The miRNA-185-5p/STIM1 Axis Regulates the Invasiveness of Nasopharyngeal Carcinoma Cell Lines by Modulating EGFR Activation-Stimulated Switch from E- to N-Cadherin
Abstract
1. Introduction
2. Results
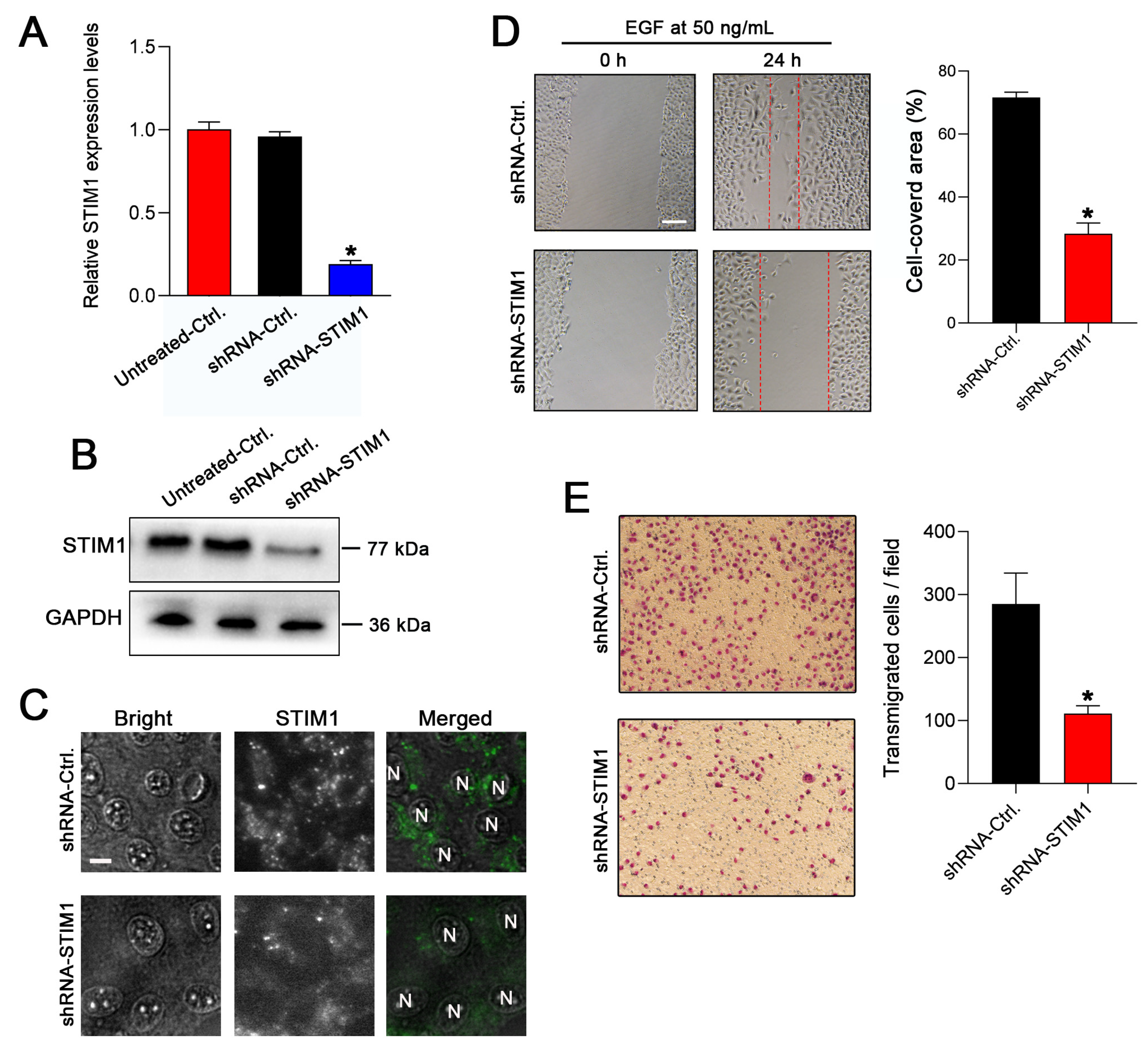
2.1. Knockdown of STIM1 Inhibits EGF-Induced Migration of 5–8F Cells
2.2. STIM1 Regulates EGFR Activation-Stimulated EMT
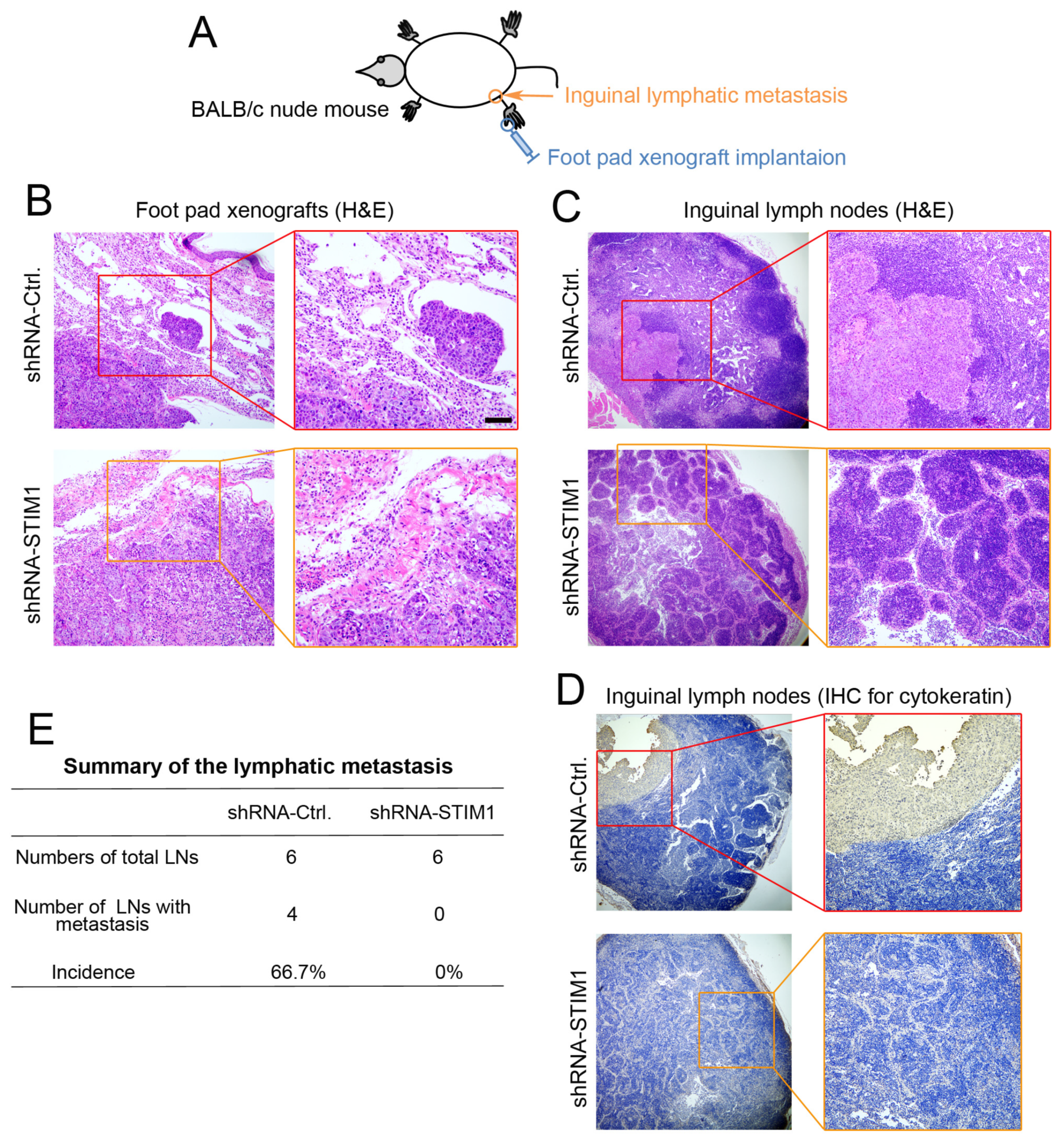
2.3. Knockdown of STIM1 Impairs Locoregional Lymphatic Metastasis
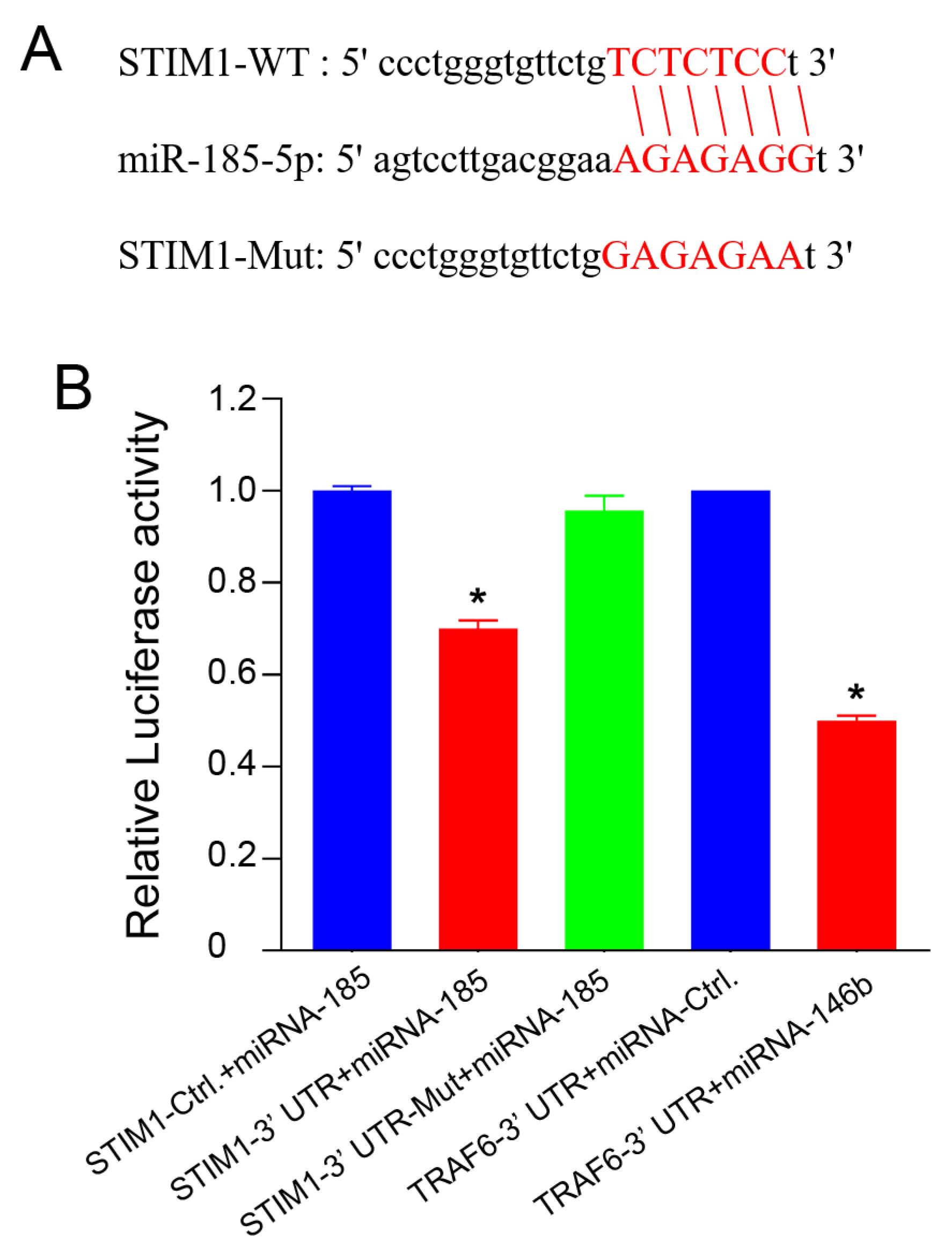
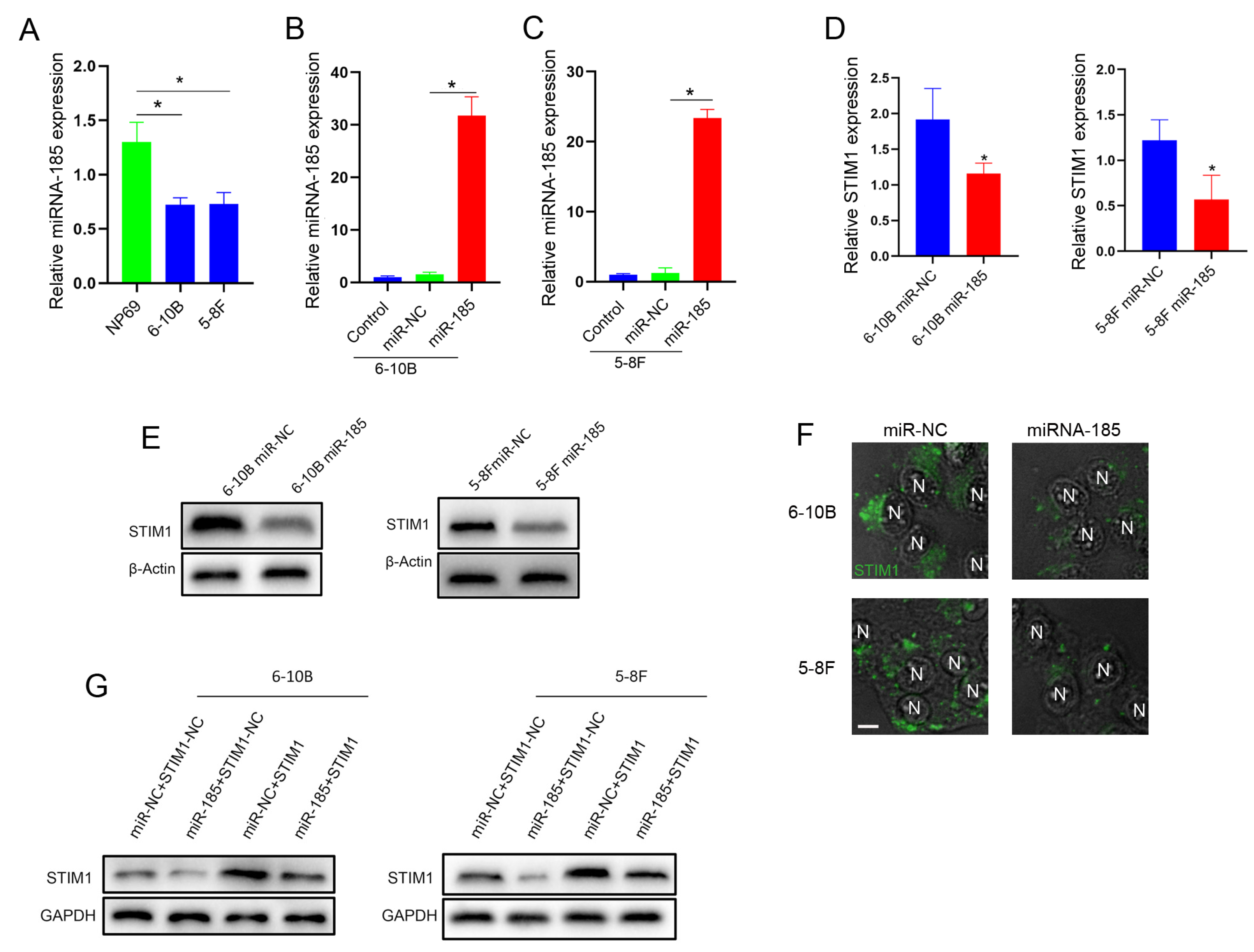
2.4. miRNA-185-5p Negatively Regulates STIM1 Expression
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Knockdown of STIM1
4.3. Wound Healing Assay
4.4. Transwell Assay
4.5. Western-Blot and Cell Immunofluorescence Staining
4.6. Inguinal Lymph Node Metastasis Model of Nude Mice
4.7. Dual-Luciferase Reporter Gene Assay
4.8. Construction of NPC Cell Lines Overexpressing miRNA-185 or Overexpressing Both miRNA-185 and STIM1
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Chen, Y.-P.; Chan, A.T.C.; Le, Q.-T.; Blanchard, P.; Sun, Y.; Ma, J. Nasopharyngeal carcinoma. Lancet 2019, 394, 64–80. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.C.W.; Hui, E.P.; Lo, K.W.; Lam, W.K.J.; Johnson, D.; Li, L.; Tao, Q.; Chan, K.C.A.; To, K.F.; King, A.D.; et al. Nasopharyngeal carcinoma: An evolving paradigm. Nat. Rev. Clin. Oncol. 2021, 18, 679–695. [Google Scholar] [CrossRef] [PubMed]
