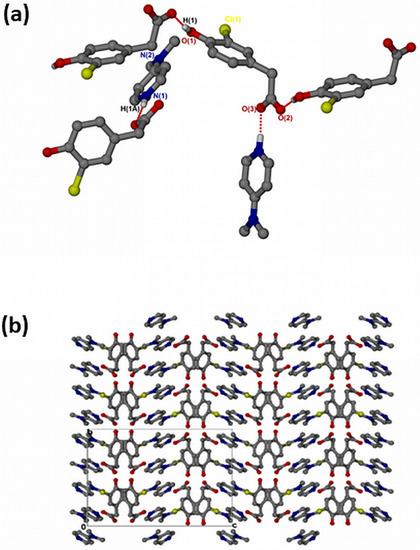
Figure 6.
(a) Hydrogen bonding in and (b) Packing diagram along [010].
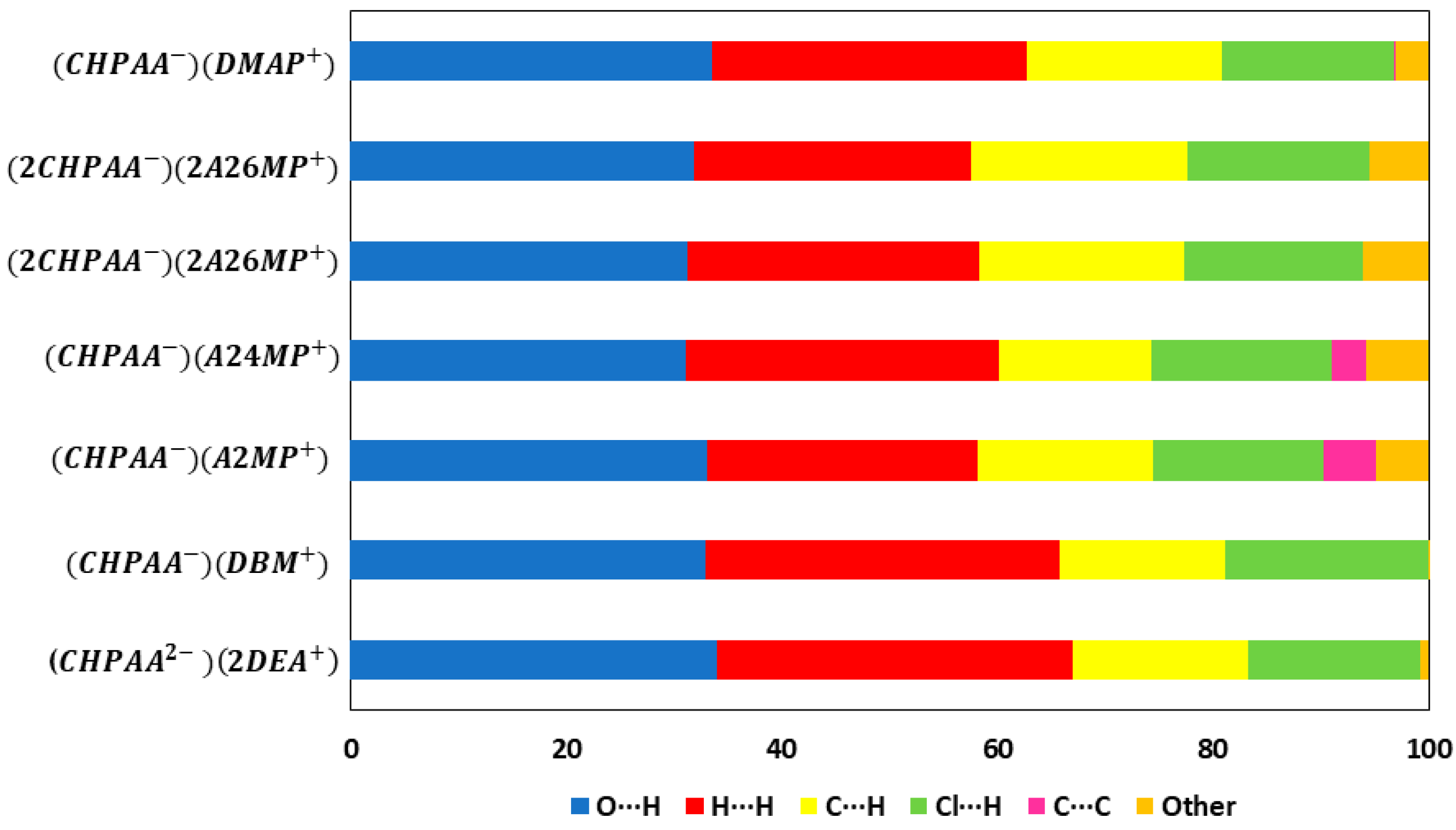
In summary, ( is the only structure that presents with a Cl···Cl contact and an N-H···Cl contact. All the structures exhibited C-H···Cl contacts with some longer than the van der Waals cut off, which is not unusual for weak interactions [5]. The findings are consistent with those found in a previous study involving CHPAA with nicotinamide, isonicotinamide, phenazine and 4,4′-bipyridine where none displayed Cl···Cl contacts and three of the structures contained C-H···Cl contacts. Crystal structures of the related compound, 2,4-dichlorophenoxyacetic acid with imidazole, 2-aminopyridine, 3-aminopyridine, isonicotinamide and pyrazinamide displayed similar C-H···Cl contacts [7]. Roy et al. studied salts of dicyclohexylamine with several substituted phenylacetic acids, including 2-, 3- and 4-chlorophenylacetic acid. In their supporting information, the authors reported the hydrogen bond metrics and tabulated