Anthocyanins in Plant Food: Current Status, Genetic Modification, and Future Perspectives
Abstract
:1. Introduction
2. Anthocyanins in Plant Food
2.1. Chemical Structure of Anthocyanins
2.2. Anthocyanins Exist in Common Plant Food
3. Biosynthesis and Regulatory of Anthocyanins in Plant Food
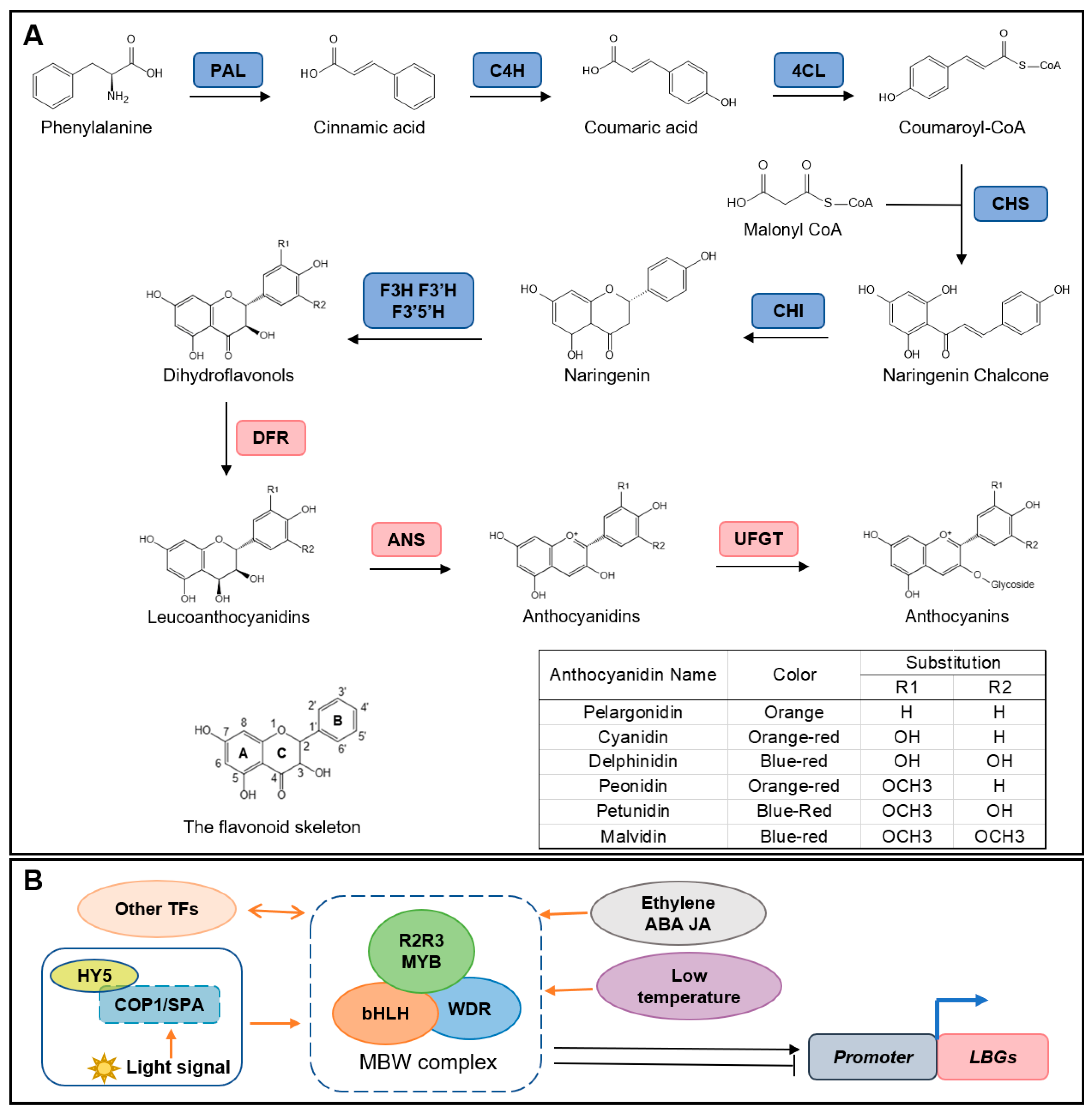
3.1. Structure Genes Involved in Biosynthesis Pathway
3.2. Other Regulators of Anthocyanin Biosynthesis
3.2.1. Transcriptional Factors Involved in Anthocyanin Biosynthesis
3.2.2. Phytohormones
3.2.3. Temperature
3.2.4. Light Signal
| Species | Gene | Type | Function | Mechanisms | Reference |
|---|---|---|---|---|---|
| Vitis vinifera | VvMYB86 | R2R3-MYB | -* | VvMYB86 represses anthocyanin biosynthesis branch in grapes by downregulating the transcript levels of VviANS and VviUFGT. | [65] |
| VvMYB2r | R2R3-MYB | + | VvMYBA2r along with VvMYCA1 and VvWDR1 form the MBW complex, which could activate the promoter of VvUFGT gene, promoting anthocyanin accumulation of grape skin. | [66] | |
| Citrus sinensis | CsRuby1 | R2R3-MYB | + | CsRuby1 encodes a MYB transcription factor that serves as the key positive regulator of anthocyanin biosynthesis. | [67] |
| Ipomoea batatas | IbMYB44 | R2R3-MYB | - | IbMYB44 interacts with IbbHLH2, IbNAC56a or IbNAC56b, competitively inhibiting the IbMYB340-IbbHLH2-IbNAC56 regulatory complex formation. | [9] |
| Solanum tuberosum | StWRKY13 | WRKY TFs | + | StWRKY13 enhances the role of StAN2 in promoting anthocyanin biosynthesis in tobacco. StWRKY13 interacts with the promotor of StCHS, StF3H, StDFR, and StANS to enhance their activities. | [68] |
| Solanum lycopersicum | SlJAF13 | bHLH | + | SlJAF13 takes part in the first MBW complex and activates SlAN1. Additionally, SlJAF13 interacts with SlMYC2, inhibiting SlMYC2 activation of SlJAZ2 transcription. | [69] |
| SlAN1 | bHLH | + | SlAN1 takes part in the second MBW complex and transcriptional activates the expression of anthocyanin biosynthesis related genes. | [70] | |
| SlAN2 (SlMYB75) | R2R3-MYB | + | SlAN2 acts as a positive regulator under high light or low temperature. SlAN2 can directly bind to the MYBPLANT and MYBPZM cis-regulatory elements and to transcriptional activate the LOXC, AADC2 and TPS genes. | [71] | |
| SlANT1 | R2R3-MYB | + | SlANT1 up-regulates structural genes in the anthocyanin pathway. | [70] | |
| SlAN2like | R2R3-MYB | + | SlAN2like activates the expression of anthocyanin biosynthetic genes and related regulatory genes. | [5] | |
| SlMYBATV | R3 MYB | - | SlMYBATV competes with Aft for binding to bHLHs, and negatively regulates anthocyanin biosynthesis. | [5] | |
| Pyrus pyrifolia | PybHLH64 | bHLH | + | PpbHLH64 interacts with PpMYB10 to form an MBW complex. | [72] |
| PyMYB10, PyMYB10.1 | R2R3-MYB | + | PpMYB10 and PpMYB10.1 interacts with PpbHLH to form the MBW complex. | [6] | |
| Pyrus | PpMYB140 | R2R3-MYB | - | PpMYB140 acts as a competitor that competes with PpMYB114 to form the MBW complex. | [51] |
| MdWRKY41 | WRKY TFs | - | MdWRKY41 downregulates the expression of MdMYB12, and interacts with MdMYB16 to form a complex, which suppresses the expression of MdANR and MdUFGT. | [73] | |
| MdbZIP44 | bZIP TFs | + | MdbZIP44 binds to MdMYB1 in response to ABA and upregulates downstream target genes to promoting anthocyanin accumulation. | [74] | |
| MdNAC52 | NAC | + | MdNAC52 binds to the promoters of MdMYB9 and MdMYB11 to promote anthocyanin accumulation. | [75] | |
| MdERF38 | ERFs | + | MdERF38 can in response to drought stress and interacts with MdMYB1 to positively regulate anthocyanin biosynthesis. | [76] | |
| MdMYB3 | R2R3 MYB | + | MdMYB3 involves in transcriptional activation of several flavonoid pathway-related genes to enhance the skin color of fruits. | [4] | |
| MdMYC2 | bHLH | + | MdMYC2 interacts with a Jasmonate signaling pathway repressor MdJAZ2, upregulating the expression of downstream genes such as MdDFR, MdUF3GT, MdF3H and MdCHS. | [77] | |
| MdMYB15L | MYB TFs | - | MdMYB15L interacts with MdbHLH33 and weakens MdbHLH33-induced anthocyanin accumulation. | [78] | |
| Actinidia chinensis | AcMYBF110 | R2R3-MYB | + | AcMYBF110 participates in the formation of the AcMYBF110-AcbHLH1-AcWDR1 complex to induce anthocyanin accumulation. | [79] |
| Fragaria × ananassa | FaRAV1 | RAV TFs | + | FaRAV1 up-regulates FaMYB10 and the genes involved in phenylpropanoid and flavonoid biosynthesis pathway. | [80] |
| FaBBX22 | B-box TFs | + | FaBBX22 positively regulates anthocyanin biosynthesis by enhancing related genes (FaPAL, FaANS, FaF3′H, FaUFGT1) and transporting gene FaRAP in a light-dependent manner. | [81] | |