- Tsao, S.W.; Yip, Y.L.; Tsang, C.M.; Pang, P.S.; Lau, V.M.; Zhang, G.; Lo, K.W. Etiological factors of nasopharyngeal carcinoma. Oral Oncol. 2014, 50, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Tao, Q.; Chan, A.T. Nasopharyngeal carcinoma: Molecular pathogenesis and therapeutic developments. Expert Rev. Mol. Med. 2007, 9, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Kam, M.K.; Teo, P.M.; Chau, R.M.; Cheung, K.Y.; Choi, P.H.; Kwan, W.H.; Leung, S.F.; Zee, B.; Chan, A.T. Treatment of nasopharyngeal carcinoma with intensity-modulated radiotherapy: The Hong Kong experience. Int. J. Radiat. Oncol. Biol. Phys. 2004, 60, 1440–1450. [Google Scholar] [CrossRef]
- Ma, J.; Liu, L.; Tang, L.; Zong, J.; Lin, A.; Lu, T.; Cui, N.; Cui, C.; Li, L. Retropharyngeal lymph node metastasis in nasopharyngeal carcinoma: Prognostic value and staging categories. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2007, 13, 1445–1452. [Google Scholar] [CrossRef]
- Pastushenko, I.; Blanpain, C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol. 2019, 29, 212–226. [Google Scholar] [CrossRef] [PubMed]
- Bakir, B.; Chiarella, A.M.; Pitarresi, J.R.; Rustgi, A.K. EMT, MET, Plasticity, and Tumor Metastasis. Trends Cell Biol. 2020, 30, 764–776. [Google Scholar] [CrossRef]
- Yip, W.K.; Seow, H.F. Activation of phosphatidylinositol 3-kinase/Akt signaling by EGF downregulates membranous E-cadherin and beta-catenin and enhances invasion in nasopharyngeal carcinoma cells. Cancer Lett. 2012, 318, 162–172. [Google Scholar] [CrossRef]
- Lo, H.W.; Hsu, S.C.; Xia, W.; Cao, X.; Shih, J.Y.; Wei, Y.; Abbruzzese, J.L.; Hortobagyi, G.N.; Hung, M.C. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 2007, 67, 9066–9076. [Google Scholar] [CrossRef]