N-H···O and C-H···O contacts for the 2-chlorophenylacetate and the 3-chlorophenylacetate salts. For the 4-chlorophenylacetate salt, the authors noted N-H···O as well as a C-H···Cl and a Cl···Cl contact [30]. N-H···O hydrogen bonds dominate for all the CHPAA structures and O-H···O for all except in the case of (. All the structures except for (CHPAA−)(DBM+) displayed C-H···O interactions. The O-H···O and N-H···O hydrogen bonds gave contact distances with average values for d(H···O) = 1.79 Å, d(H···N) = 1.89 Å, d(O···O) = 2.62 Å and d(N···O) = 2.78 Å. These were followed by the C-H···O and C-H···Cl contacts which gave average values of d(H···O) = 2.55 Å, d(H··· Cl) = 3.01 Å, d(C···O) = 3.44 Å and d(C···Cl) = 3.72 Å.
2.2. Hirshfeld Analysis
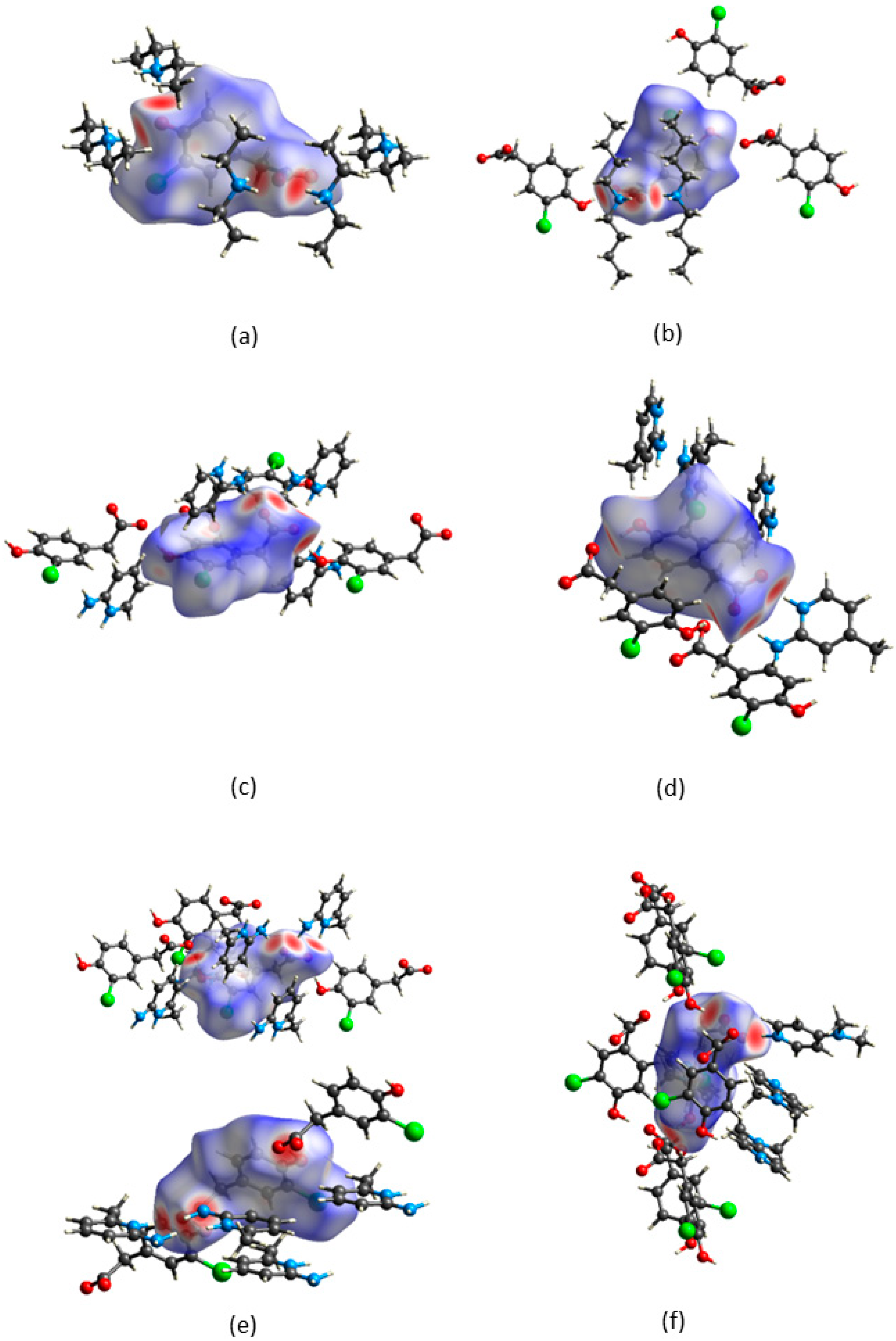
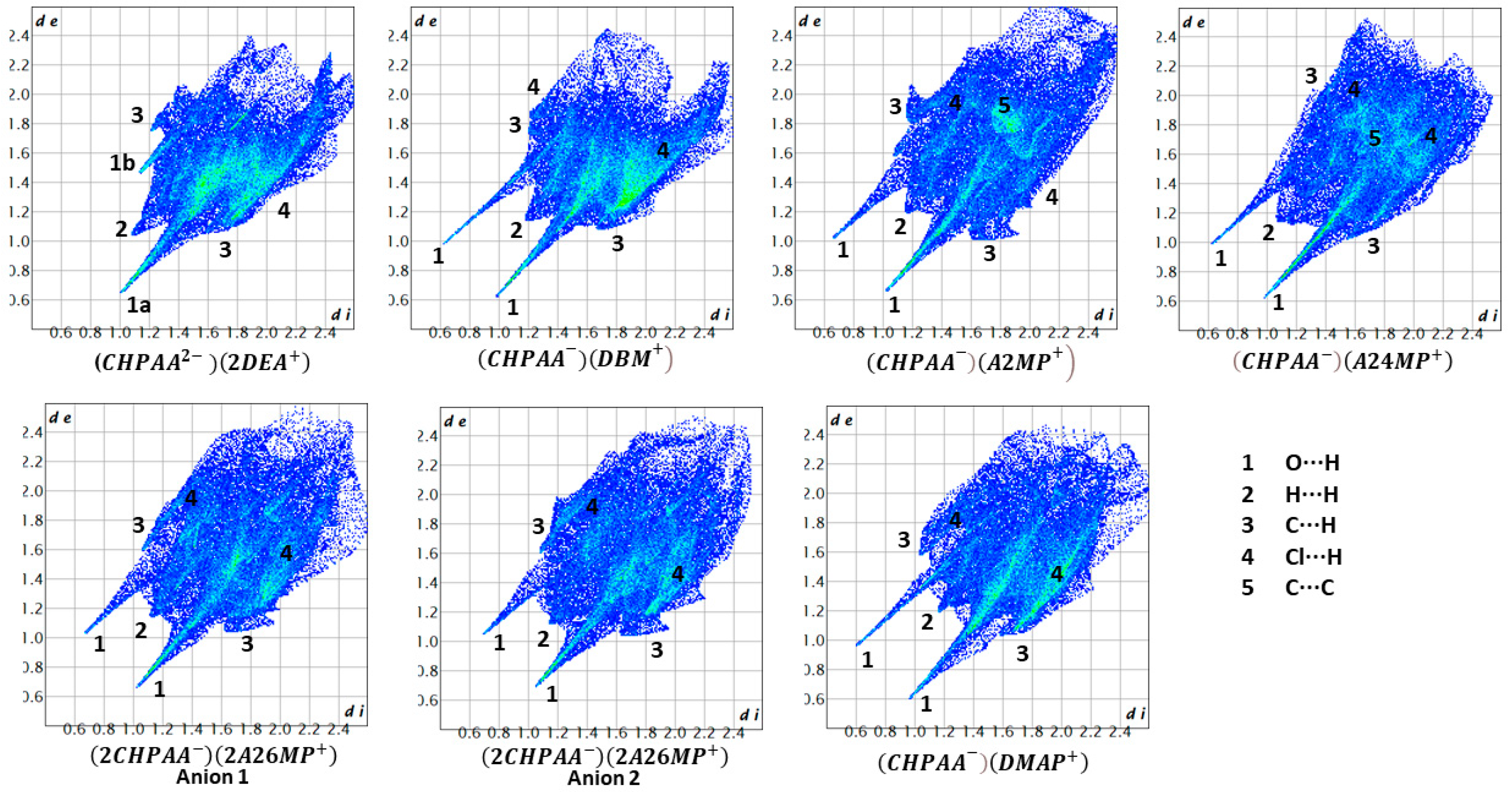
CrystalExplorer [31] was employed to generate Hirshfeld surfaces mapped with dnorm of the 3-chloro-4-hydroxyphenylacetate anion. Close contacts shorter than the sum of the van der Waals radii are highlighted in red, and the molecules involved