| Brassica rapa | BrMYBL2.1 | R3 MYB | - | BrMYBL2.1-G (from a Chinese cabbage cultivar with purple leaves) represses transcriptional activation of BrCHS and BrDFR via blocking the activity of the MBW complex. | [82] |
| Daucus carota | DcMYB6 | R2R3 MYB | + | DcMYB6 contains the conserved bHLH-interaction motif and two typical motifs of anthocyanin regulators. | [83] |
| Brassica oleracea | BoMYB2 | R2R3 MYB | + | BoMYB2 interacts with various BobHLHs to form the MBW complex, and positively regulates the LBGs in anthocyanin biosynthesis. | [84] |
| Solanum melongena | SmMYB35 | R2R3 MYB | + | SmMYB35 interacts with SmTT8 and SmTTG1 to form a MBW complex, and positively regulates SmCHS, SmF3H, SmDFR, and SmANS. | [8] |
| Oryza sativa | OsTTG1 | WD40 | + | OsTTG1 encodes a WD40 protein, and interacts with Kala4, OsC1, OsDFR and Rc. | [85] |
| OsKala4 | bHLH | + | Kala4 involved in the origin of black rice corresponds to Os04g0557500, which encodes a bHLH transcriptional factor. A structural change in the OsKala4 promoter induced ectopic expression of this bHLH protein, thus resulting in the birth of black rice. | [86] | |
| Zea mays | Zmp1, Zmp2 | R2R3 MYB | Zmp1 (ZmMYB3) and Zmp2 (ZmMYB55) encode R2R3-MYB transcription factors that accumulates flavonoid such as 3-deoxyflavonoids, flavones, and phlobaphenes. | [87,88] | |
| Zmc1, Zmpl1 | R2R3 MYB | Zmc1 (ZmMYB1) and Zmpl1 (ZmMYB2) function in the MBW complex and upregulate LBGs expression. | [87,88] |
4. Genetic Engineering to Produce Anthocyanin-Enriched Plant Foods
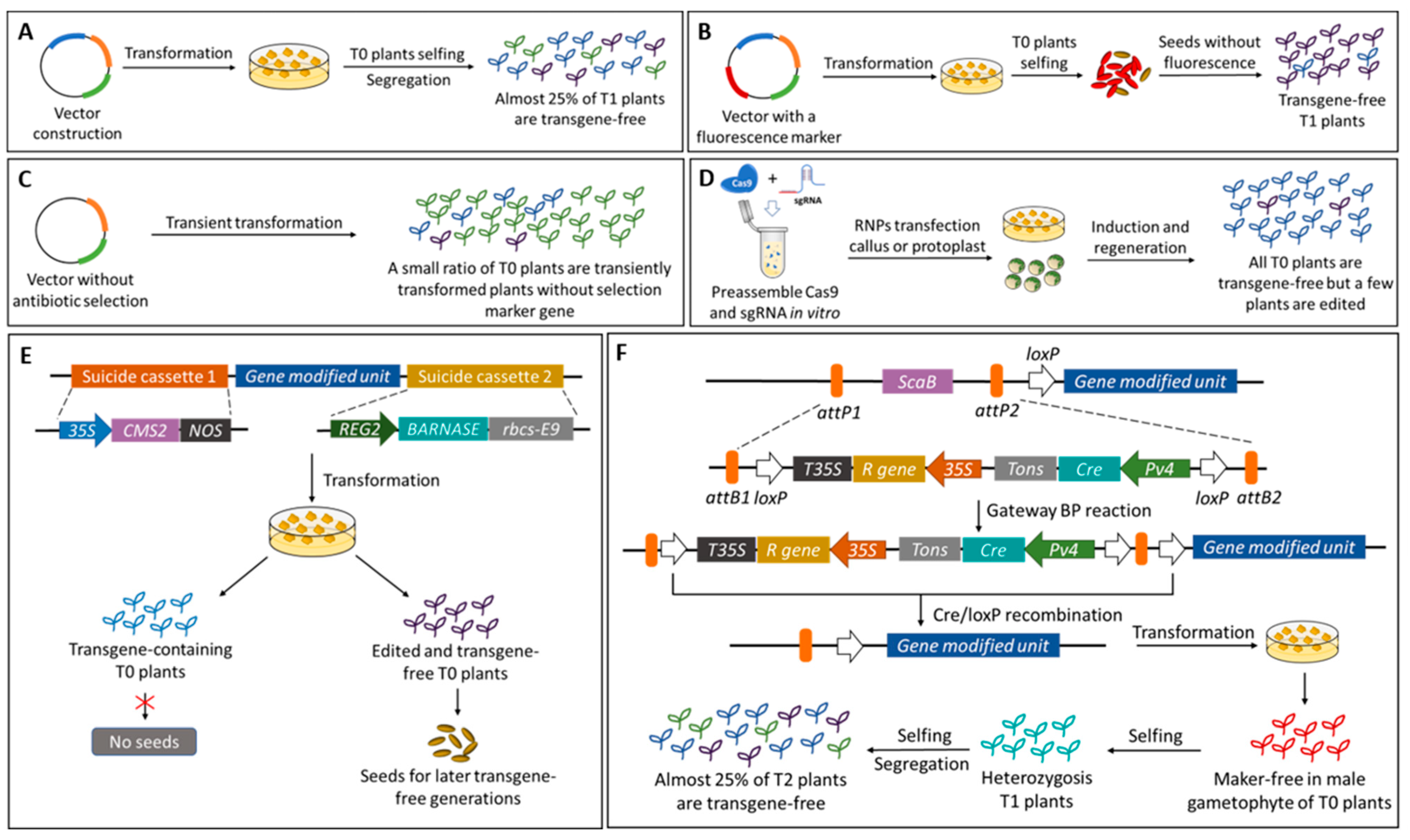
4.1. Genetic Engineering Techniques
4.2. Anthocyanins Improved Transgenic Crops
4.2.1. Tomato
4.2.2. Rice
4.2.3. Maize
4.2.4. Other Species
5. Future Perspective: Transgene/Marker-Free Anthocyanin Improved Crops
6. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shipman, E.N.; Yu, J.W.; Zhou, J.Q.; Albornoz, K.; Beckles, D.M. Can gene editing reduce postharvest waste and loss of fruit, vegetables, and ornamentals? Hortic. Res. 2021, 8, 1. [Google Scholar] [CrossRef]
- Oliveira, H.; Fernandes, A.; Bras, N.F.; Mateus, N.; de Freitas, V.; Fernandes, I. Anthocyanins as Antidiabetic Agents-In Vitro and In Silico Approaches of Preventive and Therapeutic Effects. Molecules 2020, 25, 3813. [Google Scholar] [CrossRef]
- Krga, I.; Milenkovic, D. Anthocyanins: From Sources and Bioavailability to Cardiovascular-Health Benefits and Molecular Mechanisms of Action. J. Agric. Food Chem. 2019, 67, 1771–1783. [Google Scholar] [CrossRef] [PubMed]
- Vimolmangkang, S.; Han, Y.P.; Wei, G.C.; Korban, S.S. An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 2013, 13, 176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, C.; Deng, L.; Du, M.; Zhao, J.; Chen, Q.; Huang, T.; Jiang, H.; Li, C.B.; Li, C. A transcriptional network promotes anthocyanin biosynthesis in tomato flesh. Mol. Plant 2020, 13, 42–58. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.Q.; Wang, Y.L.; Yang, S.; Xu, Y.T.; Chen, X.S. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 2010, 232, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Borovsky, Y.; Oren-Shamir, M.; Ovadia, R.; De Jong, W.; Paran, I. The A locus that controls anthocyanin accumulation in pepper encodes a MYB transcription factor homologous to Anthocyanin2 of Petunia. Theor. Appl. Genet. 2004, 109, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Li, L.Z.; Li, S.H.; Ge, H.Y.; Shi, S.L.; Li, D.L.; Liu, Y.; Chen, H.Y. A light-responsive transcription factor SmMYB35 enhances anthocyanin biosynthesis in eggplant (Solanum melongena L.). Planta 2022, 255, 12. [Google Scholar] [CrossRef]
- Wei, Z.Z.; Hu, K.D.; Zhao, D.L.; Tang, J.; Huang, Z.Q.; Jin, P.; Li, Y.H.; Han, Z.; Hu, L.Y.; Yao, G.F.; et al. MYB44 competitively inhibits the formation of the MYB340-bHLH2-NAC56 complex to regulate anthocyanin biosynthesis in purple-fleshed sweet potato. BMC Plant Biol. 2020, 20, 258. [Google Scholar] [CrossRef]
- Ferreira, V.; Matus, J.T.; Pinto-Carnide, O.; Carrasco, D.; Arroyo-Garcia, R.; Castro, I. Genetic analysis of a white-to-red berry skin color reversion and its transcriptomic and metabolic consequences in grapevine (Vitis vinifera cv. ‘Moscatel Galego’). BMC Genom. 2019, 20, 952. [Google Scholar] [CrossRef]
- Kobayashi, S.; Goto-Yamamoto, N.; Hirochika, H. Retrotransposon-induced mutations in grape skin color. Science 2004, 304, 982. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.R.; Lee, E.; Bogs, J.; McDavid, D.A.J.; Thomas, M.R.; Robinson, S.P. White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 2007, 49, 772–785. [Google Scholar] [CrossRef] [PubMed]
- Bonny, S. Genetically modified herbicide-tolerant crops, weeds, and herbicides: Overview and impact. Environ. Manag. 2016, 57, 31–48. [Google Scholar] [CrossRef] [PubMed]