- Stathopulos, P.B.; Zheng, L.; Li, G.Y.; Plevin, M.J.; Ikura, M. Structural and mechanistic insights into STIM1-mediated initiation of store-operated calcium entry. Cell 2008, 135, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Ye, J.; Luo, Y.; Weng, J.; He, Q.; Liu, F.; Li, M.; Lin, Y.; Li, Y.; Zhang, Z.; et al. EB virus promotes metastatic potential by boosting STIM1-dependent Ca2+ signaling in nasopharyngeal carcinoma cells. Cancer Lett. 2020, 478, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.J.; Sun, Y.M.; Tian, D.F.; He, Y.C.; Zeng, L.; He, Y.; Ling, C.Q.; Sun, S.H. Downregulated NM23-H1 expression is associated with intracranial invasion of nasopharyngeal carcinoma. Br. J. Cancer 2008, 98, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Clapham, D.E. Calcium signaling. Cell 2007, 131, 1047–1058. [Google Scholar] [CrossRef]
- Putney, J.W., Jr. Capacitative calcium entry: Sensing the calcium stores. J. Cell Biol. 2005, 169, 381–382. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Pan, H.; Yao, J.; Zhou, Y.; Han, W. SOCE and cancer: Recent progress and new perspectives. Int. J. Cancer 2016, 138, 2067–2077. [Google Scholar] [CrossRef]
- Cui, C.; Merritt, R.; Fu, L.; Pan, Z. Targeting calcium signaling in cancer therapy. Acta Pharm. Sin. B 2017, 7, 3–17. [Google Scholar] [CrossRef]
- Smyth, J.T.; Dehaven, W.I.; Bird, G.S.; Putney, J.W., Jr. Ca2+-store-dependent and -independent reversal of Stim1 localization and function. J. Cell Sci. 2008, 121, 762–772. [Google Scholar] [CrossRef]