are also represented (Figure 7). The bright red regions are associated with N-H···O and O-H···O contacts, whereas the lighter red areas are due to C-H···O contacts. The 2D fingerprint plots are shown in Figure 8. The main features of the packing for all the structures involve O···H, H···H, C···H and Cl···H contacts. The highest percentages were found for O···H (31.1–33.9%) and H···H (25.2–33%). The dominance of O···H contacts is consistent with the presence of N-H···O, O-H···O and C-H···O hydrogen bonds in the structures. Although there are no O-H···O hydrogen bonds in ( due to the deprotonation of both the carboxylic acid and the free hydroxyl, there are several N-H···O and C-H···O hydrogen bonds in this structure. In the 2D fingerprint plot for (, the long spike labelled 1a is due to contacts between N-H of the DEA+ and the oxygen of CHPAA2−. Contacts between C-H of CHPAA2− and oxygen outside the Hirshfeld surface result in the short spike labelled 1b. For all the other structures, the two spikes labelled 1 have similar lengths due to the presence of the free hydroxyl group resulting in reciprocal O···H contacts. All the structures displayed significant C···H (14.1–20%) and Cl···H (15.8–18.8%) contacts. There are prominent wings on the fingerprint plots forand due to C-H···π interactions. C···C contacts were observed for : 4.8% and : 3.2% which is attributed to the π···π stacking between CHPAA− anions in these structures. Only ( displayed Cl···Cl contacts, although it was a very small percentage (0.8%). This is consistent with CHPAA containing one chlorine atom. A quantitative summary of the contributions is given in Table S3 and Figure 9.
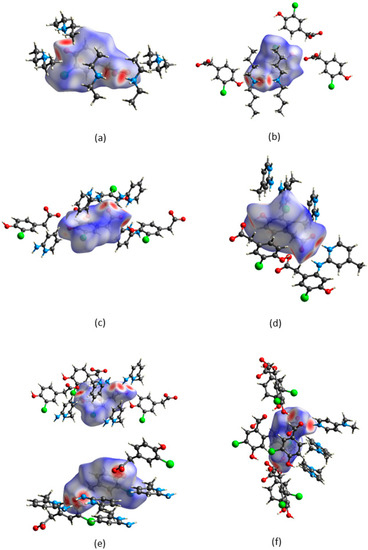
Figure 7.
Hirshfeld surfaces mapped over dnorm for (a) (CHPAA2−)(2DEA+), (b) (CHPAA−)(DBM+), (c) (CHPAA−)(A2MP+), (d) (CHPAA−)(A24MP+), (e) (2CHPAA−)(2A26MP+) and (f) (CHPAA−)(DMAP+).
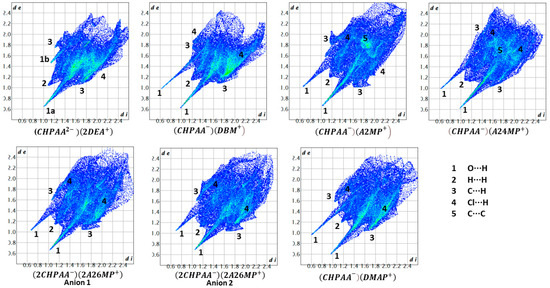
Figure 8.
Fingerprint plots for the different salts.
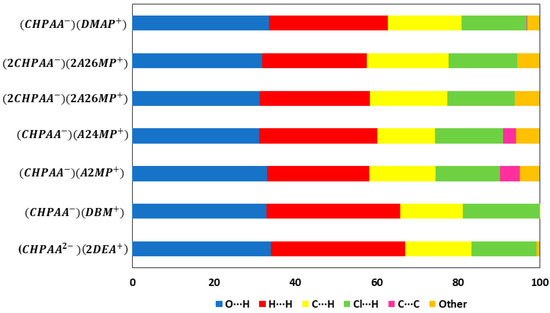
Figure 9.
Graph showing the percentage contributions for various contacts in the different salt structures.