- Mattioli, R.; Francioso, A.; Mosca, L.; Silva, P. Anthocyanins: A comprehensive review of their chemical properties and health effects on cardiovascular and neurodegenerative diseases. Molecules 2020, 25, 3809. [Google Scholar] [CrossRef]
- Liu, J.Y.; Osbourn, A.; Ma, P.D. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.J.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Yin, K.Q.; Gao, C.X.; Qiu, J.L. Progress and prospects in plant genome editing. Nat. Plants 2017, 3, 17107. [Google Scholar] [CrossRef]
- Delaney, B.; Goodman, R.E.; Ladics, G.S. Food and feed safety of genetically engineered food crops. Toxicol. Sci. 2018, 162, 361–371. [Google Scholar] [CrossRef] [Green Version]
- Nap, J.P.; Metz, P.L.J.; Escaler, M.; Conner, A.J. The release of genetically modified crops into the environment—Part I. Overview of current status and regulations. Plant J. 2003, 33, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Liang, J.; Yang, X.; Jiao, Y.; Wang, D.; Zhao, Q.; Sun, Y.; Li, Y.; Wu, K. The evolution of China’s regulation of agricultural biotechnology. Abiotech 2022, 3, 237–249. [Google Scholar] [CrossRef]
- Gu, X.; Liu, L.; Zhang, H. Transgene-free Genome Editing in Plants. Front. Genome Ed. 2021, 3, 805317. [Google Scholar] [CrossRef] [PubMed]
- Alappat, B.; Alappat, J. Anthocyanin pigments: Beyond aesthetics. Molecules 2020, 25, 5500. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef]
- Fang, J.; Jogaiah, S.; Guan, L.; Sun, X.; Abdelrahman, M. Coloring biology in grape skin: A prospective strategy for molecular farming. Physiol. Plant 2018, 164, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Colanero, S.; Perata, P.; Gonzali, S. What’s behind purple tomatoes? Insight into the mechanisms of anthocyanin synthesis in tomato fruits. Plant Physiol. 2020, 182, 1841–1853. [Google Scholar] [CrossRef] [Green Version]
- Lila, M.A.; Burton-Freeman, B.; Grace, M.; Kalt, W. Unraveling anthocyanin bioavailability for human health. Annu. Rev. Food Sci. Technol. 2016, 7, 375–393. [Google Scholar] [CrossRef]
- Chaves, V.C.; Boff, L.; Vizzotto, M.; Calvete, E.; Reginatto, F.H.; Simoes, C.M. Berries grown in Brazil: Anthocyanin profiles and biological properties. J. Sci. Food Agric. 2018, 98, 4331–4338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponder, A.; Hallmann, E.; Kwolek, M.; Srednicka-Tober, D.; Kazimierczak, R. Genetic differentiation in anthocyanin content among berry fruits. Curr. Issues Mol. Biol. 2021, 43, 36–51. [Google Scholar] [CrossRef]
- Kapusta, I.; Cebulak, T.; Oszmianski, J. The anthocyanins profile of red grape cultivars growing in south-east Poland (Subcarpathia region). J. Food Meas. Charact. 2017, 11, 1863–1873. [Google Scholar] [CrossRef]
- Lao, F.; Giusti, M.M. Quantification of purple corn (Zea mays L.) anthocyanins using spectrophotometric and hplc approaches: Method comparison and correlation. Food Anal. Methods 2016, 9, 1367–1380. [Google Scholar] [CrossRef]
- Mustafa, A.M.; Angeloni, S.; Abouelenein, D.; Acquaticci, L.; Xiao, J.B.; Sagratini, G.; Maggi, F.; Vittori, S.; Caprioli, G. A new HPLC-MS/MS method for the simultaneous determination of 36 polyphenols in blueberry, strawberry and their commercial products and determination of antioxidant activity. Food Chem. 2022, 367, 130743. [Google Scholar] [CrossRef] [PubMed]
- Legua, P.; Modica, G.; Porras, I.; Conesa, A.; Continella, A. Bioactive compounds, antioxidant activity and fruit quality evaluation of eleven blood orange cultivars. J. Sci. Food Agric. 2022, 102, 2960–2971. [Google Scholar] [CrossRef]
- Chen, S.S.; Hu, N.; Wang, H.L.; Wu, Y.N.; Li, G.L. Bioactivity-guided isolation of the major anthocyanin from Lycium ruthenicum Murr. fruit and its antioxidant activity and neuroprotective effects in vitro and in vivo. Food Funct. 2022, 13, 3247–3257. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.F.; Hu, Z.Q.; Yu, Y.H.; Mou, R.X.; Zhu, Z.W.; Beta, T. Phenolic acids, anthocyanins, proanthocyanidins, antioxidant activity, minerals and their correlations in non-pigmented, red, and black rice. Food Chem. 2018, 239, 733–741. [Google Scholar] [CrossRef]
- Tu, M.X.; Fang, J.H.; Zhao, R.K.; Liu, X.Y.; Yin, W.C.; Wang, Y.; Wang, X.H.; Wang, X.P.; Fang, Y.L. CRISPR/Cas9-mediated mutagenesis of VvbZIP36 promotes anthocyanin accumulation in grapevine (Vitis vinifera). Hortic. Res. 2022, 9, uhac022. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.T.; Han, R.P.; Yu, J.W.; Zhu, M.K.; Zhang, Y.; Gong, Y.; Li, Z.Y. Anthocyanins accumulation and molecular analysis of correlated genes by metabolome and transcriptome in green and purple asparaguses (Asparagus officinalis, L.). Food Chem. 2019, 271, 18–28. [Google Scholar] [CrossRef]