- Chen, Y.F.; Chiu, W.T.; Chen, Y.T.; Lin, P.Y.; Huang, H.J.; Chou, C.Y.; Chang, H.C.; Tang, M.J.; Shen, M.R. Calcium store sensor stromal-interaction molecule 1-dependent signaling plays an important role in cervical cancer growth, migration, and angiogenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 15225–15230. [Google Scholar] [CrossRef]
- Xia, J.; Wang, H.; Huang, H.; Sun, L.; Dong, S.; Huang, N.; Shi, M.; Bin, J.; Liao, Y.; Liao, W. Elevated Orai1 and STIM1 expressions upregulate MACC1 expression to promote tumor cell proliferation, metabolism, migration, and invasion in human gastric cancer. Cancer Lett. 2016, 381, 31–40. [Google Scholar] [CrossRef]
- Nguyen, N.T.; Han, W.; Cao, W.M.; Wang, Y.; Wen, S.; Huang, Y.; Li, M.; Du, L.; Zhou, Y. Store-Operated Calcium Entry Mediated by ORAI and STIM. Compr. Physiol. 2018, 8, 981–1002. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Wei, J.; Luo, Y.; Deng, Y.; Que, T.; Zhang, X.; Liu, F.; Zhang, J.; Luo, X. Epstein-Barr Virus Promotes Tumor Angiogenesis by Activating STIM1-Dependent Ca2+ Signaling in Nasopharyngeal Carcinoma. Pathogens 2021, 10, 1275. [Google Scholar] [CrossRef] [PubMed]
- Liou, J.; Kim, M.L.; Heo, W.D.; Jones, J.T.; Myers, J.W.; Ferrell, J.E., Jr.; Meyer, T. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. CB 2005, 15, 1235–1241. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.M.; Buchanan, J.; Luik, R.M.; Lewis, R.S. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J. Cell Biol. 2006, 174, 803–813. [Google Scholar] [CrossRef]