2.3. Thermal Analysis
Differential scanning calorimetry (DSC) results are presented for all the salts, and thermogravimetric analysis (TGA) was completed for (and The thermal analysis curves are given in Figure S1, and the results are in Tables S4 and S5. Two mass loss steps were observed for (. The first mass loss of 10.5% observed at 315 K is due to the loss of ½DEA, and the second mass loss at 396 K for the remaining DEA overlaps with the decomposition of CHPAA. The DSC curve of ( shows two endothermic peaks. The first peak at 316 K corresponds to the partial decomposition of DEA and is in agreement with the TGA result. The second peak at 391 K is due to the melt of CHPAA and the loss of the remaining DEA. For , a continuous mass loss was observed at 400 K due to the loss of DBM and the melt of the CHPAA. The higher temperature at which DBM was released correlates to its higher normal boiling point (433 K) compared to DEA (normal boiling point 329 K). The remaining salts involving the aromatic amines had melting points higher than the starting materials. melt occurred at = 423.3 K (CHPAA: = 378.3 K and A2MP: = 331.4 K). For the melting point is = 410.7 K (A24MP: = 365.8 K). In the case of , = 414.8 K (A26MP: = 306.7 K). showed a melting peak at = 427.6 K (DMAP+: Tonset = 383.9 K). Thus, salt formation with the aromatic amines resulted in increased thermal stability with the highest melting point observed for .
2.4. Infrared Spectroscopy
FTIR spectroscopy is often used in screening new solid forms to distinguish between co-crystal and salt formation [32,33]. It is particularly useful in the study of carboxylic acids as after salt formation, the characteristic -OH stretching band of the -COOH group will shift to form COO− stretching bands. The CHPAA spectrum displayed a stretching peak at around 3431 cm−1 that is assigned to the carboxylic acid-free OH. This peak is absent or shifted in all the new solid forms, which is indicative of salt formation. CHPAA also contains one major stretching peak at 1694 cm−1 assigned to C=O. This peak was absent in all the new solid forms. The peaks at 1545–1571 cm−1 in the IR spectra of salts with aromatic amines were assigned to the COO− group. In the salts formed from linear amines, the peak due to the carboxylate appeared at approximately 1640 cm−1. Furthermore, the band due to the NH stretch in A2MP (3444 cm−1), A24MP (3429 cm−1) and A26MP (3461 cm−1) either disappeared or diminished in the IR spectra of the resulting salts. This can be attributed to the −NH2 group’s involvement in hydrogen bonding in the A2MP, A24MP and A26MP salts. The FTIR spectra are given in Figures S2 and S3.
2.5. Grinding and Slurry Experiments
PXRD analyses of all compounds obtained from the grinding and slurry experiments were performed to determine whether the salts obtained by the slow evaporation technique can also be prepared using other techniques. The calculated PXRD patterns obtained from LAZYPULVERIX [34] were compared to the PXRD of the CHPAA starting material, as well as those obtained from the various preparation methods (Figure S4). The calculated PXRD pattern of ( was a good match with the slurry PXRD pattern. The PXRD pattern of the ground material still contained peaks found in the starting material, indicating that the reaction was incomplete. For CHPAA and DBM, the PXRD patterns of the slurry and the ground product were a perfect match, with additional peaks present compared to that of the calculated pattern, indicating an incomplete reaction. All PXRD patterns of the resulting salts of CHPAA with A2MP, A24MP and A26MP were similar to those of the calculated PXRD patterns, indicating that the slurry and ground preparations were successful in forming the required salts. In the case of CHPAA and DMAP, the grinding and slurry PXRD patterns were similar but contained additional peaks not found in the calculated PXRD pattern, which can be ascribed to a partial reaction.
3. Materials and Methods
All chemicals were purchased from Sigma Aldrich (Schnelldorf, Germany) and were used as received.
3.1. Crystallisation
( and : CHPAA (30 mg, 0.161 mmol) was dissolved in 3 mL of the respective amine and a few drops of ethanol with heating to form solutions. The solutions were sealed and kept at room temperature to evaporate. ( formed yellow-green crystals and presented as dark yellow crystals after a few weeks.
: CHPAA (30 mg, 0.161 mmol) and A2MP (15.13 mg, 0.161 mmol) were dissolved in ethanol and a few drops of chloroform to form dilute solutions with heating. The solutions were sealed and kept at room temperature to evaporate. After 1 week, dark yellow crystals were obtained.
, : CHPAA (30 mg, 0.161 mmol) and the respective amine (17.385 mg, 0.161 mmol) were dissolved in excess ethanol with heating to form solutions. Slow evaporation at room temperature resulted in yellow-green crystals for and colourless crystals for .
: CHPAA (30 mg, 0.161 mmol) and DMAP (19.633 mg, 0.161 mmol) were dissolved in excess ethylacetate with heating. Yellow-green crystals were obtained after one week of slow evaporation at room temperature.
3.2. Grinding and Slurry Experiments
( and : CHPAA and the respective amine were mixed with the addition of a few drops of ethanol. The mixtures were manually ground for 15 min using a mortar and pestle. For the slurry experiments, 30 mg of CHPAA and an excess of amine, together with a few drops of ethanol, were heated until a solution was obtained. The solution was removed from the heat and stirred continuously until a powder formed. The resulting powder was filtered and left to dry at ambient temperature.
, and : The grinding and slurry experiments were performed in a similar manner to the above using equimolar mixtures of the acid and the amine.