- Gonzali, S.; Mazzucato, A.; Perata, P. Purple as a tomato: Towards high anthocyanin tomatoes. Trends Plant Sci. 2009, 14, 237–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef]
- Albert, N.W.; Davies, K.M.; Lewis, D.H.; Zhang, H.; Montefiori, M.; Brendolise, C.; Boase, M.R.; Ngo, H.; Jameson, P.E.; Schwinn, K.E. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 2014, 26, 962–980. [Google Scholar] [CrossRef] [Green Version]
- Strygina, K.V.; Kochetov, A.V.; Khlestkina, E.K. Genetic control of anthocyanin pigmentation of potato tissues. BMC Genet. 2019, 20, 27. [Google Scholar] [CrossRef]
- D’Amelia, V.; Aversano, R.; Batelli, G.; Caruso, I.; Moreno, M.C.; Castro-Sanz, A.B.; Chiaiese, P.; Fasano, C.; Palomba, F.; Carputo, D. High AN1 variability and interaction with basic helix-loop-helix co-factors related to anthocyanin biosynthesis in potato leaves. Plant J. 2014, 80, 527–540. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.H.; Lin-Wang, K.; Espley, R.V.; Wang, L.; Yang, H.; Yu, B.; Dare, A.; Gasic, E.V.; Wang, J.; Zhang, J.; et al. Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 2016, 67, 2159–2176. [Google Scholar] [CrossRef] [PubMed]
- Ning, Z.; Hu, K.; Zhou, Z.; Zhao, D.; Tang, J.; Wang, H.; Li, L.; Ding, C.; Chen, X.; Yao, G.; et al. IbERF71, with IbMYB340 and IbbHLH2, coregulates anthocyanin accumulation by binding to the IbANS1 promoter in purple-fleshed sweet potato (Ipomoea batatas L.). Plant Cell Rep. 2021, 40, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Mattoo, A.K.; Dwivedi, S.L.; Dutt, S.; Singh, B.; Garg, M.; Ortiz, R. Anthocyanin-rich vegetables for human consumption-focus on potato, sweetpotato and tomato. Int. J. Mol. Sci. 2022, 23, 2634. [Google Scholar] [CrossRef]
- Kiferle, C.; Fantini, E.; Bassolino, L.; Povero, G.; Spelt, C.; Buti, S.; Giuliano, G.; Quattrocchio, F.; Koes, R.; Perata, P.; et al. Tomato R2R3-MYB proteins SlANT1 and SlAN2: Same protein activity, different roles. PLoS ONE 2015, 10, e0136365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhi, J.J.; Liu, X.X.; Li, D.J.; Huang, Y.G.M.; Yan, S.S.; Cao, B.H.; Qiu, Z.K. CRISPR/Cas9-mediated SlAN2 mutants reveal various regulatory models of anthocyanin biosynthesis in tomato plant. Plant Cell Rep. 2020, 39, 799–809. [Google Scholar] [CrossRef] [PubMed]
- Colanero, S.; Tagliani, A.; Perata, P.; Gonzali, S. Alternative splicing in the anthocyanin fruit gene encoding an r2r3 myb transcription factor affects anthocyanin biosynthesis in tomato fruits. Plant Commun. 2020, 1, 100006. [Google Scholar] [CrossRef]
- Yan, S.; Chen, N.; Huang, Z.; Li, D.; Zhi, J.; Yu, B.; Liu, X.; Cao, B.; Qiu, Z. Anthocyanin Fruit encodes an R2R3-MYB transcription factor, SlAN2-like, activating the transcription of SlMYBATV to fine-tune anthocyanin content in tomato fruit. New Phytol. 2020, 225, 2048–2063. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Li, Y.Y.; Song, L.Q.; Zhao, L.L.; You, C.X.; Hao, Y.J. EIN3-LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 act in a regulatory loop that synergistically modulates ethylene biosynthesis and anthocyanin accumulation. Plant Physiol. 2018, 178, 808–823. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.L.; Liu, X.X.; Huang, Y.G.M.; Xia, Z.L.; Lian, Z.L.; Qian, L.J.; Yan, S.S.; Cao, B.H.; Qiu, Z.K. Ethylene inhibits anthocyanin biosynthesis by repressing the R2R3-MYB regulator SlAN2-like in tomato. Int. J. Mol. Sci. 2022, 23, 7648. [Google Scholar] [CrossRef]
- Ni, J.B.; Premathilake, A.T.; Gao, Y.H.; Yu, W.J.; Tao, R.Y.; Teng, Y.W.; Bai, S.L. Ethylene-activated PpERF105 induces the expression of the repressor-type R2R3-MYB gene PpMYB140 to inhibit anthocyanin biosynthesis in red pear fruit. Plant J. 2021, 105, 167–181. [Google Scholar] [CrossRef]
- An, J.P.; Zhang, X.W.; Liu, Y.J.; Wang, X.F.; You, C.X.; Hao, Y.J. ABI5 regulates ABA-induced anthocyanin biosynthesis by modulating the MYB1-bHLH3 complex in apple. J. Exp. Bot. 2021, 72, 1460–1472. [Google Scholar] [CrossRef]
- Oraei, M.; Panahirad, S.; Zaare-Nahandi, F.; Gohari, G. Pre-veraison treatment of salicylic acid to enhance anthocyanin content of grape (Vitis vinifera L.) berries. J. Sci. Food Agric. 2019, 99, 5946–5952. [Google Scholar] [CrossRef]