- Yang, N.; Tang, Y.; Wang, F.; Zhang, H.; Xu, D.; Shen, Y.; Sun, S.; Yang, G. Blockade of store-operated Ca2+ entry inhibits hepatocarcinoma cell migration and invasion by regulating focal adhesion turnover. Cancer Lett. 2013, 330, 163–169. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, X.; Feng, B.; Liu, N.; Wu, Q.; Han, Y.; Nie, Y.; Wu, K.; Shi, Y.; Fan, D. STIM1, a direct target of microRNA-185, promotes tumor metastasis and is associated with poor prognosis in colorectal cancer. Oncogene 2015, 34, 4808–4820. [Google Scholar] [CrossRef]
- Chen, Y.F.; Chen, Y.T.; Chiu, W.T.; Shen, M.R. Remodeling of calcium signaling in tumor progression. J. Biomed. Sci. 2013, 20, 23. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, J.J.; Huang, X.Y. Orai1 and STIM1 are critical for breast tumor cell migration and metastasis. Cancer Cell 2009, 15, 124–134. [Google Scholar] [CrossRef]
- Zhang, J.; Wei, J.; Kanada, M.; Yan, L.; Zhang, Z.; Watanabe, H.; Terakawa, S. Inhibition of store-operated Ca2+ entry suppresses EGF-induced migration and eliminates extravasation from vasculature in nasopharyngeal carcinoma cell. Cancer Lett. 2013, 336, 390–397. [Google Scholar] [CrossRef]
- Singh, M.; Yelle, N.; Venugopal, C.; Singh, S.K. EMT: Mechanisms and therapeutic implications. Pharmacol. Ther. 2018, 182, 80–94. [Google Scholar] [CrossRef]
- Lu, W.; Kang, Y. Epithelial-Mesenchymal Plasticity in Cancer Progression and Metastasis. Dev. Cell 2019, 49, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Miao, Y.; Zheng, X.; Gong, Y.; Zhang, J.; Zou, F.; Cai, C. STIM1 and STIM2 differently regulate endogenous Ca2+ entry and promote TGF-beta-induced EMT in breast cancer cells. Biochem. Biophys. Res. Commun. 