3.3. Crystal Structure Analysis
Suitable crystals were selected for single crystal X-ray diffraction analysis at 173 (2) K. Diffraction data for all compounds were collected on a Bruker APEX II diffractometer (Bruker, Karlsruhe, Germany, with a graphite monochromated MoKα (λ = 0.71073 Å at 173 K). SADABS [35] was used to correct the intensities collected for absorption. The structures were solved by direct methods using SHELX-97 [36] and refined using full-matrix least squares methods in SHELXL. The graphical interface used was X-SEED [37]. All hydrogen atoms were placed geometrically with a riding model for their isotropic temperature factors except for those involved in hydrogen bonding, which were found in the electron density map and refined isotropically.
3.4. Powder X-ray Diffraction
A D2 PHASER Bruker diffractometer with Cu-Kα radiation (1.54184 Å) was used for PXRD. The voltage tube and current were at 30 kV and 10 mA max, respectively, with a scintillation counter 1-dim LYNXEYE detector. The scanning process of each sample was between 4–50° 2θ.
3.5. Infrared Spectroscopy
Spectra were obtained from the universal attenuated total reflectance (UTAR) infrared spectrometer Perkin Elmer spectrum two. Sample spectra were measured over the range of 4000–400 cm−1.
3.6. Thermal Analysis
DSC analyses were performed on a Perkin-Elmer 6 system with a purge of nitrogen at 20 mL min−1. These analyses were conducted from 303–573 K with a heating rate of 10 K min−1.
Samples of 2–5 mg were removed from the mother liquor, dried with filter paper, then crushed to a fine powder and placed in a vented pan for the DSC analysis.
4. Conclusions
Salts of CHPAA with selected linear and aromatic amines were successfully prepared. The formation of the new solid forms was validated using DSC analysis, and in the case of the aromatic amines, the resulting salts demonstrated enhanced thermal stability compared to the starting materials. FTIR spectroscopy confirmed salt formation. The salt structures of CHPAA, except for ( demonstrated N-H···O and O-H···O hydrogen bonds as the major interactions. In the case of (, N-H···O contacts dominated. All the structures exhibited C-H···Cl contacts, which shows the significance of this interaction in stabilizing the structures. C-H···O contacts were found in all the structures except for (CHPAA−)(DBM+). The Cl···Cl halogen bond was only found in (, which was also the only structure that presented with an N-H···Cl contact. CHPAA utilised all its hydrogen bond donor and acceptor sites in the formation of the salts.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules28196965/s1, Figure S1: TG and DSC curves for (a) (CHPAA2−)(2DEA+) and (b) (CHPAA−)(DBM+); DSC curves for (c) (CHPAA−)(A2MP+), (d) (CHPAA−)(A24MP+), (e) (2CHPAA−)(2A26MP+) and (f) (CHPAA−)(DMAP+); Figure S2: FTIR spectroscopy of (a) CHPAA, (b) (CHPAA2−)(2DEA+) and (c) (CHPAA−)(2DBM+); Figure S3: FTIR spectroscopy of (a) CHPAA, (b) A2MP, (c) (CHPAA−)(A2MP+), (d) A24MP, (e) (CHPAA−)(A24MP+), (f) A26MP, (g) (2CHPAA−)(2A26MP+)CHPAA), (h) DMAP and (i) (CHPAA−)(DMAP+); Figure S4: PXRD analyses for (a) (CHPAA2−)(2DEA+), (b) (CHPAA−)(DBM+), (c) (CHPAA−)(A2MP+), (d) (CHPAA−)(A24MP+), (e) (2CHPAA−)(2A26MP+) and (f) (CHPAA−)(DMAP+); Table S1: Summary of crystallographic data; Table S2: Summary of hydrogen bonds; Table S3: Summary of the various interactions; Table S4: DSC and TGA results of CHPAA linear salts; Table S5: DSC results of CHPAA aromatic salts and starting materials.
Author Contributions
Conceptualization, A.J.; methodology, A.J. and R.R.N.T.; software, A.J. and R.R.N.T.; formal analysis, A.J. and R.R.N.T.; investigation, R.R.N.T.; data curation, A.J.; writing—original draft preparation, A.J. and R.R.N.T.; writing—review and editing, A.J.; visualization, A.J. and R.R.N.T.; supervision, A.J.; project administration, A.J.; funding acquisition, A.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Research Foundation of South Africa, grant number 81245.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The supplementary data containing crystal data tables, hydrogen bonds, thermal analysis, FTIR spectroscopy and PXRD results can be accessed online at: https://www.mdpi.com/article/10.3390/molecules28196965/s1. The crystallographic data have been deposited with the CCDC, deposition numbers 2286796-2286801. These data can be obtained free of charge from the Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/data_request/cif.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Sample Availability
Samples of the compounds are not available from the authors.