- Lin-Wang, K.; Micheletti, D.; Palmer, J.; Volz, R.; Lozano, L.; Espley, R.; Hellens, R.P.; Chagné, D.; Rowan, D.D.; Troggio, M.; et al. High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Environ. 2011, 34, 1176–1190. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.H.; Lin-Wang, K.; Espley, R.; Wang, L.; Li, Y.; Liu, Z.; Zhou, P.; Zeng, L.; Zhang, X.; Zhang, J.; et al. StMYB44 negatively regulates anthocyanin biosynthesis at high temperatures in tuber flesh of potato. J. Exp. Bot. 2019, 70, 3809–3824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Z.H.; Li, W.F.; Mao, J.; Li, W.; Zuo, C.W.; Zhao, X.; Dawuda, M.M.; Shi, X.Y.; Chen, B.H. Synthesis of light-inducible and light-independent anthocyanins regulated by specific genes in grape ‘Marselan’ (V-vinifera L.). PeerJ 2019, 7, e6521. [Google Scholar] [CrossRef] [Green Version]
- Kataoka, I.; Beppu, K.; Yanagi, T.; Okamoto, K. Light components contributing to accumulation of anthocyanins in ‘Gros Colman’ grape berries. Vitic.-Living Limit. 2004, 640, 333–339. [Google Scholar]
- Guo, X.; Wang, Y.; Zhai, Z.; Huang, T.; Zhao, D.; Peng, X.; Feng, C.; Xiao, Y.; Li, T. Transcriptomic analysis of light-dependent anthocyanin accumulation in bicolored cherry fruits. Plant Physiol. Biochem. 2018, 130, 663–677. [Google Scholar] [CrossRef] [PubMed]
- Abbas, N.; Maurya, J.P.; Senapati, D.; Gangappa, S.N.; Chattopadhyay, S. Arabidopsis CAM7 and HY5 Physically Interact and Directly Bind to the HY5 Promoter to Regulate Its Expression and Thereby Promote Photomorphogenesis. Plant Cell 2014, 26, 1036–1052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Xu, P.; Chen, G.; Wu, J.; Liu, Z.; Lian, H. FvbHLH9 Functions as a Positive Regulator of Anthocyanin Biosynthesis by Forming a HY5-bHLH9 Transcription Complex in Strawberry Fruits. Plant Cell Physiol. 2020, 61, 826–837. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Jeong, C.Y.; Kang, G.H.; Yoo, S.D.; Hong, S.W.; Lee, H. MYBD employed by HY5 increases anthocyanin accumulation via repression of MYBL2 in Arabidopsis. Plant J. 2015, 84, 1192–1205. [Google Scholar] [CrossRef]
- Maier, A.; Hoecker, U. COP1/SPA ubiquitin ligase complexes repress anthocyanin accumulation under low light and high light conditions. Plant Signal. Behav. 2015, 10, e970440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, X.D.; Zhou, C.M.; Xu, P.B.; Luo, Q.; Lian, H.L.; Yang, H.Q. Red-Light-Dependent Interaction of phyB with SPA1 Promotes COP1-SPA1 Dissociation and Photomorphogenic Development in Arabidopsis. Mol. Plant 2015, 8, 467–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.Y.; Ma, X.; Gao, X.; Wu, W.L.; Zhou, B. Light induced regulation pathway of anthocyanin biosynthesis in plants. Int. J. Mol. Sci. 2021, 22, 11116. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Yu, K.J.; Shi, Y.; Wang, J.; Duan, C.Q. Transcription factor VviMYB86 oppositely regulates proanthocyanidin and anthocyanin biosynthesis in grape berries. Front. Plant Sci. 2021, 11, 613677. [Google Scholar] [CrossRef]
- Jiu, S.T.; Guan, L.; Leng, X.; Zhang, K.; Haider, M.S.; Yu, X.; Zhu, X.; Zheng, T.; Ge, M.; Wang, C.; et al. The role of VvMYBA2r and VvMYBA2w alleles of the MYBA2 locus in the regulation of anthocyanin biosynthesis for molecular breeding of grape (Vitis spp.) skin coloration. Plant Biotechnol. J. 2021, 19, 1216–1239. [Google Scholar] [CrossRef]
- Butelli, E.; Licciardello, C.; Zhang, Y.; Liu, J.J.; Mackay, S.; Bailey, P.; Reforgiato-Recupero, G.; Martin, C. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 2012, 24, 1242–1255. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.L.; Zhang, Z.H.; Zhao, Y.A.; Guo, D.L.; Zhao, X.J.; Gao, W.; Zhang, J.P.; Song, B.T. StWRKY13 promotes anthocyanin biosynthesis in potato (Solanum tuberosum) tubers. Funct. Plant Biol. 2022, 49, 102–114. [Google Scholar] [CrossRef]
- Chen, Y.Z.; Kim, P.; Kong, L.; Wang, X.; Tan, W.; Liu, X.; Chen, Y.; Yang, J.; Chen, B.; Song, Y.; et al. A dual-function transcription factor, SlJAF13, promotes anthocyanin biosynthesis in tomato. J. Exp. Bot. 2022, 73, 5559–5580. [Google Scholar] [CrossRef]
- Kang, S.I.; Rahim, M.A.; Afrin, K.S.; Jung, H.J.; Kim, H.T.; Park, J.I.; Nou, I.S. Expression of anthocyanin biosynthesis-related genes reflects the peel color in purple tomato. Hortic. Environ. Biotechnol. 2018, 59, 435–445. [Google Scholar] [CrossRef]
- Jian, W.; Cao, H.; Yuan, S.; Liu, Y.; Lu, J.; Lu, W.; Li, N.; Wang, J.; Zou, J.; Tang, N.; et al. SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic. Res. 2019, 6, 22. [Google Scholar] [CrossRef]
- Tao, R.Y.; Yu, W.; Gao, Y.; Ni, J.; Yin, L.; Zhang, X.; Li, H.; Wang, D.; Bai, S.; Teng, Y. Light-induced basic/helix-loop-helix64 enhances anthocyanin biosynthesis and undergoes CONSTITUTIVELY PHOTOMORPHOGENIC1-mediated degradation in pear. Plant Physiol. 2020, 184, 1684–1701. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.L.; Jiang, H.; Wang, S.; Wang, Y.; Yu, L.; Zou, Q.; Liu, W.; Jiang, S.; Wang, N.; Zhang, Z.; et al. The MdHY5-MdWRKY41-MdMYB transcription factor cascade regulates the anthocyanin and proanthocyanidin biosynthesis in red-fleshed apple. Plant Sci. 2021, 306, 110848. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Yao, J.F.; Xu, R.R.; You, C.X.; Wang, X.F.; Hao, Y.J. Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation. Plant Cell Environ. 2018, 41, 2678–2692. [Google Scholar] [CrossRef]