2017, 488, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Lambert, P.F.; Rapraeger, A.C.; Anderson, R.A. Stress-Induced EGFR Trafficking: Mechanisms, Functions, and Therapeutic Implications. Trends Cell Biol. 2016, 26, 352–366. [Google Scholar] [CrossRef] [PubMed]
- Yewale, C.; Baradia, D.; Vhora, I.; Patil, S.; Misra, A. Epidermal growth factor receptor targeting in cancer: A review of trends and strategies. Biomaterials 2013, 34, 8690–8707. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, R.I.; Gee, J.M.; Harper, M.E. EGFR and cancer prognosis. Eur. J. Cancer 2001, 37, S9–S15. [Google Scholar] [CrossRef]
- Ciardiello, F.; Tortora, G. EGFR antagonists in cancer treatment. N. Engl. J. Med. 2008, 358, 1160–1174. [Google Scholar] [CrossRef]
- Mishra, S.; Yadav, T.; Rani, V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit. Rev. Oncol. /Hematol. 2016, 98, 12–23. [Google Scholar] [CrossRef]
- Chen, L.; Heikkinen, L.; Wang, C.; Yang, Y.; Sun, H.; Wong, G. Trends in the development of miRNA bioinformatics tools. Brief. Bioinform. 2019, 20, 1836–1852. [Google Scholar] [CrossRef]
- Yu, J.; Lei, R.; Zhuang, X.; Li, X.; Li, G.; Lev, S.; Segura, M.F.; Zhang, X.; Hu, G. MicroRNA-182 targets SMAD7 to potentiate TGFbeta-induced epithelial-mesenchymal transition and metastasis of cancer cells. Nat. Commun. 2016, 7, 13884. [Google Scholar] [CrossRef]
- Kim, J.; Yao, F.; Xiao, Z.; Sun, Y.; Ma, L. MicroRNAs and metastasis: Small RNAs play big roles. Cancer Metastasis Rev. 2018, 37, 5–15. [Google Scholar] [CrossRef]