References
- Wathoni, N.; Sari, W.A.; Elamin, K.M.; Mohammed, A.F.A.; Suharyani, I. A Review of Coformer Utilization in Multicomponent Crystal Formation. Molecules 2022, 27, 8693. [Google Scholar] [CrossRef]
- Putra, O.D.; Uekusa, H. Pharmaceutical Multicomponent Crystals: Structure, Design, and Properties. In Advances in Organic Crystal Chemistry; Springer: Berlin, Germany, 2020; pp. 153–184. [Google Scholar]
- Khan, R.A. Natural products chemistry: The emerging trends and prospective goals. Saudi Pharm. J. 2018, 26, 739–753. [Google Scholar] [CrossRef]
- Kim, S.; Lim, S.W.; Choi, J. Drug discovery inspired by bioactive small molecules from nature. Anim. Cells Syst. 2022, 26, 254–265. [Google Scholar] [CrossRef]
- Xiao, Y.; Wu, C.; Cui, P.; Zhou, L.; Yin, Q. Pursuing Green and Efficient Agriculture from Molecular Assembly: A Review of Solid-State Forms on Agrochemicals. J. Agric. Food Chem. 2023, 71, 10500–10524. [Google Scholar] [CrossRef]
- da Silva, A.K.; Diniz, L.F.; Tenorio, J.C.; Nazário, C.E.; Ribeiro, C.; Carvalho, P.S., Jr. Driving a sustainable application of s-triazine ametryn and atrazine herbicides through multicomponent crystals with improved solubility. CrystEngComm 2021, 23, 4252–4263. [Google Scholar] [CrossRef]
- Fang, L.; Xiao, Y.; Zhang, C.; Gao, Z.; Wu, S.; Gong, J.; Rohani, S. Intermolecular interactions and solubility behavior of multicomponent crystal forms of 2,4-D: Design, structure analysis, and solid-state characterization. CrystEngComm 2021, 23, 7615–7627. [Google Scholar] [CrossRef]
- Desiraju, G.R.; Steiner, T. The Weak Hydrogen Bond: In Structural Chemistry and Biology, IUCR Monographs on Crystallography; Oxford University Press: Oxford, UK, 1999; Volume 9. [Google Scholar]
- Desiraju, G.R. Supramolecular Synthons in Crystal Engineering—A New Organic Synthesis. Angew. Chem. 1995, 34, 2311–2327. [Google Scholar] [CrossRef]
- Etter, M.C. Encoding and decoding hydrogen-bond patterns of organic compounds. Acc. Chem. Res. 1990, 23, 120–126. [Google Scholar] [CrossRef]
- Shattock, T.R.; Arora, K.K.; Vishweshwar, P.; Zaworotko, M.J. Hierarchy of supramolecular synthons: Persistent carboxylic acid·pyridine hydrogen bonds in cocrystals that also contain a hydroxyl moiety. Cryst. Growth Des. 2008, 8, 4533–4545. [Google Scholar] [CrossRef]
- Bis, J.A.; Zaworotko, M.J. The 2-Aminopyridinium-carboxylate Supramolecular Heterosynthon: A Robust Motif for Generation of Multiple-Component Crystals. Cryst. Growth Des. 2005, 5, 1169–1179. [Google Scholar] [CrossRef]
- Jacobs, A.; Bàthori, N.B.; Nassimbeni, L.R.; Sebogisi, B.K. Salts of (+)-deoxycholic acid with amines: Structure, thermal stability, kinetics of salt formation, decomposition and chiral resolution. CrystEngComm 2013, 15, 931–939. [Google Scholar] [CrossRef]
- Davis, R.A.; Watters, D.; Healy, P.C. The isolation and synthesis of 3-chloro-4-hydroxyphenylacetamide produced by a plant-associated microfungus of the genus Xylaria. Tetrahedron Lett. 2005, 46, 919–921. [Google Scholar] [CrossRef]
- Kumar, R.; Sadowski, M.C.; Levrier, C.; Nelson, C.C.; Jones, A.J.; Holleran, J.P.; Avery, V.M.; Healy, P.C.; Davis, R.A. Design and Synthesis of a Screening Library Using the Natural Product Scaffold 3-Chloro-4-hydroxyphenylacetic Acid. J. Nat. Prod. 2015, 78, 914–918. [Google Scholar] [CrossRef]
- Davis, R.A.; Pierens, G.K.; Parsons, P.G. Synthesis and spectroscopic characterisation of a combinatorial library based on the fungal natural product 3-chloro-4-hydroxyphenylacetamide. Magn. Reson Chem. 2007, 45, 442–445. [Google Scholar] [CrossRef]
- Ngoma Tchibouanga, R.R.; Jacobs, A. 3-Chloro-4-hydroxyphenylacetic acid co-crystals with nicotinamide, isonicotinamide, phenazine and 4,4′-bipyridine: An investigation of synthon motifs. J. Mol. Struct. 2020, 1204, 127195. [Google Scholar] [CrossRef]
- Groom, C.R.; Bruno, I.J.; Lightfoot, M.P.; Ward, S.C. The Cambridge Structural Database. Acta Crystallogr. 2016, B72, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Chemicalize Was Used for the Prediction of pKa Values. Developed by ChemAxon. Available online: https://chemicalize.com/welcome (accessed on 28 August 2018).