- Sun, Q.G.; Jiang, S.; Zhang, T.; Xu, H.; Fang, H.; Zhang, J.; Su, M.; Wang, Y.; Zhang, Z.; Wang, N.; et al. Apple NAC transcription factor MdNAC52 regulates biosynthesis of anthocyanin and proanthocyanidin through MdMYB9 and MdMYB11. Plant Sci. 2019, 289, 110286. [Google Scholar] [CrossRef]
- An, J.P.; Zhang, X.W.; Bi, S.Q.; You, C.X.; Wang, X.F.; Hao, Y.J. The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple. Plant J. 2020, 101, 573–589. [Google Scholar] [CrossRef]
- An, J.P.; Li, H.H.; Song, L.Q.; Su, L.; Liu, X.; You, C.X.; Wang, X.F.; Hao, Y.J. The molecular cloning and functional characterization of MdMYC2, a bHLH transcription factor in apple. Plant Physiol. Biochem. 2019, 135, 612. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.F.; Yang, G.X.; Zhang, J.; Wang, Y.C.; Zhang, T.L.; Wang, N.; Jiang, S.H.; Zhang, Z.Y.; Chen, X.S. Overexpression of a repressor MdMYB15L negatively regulates anthocyanin and cold tolerance in red-fleshed callus. Biochem. Biophys. Res. Commun. 2018, 500, 405–410. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, K.; Qi, Y.; Lv, G.; Ren, X.; Liu, Z.; Ma, F. Transcriptional Regulation of Anthocyanin Synthesis by MYB-bHLH-WDR Complexes in Kiwifruit (Actinidia chinensis). J. Agric. Food Chem. 2021, 69, 3677–3691. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Shi, Y.; Ma, Y.; Yang, X.; Yin, X.; Zhang, Y.; Xiao, Y.; Liu, W.; Li, Y.; Li, S.; et al. The strawberry transcription factor FaRAV1 positively regulates anthocyanin accumulation by activation of FaMYB10 and anthocyanin pathway genes. Plant Biotechnol. J. 2020, 18, 2267–2279. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Ye, Y.; Wang, Y.; Jiang, L.; Yue, M.; Tang, L.; Jin, M.; Zhang, Y.; Lin, Y.; Tang, H. B-Box Transcription factor FaBBX22 promotes light-induced anthocyanin accumulation in strawberry (Fragaria x ananassa). Int. J. Mol. Sci. 2022, 23, 7757. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, D.H.; Lee, J.Y.; Lim, S.H. The R3-Type MYB transcription factor BrMYBL2.1 negatively regulates anthocyanin biosynthesis in Chinese cabbage (Brassica rapa L.) by repressing MYB-bHLH-WD40 complex activity. Int. J. Mol. Sci. 2022, 23, 3382. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.S.; Feng, K.; Que, F.; Wang, F.; Xiong, A.S. A MYB transcription factor, DcMYB6, is involved in regulating anthocyanin biosynthesis in purple carrot taproots. Sci. Rep. 2017, 7, 45324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiu, L.W.; Li, L. Characterization of the regulatory network of BoMYB2 in controlling anthocyanin biosynthesis in purple cauliflower. Planta 2012, 236, 1153–1164. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.H.; Wang, J.; Xia, X.; Zhang, Z.; He, J.; Nong, B.; Luo, T.; Feng, R.; Wu, Y.; Pan, Y.; et al. OsTTG1, a WD40 repeat gene, regulates anthocyanin biosynthesis in rice. Plant J. 2021, 107, 198–214. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, T.; Maeda, H.; Oguchi, T.; Yamaguchi, T.; Tanabe, N.; Ebana, K.; Yano, M.; Ebitani, T.; Izawa, T. The birth of a black rice gene and its local spread by introgression. Plant Cell 2015, 27, 2401–2414. [Google Scholar] [CrossRef] [Green Version]
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef]
- Peniche-Pavia, H.A.; Guzman, T.J.; Magana-Cerino, J.M.; Gurrola-Diaz, C.M.; Tiessen, A. Maize Flavonoid Biosynthesis, Regulation, and Human Health Relevance: A Review. Molecules 2022, 27, 5166. [Google Scholar] [CrossRef]
- Chilton, M.D.; Drummond, M.H.; Merlo, D.J.; Sciaky, D.; Montoya, A.L. Stable incorporation of plasmid DNA into higher plant cells: The molecular basis of crown gall tumorigenesis. Cell 1977, 11, 263–271. [Google Scholar] [CrossRef]
- Fraley, R.T.; Rogers, S.G.; Horsch, R.B.; Sanders, P.R.; Flick, J.S.; Adams, S.P.; Bittner, M.L.; A Brand, L.; Fink, C.L.; Fry, J.S.; et al. Expression of bacterial genes in plant-cells. Proc. Natl. Acad. Sci. USA 1983, 80, 4803–4807. [Google Scholar] [CrossRef] [Green Version]
- Herrera-Estrella, L.; Ann Depicker, A.; Montagu, M.V.; Schen, J. Expression of chimaeric genes transfered into plant cells using a Ti-plasmid-derived vector. Nature 1983, 303, 209–213. [Google Scholar] [CrossRef]
- Kumar, K.; Gambhir, G.; Dass, A.; Tripathi, A.K.; Singh, A.; Jha, A.K.; Yadava, P.; Choudhary, M.; Rakshit, S. Genetically modified crops: Current status and future prospects. Planta 2020, 251, 91. [Google Scholar] [CrossRef] [PubMed]