- Annese, T.; Tamma, R.; De Giorgis, M.; Ribatti, D. microRNAs Biogenesis, Functions and Role in Tumor Angiogenesis. Front. Oncol. 2020, 10, 581007. [Google Scholar] [CrossRef]
- Fridrichova, I.; Zmetakova, I. MicroRNAs Contribute to Breast Cancer Invasiveness. Cells 2019, 8, 1361. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Lai, M. The microRNA network and tumor metastasis. Oncogene 2010, 29, 937–948. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Park, S.; Kim, H.; Choi, Y.J.; Kim, S.Y.; Sung, K.J.; Sung, Y.H.; Choi, C.M.; Yun, M.; Yi, Y.S.; et al. Tumor-derived exosomal miR-619-5p promotes tumor angiogenesis and metastasis through the inhibition of RCAN1. Cancer Lett. 2020, 475, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Liu, L.; Zhu, Q.; Wu, Y.; Tian, B.; Cui, L.; Liu, Y.; Li, X. MicroRNA-185 inhibits angiogenesis in human microvascular endothelial cells through targeting stromal interaction molecule. Cell Biol. Int. 2016, 40, 318–328. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Zhang, L.; Song, L.; Ma, Y.; Huang, B.; Liang, Q.; Zeng, Y. Differentially expressed gene in nasopharyngeal carcinoma cell lines with various metastatic potentialities. Zhonghua Zhong Liu Za Zhi 2002, 24, 430–434. [Google Scholar]
- Ren, X.; Yang, X.; Cheng, B.; Chen, X.; Zhang, T.; He, Q.; Li, B.; Li, Y.; Tang, X.; Wen, X.; et al. HOPX hypermethylation promotes metastasis via activating SNAIL transcription in nasopharyngeal carcinoma. Nat. Commun. 2017, 8, 14053. [Google Scholar] [CrossRef]