- Cruz-Cabeza, A.J. Acid–base crystalline complexes and the pKa rule. CrystEngComm 2012, 14, 6362–6365. [Google Scholar] [CrossRef]
- Etter, M.C.; MacDonald, J.C.; Bernstein, J. Graph-set analysis of hydrogen-bond patterns. Acta Crystallogr. B 1990, 46, 256–262. [Google Scholar] [CrossRef]
- Bondi, A. van der Waals volumes and radii. J. Phys. Chem. 1964, 68, 441–451. [Google Scholar] [CrossRef]
- Nyburg, S.C.; Wong-Ng, W. Potential energy interactions in solid dichlorine. Inorg. Chem. 1979, 18, 2790–2791. [Google Scholar] [CrossRef]
- Nyburg, S.C.; Wong-Ng, W. Anisotropic atom–atom forces and the space group of solid chlorine. Proc. R. Soc. Lond. A. Math. Phys. Sci. 1979, 367, 29–45. [Google Scholar]
- Williams, D.E.; Hsu, L.Y. Transferability of nonbonded Cl⋯ Cl potential energy function to crystalline chlorine. Acta Crystallogr. Sect. A Found. Crystallogr. 1985, 41, 296–301. [Google Scholar] [CrossRef]
- Desiraju, G.R.; Parthasarathy, R. The nature of halogen…halogen interactions: Are short halogen contacts due to specific attractive forces or due to close packing of nonspherical atoms? J. Am. Chem. Soc. 1989, 111, 8725–8726. [Google Scholar] [CrossRef]
- Price, S.L.; Stone, A.J.; Lucas, J.; Rowland, R.S.; Thornley, A.E. The nature of -Cl···Cl- intermolecular interactions. J. Am. Chem. Soc. 1994, 116, 4910–4918. [Google Scholar] [CrossRef]
- Hathwar, V.R.; Guru Row, T.N. Nature of Cl ··· Cl Intermolecular Interactions via Experimental and Theoretical Charge Density Analysis: Correlation of Polar Flattening Effects with Geometry. J. Phys. Chem. A 2010, 114, 13434–13441. [Google Scholar] [CrossRef] [PubMed]
- Hathwar, V.R.; Roopani, S.M.; Subashini, R.; Khan, F.N.; Guru Row, T.N. Analysis of Cl…Cl and C–H…Cl intermolecular interactions involving chlorine in substituted 2-chloroquinoline derivatives. J. Chem. Sci. 2010, 122, 677–685. [Google Scholar] [CrossRef]
- Roy, R.; Adalder, T.K.; Dastidar, P. Supramolecular Gels Derived from the Salts of Variously Substituted Phenylacetic Acid and Dicyclohexylamine: Design, Synthesis, Structures, and Dye Adsorption. Chem. Asian J. 2018, 13, 552–559. [Google Scholar] [CrossRef]
- Spackman, P.R.; Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer: A program for Hirshfeld surface analysis, visualization and quantitative analysis of molecular crystals. J. Appl. Cryst. 2021, 54, 1006–1011. [Google Scholar] [CrossRef]
- da Silva, C.C.; Guimarães, F.F.; Ribeiro, L.; Martins, F.T. Salt or cocrystal of salt? Probing the nature of multicomponent crystal forms with infrared spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2016, 167, 89–95. [Google Scholar] [CrossRef]
- Chadha, R.; Saini, A.; Khullar, S.; Singh Jain, D.; Mandal, S.K.; Guru Row, T.N. Crystal Structures and Physicochemical Properties of Four New Lamotrigine Multicomponent Forms. Cryst. Growth Des. 2013, 13, 858–870. [Google Scholar] [CrossRef]
- Yvon, K.; Jeitschko, W.; Parthe, E. LAZY PULVERIX, a computer program, for calculating X-ray and neutron diffraction powder patterns. J. Appl. Cryst. 1997, 10, 73–74. [Google Scholar] [CrossRef]
- Sheldrick, G. SADABS, Version 2.05; University of Göttingen: Göttingen, Germany, 2007. [Google Scholar]
- Sheldrick, G. A short history of SHELX. Acta Crystallogr. A 2008, 64, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Barbour, L.J. X-Seed 4: Updates to a program for small-molecule supramolecular crystallography. J. Appl. Cryst. 2020, 53, 1141–1146. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).