- Klumper, W.; Qaim, M. A meta-analysis of the impacts of genetically modified crops. PLoS ONE 2014, 9, e111629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shehryar, K.; Khan, R.S.; Iqbal, A.; Hussain, S.A.; Imdad, S.; Bibi, A.; Hamayun, L.; Nakamura, I. Transgene stacking as effective tool for enhanced disease resistance in plants. Mol. Biotechnol. 2020, 62, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.L.; Yu, S.; Zeng, D.; Liu, H.; Wang, H.; Yang, Z.; Xie, X.; Shen, R.; Tan, J.; Li, H.; et al. Development of “Purple Endosperm Rice” by engineering anthocyanin biosynthesis in the endosperm with a high-efficiency transgene stacking system. Mol. Plant 2017, 10, 918–929. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.C.; Han, J.; Tan, J.; Yang, Y.; Li, S.; Gou, Y.; Luo, Y.; Li, T.; Xiao, W.; Xue, Y.; et al. Efficient assembly of long DNA fragments and multiple genes with improved nickase-based cloning and Cre/loxP recombination. Plant Biotechnol. J. 2022, 20, 1983–1995. [Google Scholar] [CrossRef]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide-sequence of the iap gene, responsible for alkaline-phosphatase isozyme conversion in Escherichia-coli, and identification of the gene-product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef] [Green Version]
- Moon, S.B.; Kim, D.; Ko, J.H.; Kim, Y.S. Recent advances in the CRISPR genome editing tool set. Exp. Mol. Med. 2019, 51, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Pan, C.T.; Wu, X.; Markel, K.; Malzahn, A.A.; Kundagrami, N.; Sretenovic, S.; Zhang, Y.; Cheng, Y.; Shih, P.M.; Qi, Y. CRISPR-Act3.0 for highly efficient multiplexed gene activation in plants. Nat. Plants 2021, 7, 942–953. [Google Scholar] [CrossRef]
- Li, Z.X.; Zhang, D.D.; Xiong, X.Y.; Yan, B.Y.; Xie, W.; Sheen, J.; Li, J.F. A potent Cas9-derived gene activator for plant and mammalian cells. Nat. Plants 2017, 3, 930–936. [Google Scholar] [CrossRef] [Green Version]
- Lowder, L.G.; Zhou, J.P.; Zhang, Y.X.; Malzahn, A.; Zhong, Z.H.; Hsieh, T.F.; Voytas, D.F.; Zhang, Y.; Qi, Y.P. Robust transcriptional activation in plants using multiplexed CRISPR-Act2.0 and mTALE-Act systems. Mol. Plant 2018, 11, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Waltz, E. Food firms test fry Pioneer’s trans fat-free soybean oil. Nat. Biotechnol. 2010, 28, 769–770. [Google Scholar] [CrossRef] [PubMed]
- Waltz, E. USDA approves next-generation GM potato. Nat. Biotechnol. 2015, 33, 12–13. [Google Scholar] [CrossRef]
- Waltz, E. Nonbrowning GM apple cleared for market. Nat. Biotechnol. 2015, 33, 326–327. [Google Scholar] [CrossRef]
- Waltz, E. GABA-enriched tomato is first CRISPR-edited food to enter market. Nat. Biotechnol. 2022, 40, 9–11. [Google Scholar] [CrossRef]
- Nonaka, S.; Arai, C.; Takayama, M.; Matsukura, C.; Ezura, H. Efficient increase of-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Sci. Rep. 2017, 7, 7057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schijlen, E.; de Vos, C.H.R.; Jonker, H.; van den Broeck, H.; Molthoff, J.; van Tunen, A.; Martens, S.; Bovy, A. Pathway engineering for healthy phytochemicals leading to the production of novel flavonoids in tomato fruit. Plant Biotechnol. J. 2006, 4, 433–444. [Google Scholar] [CrossRef]
- Muir, S.R.; Collins, G.J.; Robinson, S.; Hughes, S.; Bovy, A.; De Vos, C.H.R.; van Tunen, A.J.; Verhoeyen, M.E. Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nat. Biotechnol. 2001, 19, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Bovy, A.; de Vos, R.; Kemper, M.; Schijlen, E.; Almenar Pertejo, M.; Muir, S.; Collins, G.; Robinson, S.; Verhoeyen, M.; Hughes, S.; et al. High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell 2002, 14, 2509–2526. [Google Scholar] [CrossRef] [Green Version]
- Mathews, H.; Clendennen, S.K.; Caldwell, C.G.; Liu, X.L.; Connors, K.; Matheis, N.; Schuster, D.K.; Menasco, D.J.; Wagoner, W.; Lightner, J.; et al. Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 2003, 15, 1689–1703. [Google Scholar] [CrossRef] [Green Version]
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.-P.; Matros, A.; Peterek, S.; Schijlen, E.G.W.M.; Hall, R.D.; Bovy, A.G.; Luo, J.; et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 2008, 26, 1301–1308. [Google Scholar] [CrossRef]
- Qiu, Z.; Wang, H.; Li, D.; Yu, B.; Hui, Q.; Yan, S.; Huang, Z.; Cui, X.; Cao, B. Identification of Candidate HY5-Dependent and -Independent Regulators of Anthocyanin Biosynthesis in Tomato. Plant Cell Physiol. 2019, 60, 643–656. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.D.; Al-Babili, S.; Kloti, A.; Zhang, J.; Lucca, P.; Beyer, P.; Potrykus, I. Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 2000, 287, 303–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.Q.; Li, S.Z.; Yang, W.Z.; Mu, B.N.; Jiao, Y.; Zhou, X.J.; Zhang, C.Y.; Fan, Y.L.; Chen, R.M. Synthesis of Seed-Specific Bidirectional Promoters for Metabolic Engineering of Anthocyanin-Rich Maize. Plant Cell Physiol. 2018, 59, 1942–1955. [Google Scholar] [CrossRef] [Green Version]