- Cao, S.; Cui, Y.; Xiao, H.; Mai, M.; Wang, C.; Xie, S.; Yang, J.; Wu, S.; Li, J.; Song, L.; et al. Upregulation of flotillin-1 promotes invasion and metastasis by activating TGF-β signaling in nasopharyngeal carcinoma. Oncotarget 2016, 7, 4252–4264. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, Y.; Ye, J.; Deng, Y.; Huang, Y.; Liu, X.; He, Q.; Chen, Y.; Li, Q.; Lin, Y.; Liang, R.; et al. The miRNA-185-5p/STIM1 Axis Regulates the Invasiveness of Nasopharyngeal Carcinoma Cell Lines by Modulating EGFR Activation-Stimulated Switch from E- to N-Cadherin. Molecules 2023, 28, 818. https://doi.org/10.3390/molecules28020818
Luo Y, Ye J, Deng Y, Huang Y, Liu X, He Q, Chen Y, Li Q, Lin Y, Liang R, et al. The miRNA-185-5p/STIM1 Axis Regulates the Invasiveness of Nasopharyngeal Carcinoma Cell Lines by Modulating EGFR Activation-Stimulated Switch from E- to N-Cadherin. Molecules. 2023; 28(2):818. https://doi.org/10.3390/molecules28020818
Chicago/Turabian StyleLuo, Yue, Jiaxiang Ye, Yayan Deng, Yujuan Huang, Xue Liu, Qian He, Yong Chen, Qiuyun Li, Yan Lin, Rong Liang, and et al. 2023. "The miRNA-185-5p/STIM1 Axis Regulates the Invasiveness of Nasopharyngeal Carcinoma Cell Lines by Modulating EGFR Activation-Stimulated Switch from E- to N-Cadherin" Molecules 28, no. 2: 818. https://doi.org/10.3390/molecules28020818
APA StyleLuo, Y., Ye, J., Deng, Y., Huang, Y., Liu, X., He, Q., Chen, Y., Li, Q., Lin, Y., Liang, R., Li, Y., Wei, J., & Zhang, J. (2023). The miRNA-185-5p/STIM1 Axis Regulates the Invasiveness of Nasopharyngeal Carcinoma Cell Lines by Modulating EGFR Activation-Stimulated Switch from E- to N-Cadherin. Molecules, 28(2), 818. https://doi.org/10.3390/molecules28020818