- D’Amelia, V.; Villano, C.; Batelli, G.; Çobanoğlu, Ö.; Carucci, F.; Melito, S.; Chessa, M.; Chiaiese, P.; Aversano, R.; Carputo, D. Genetic and epigenetic dynamics affecting anthocyanin biosynthesis in potato cell culture. Plant Sci. 2020, 298, 110597. [Google Scholar] [CrossRef] [PubMed]
- Ming, M.L.; Long, H.; Ye, Z.; Pan, C.; Chen, J.; Tian, R.; Sun, C.; Xue, Y.; Zhang, Y.; Li, J.; et al. Highly efficient CRISPR systems for loss-of-function and gain-of-function research in pear calli. Hortic. Res. 2022, 9, uhac148. [Google Scholar] [CrossRef]
- He, Y.; Zhao, Y. Technological breakthroughs in generating transgene-free and genetically stable CRISPR-edited plants. Abiotech 2020, 1, 88–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowe, B.A.; Prakash, N.S.; Way, M.; Mann, M.T.; Spencer, T.M.; Boddupalli, R.S. Enhanced single copy integration events in corn via particle bombardment using low quantities of DNA. Transgenic Res. 2009, 18, 831–840. [Google Scholar] [CrossRef]
- He, Y.B.; Wang, R.C.; Dai, X.H.; Zhao, Y.D. On Improving CRISPR for Editing Plant Genes: Ribozyme-Mediated Guide RNA Production and Fluorescence-Based Technology for Isolating Transgene-Free Mutants Generated by CRISPR. Gene Ed. Plants 2017, 149, 151–166. [Google Scholar]
- Gao, X.H.; Chen, J.L.; Dai, X.H.; Zhang, D.; Zhao, Y.D. An Effective Strategy for Reliably Isolating Heritable and Cas9-Free Arabidopsis Mutants Generated by CRISPR/Cas9-Mediated Genome Editing. Plant Physiol. 2016, 171, 1794–1800. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.Z.; Li, W.; Katin-Grazzini, L.; Ding, J.; Gu, X.; Li, Y.; Gu, T.; Wang, R.; Lin, X.; Deng, Z.; et al. A method for the production and expedient screening of CRISPR/Cas9-mediated non-transgenic mutant plants. Hortic. Res. 2018, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalán, C.; Cho, S.W.; Kim, H.; Kim, S.-G.; Kim, S.-T.; Choe, S.; Kim, J.-S. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1164. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Choe, S. DNA-free genome editing with preassembled CRISPR/Cas9 ribonucleoproteins in plants. Transgenic Res. 2019, 28, 61–64. [Google Scholar] [CrossRef] [PubMed]
- He, Y.B.; Zhu, M.; Wang, L.H.; Wu, J.H.; Wang, Q.Y.; Wang, R.C.; Zhao, Y.D. Programmed self-elimination of the CRISPR/Cas9 construct greatly accelerates the isolation of edited and transgene-free rice plants. Mol. Plant 2018, 11, 1210–1213. [Google Scholar] [CrossRef] [PubMed]
| Species | Part for Determination | Representative Compounds | Reference |
|---|---|---|---|
| Fragaria × ananassa | Receptacle | Pelargonidin-3-O-glucosides | [31] |
| Rubus | Fruit | Cyanidin-3-O-glucosides | [28] |
| Vaccinium | Fruit | Delphindin-3-O-galactoside, cyanidin-3-O-glucoside, petunidin-3-O-glucoside, malvidin-3-O-galactoside | [31] |
| Ribes nigrum | Fruit | Delphinidin-3-O-glucoside, delphinidin-3-O-rutinoside | [28] |
| Vitis vinifera | Peel | Delphinidin-3-O-glucoside, malvidin-3-O-glucoside, petunidin-3-O-glucoside, malvidin-3-O-glucoside-5-O-glucoside | [29] |
| Citrus sinensis | Pulp | Cyanidin-3-O-glucoside, cyanidin-3-O-(6’’-malonylglucoside) | [32] |
| Lycium ruthenicum | Fruit | Petunidin-3,5-O-diglucoside | [33] |
| Oryza sativa | Seed | Cyanidin-3-O-glucoside, peonidin-3-O-glucoside | [34] |
| Zea mays | Fruit | Cyanidin-3-O-glucoside, pelargonidin-3-O-glucoside, peonidin-3-O-glucoside, cyanidin-3-O-(6″-malonylglucoside), pelargonidin-3-O-(6″-malonylglucoside), peonidin-3-O-(6″-malonylglucoside) | [30] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, P.; Zhu, H. Anthocyanins in Plant Food: Current Status, Genetic Modification, and Future Perspectives. Molecules 2023, 28, 866. https://doi.org/10.3390/molecules28020866
Zhang P, Zhu H. Anthocyanins in Plant Food: Current Status, Genetic Modification, and Future Perspectives. Molecules. 2023; 28(2):866. https://doi.org/10.3390/molecules28020866
Chicago/Turabian StyleZhang, Peiyu, and Hongliang Zhu. 2023. "Anthocyanins in Plant Food: Current Status, Genetic Modification, and Future Perspectives" Molecules 28, no. 2: 866. https://doi.org/10.3390/molecules28020866
APA StyleZhang, P., & Zhu, H. (2023). Anthocyanins in Plant Food: Current Status, Genetic Modification, and Future Perspectives. Molecules, 28(2), 866. https://doi.org/10.3390/molecules28020866








