Synthesis and Structural Characterization of Novel Dimers of Dipyridothiazine as Promising Antiproliferative Agents
Abstract
:1. Introduction
2. Results and Discussion
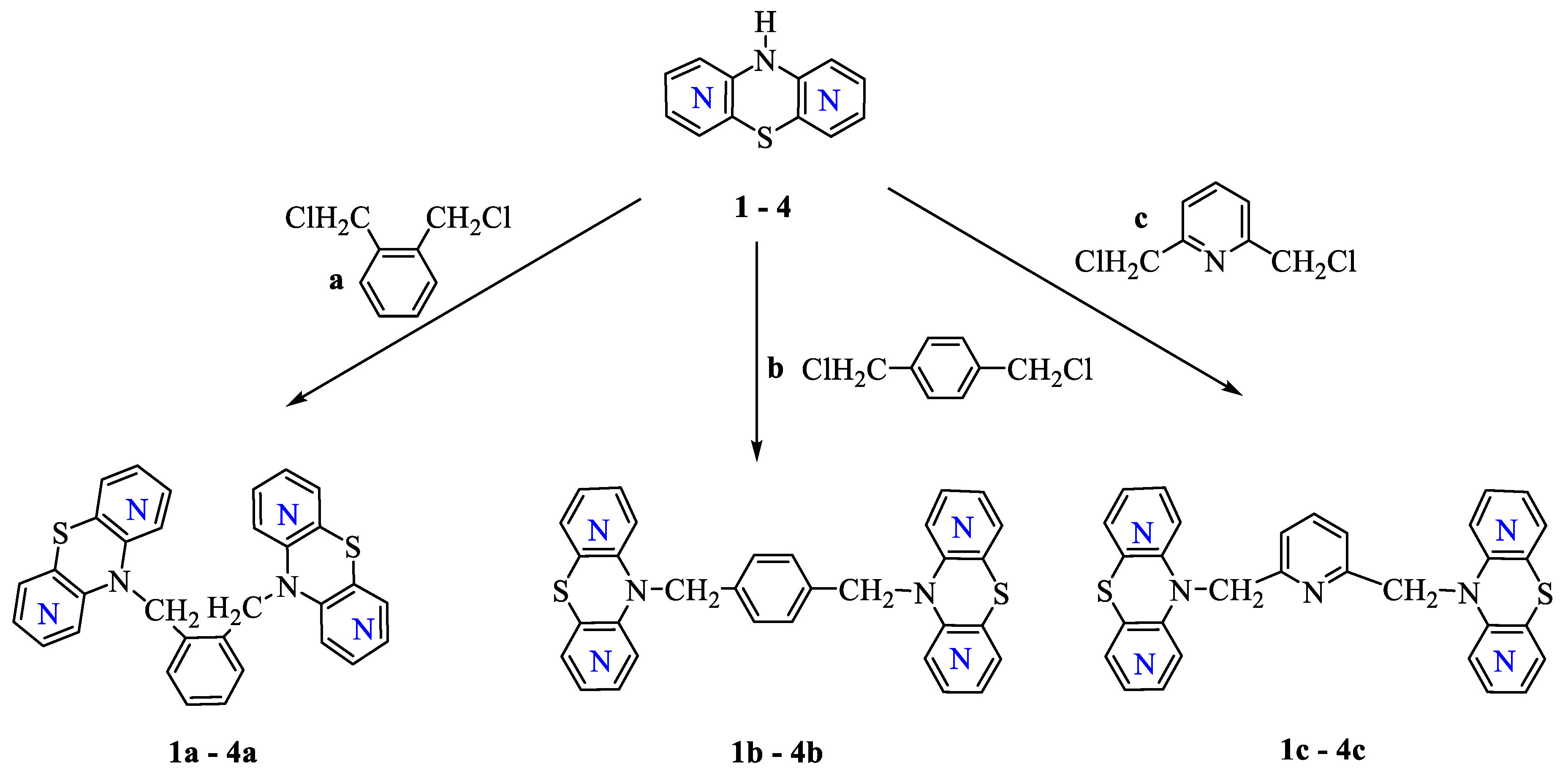
2.1. Chemistry Part
2.2. Structural 1H, 13C NMR and HRMS Study
2.3. In Silico Target Prediction
2.4. Biology Part
3. Materials and Methods
3.1. Chemistry
3.1.1. General Procedure for Synthesis of Compounds (1a,b,c–4a,b,c)
Dimers of 1,6-Diazaphenothiazines 1a,b,c
Dimers of 1,8-Diazaphenothiazines 2a,b,c
Dimers of 2,7-Diazaphenothiazines 3a,b,c
Dimers of 3,6-Diazaphenothiazines 4a,b,c
3.2. In Silico Target Prediction
3.3. Biological Evaluation
3.3.1. Human Cancer Cell Lines
3.3.2. MTT Cell Viability Assay
3.3.3. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Matthews, H.K.; Bertoli, C.; de Bruin, R.A.M. Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 2022, 23, 74–88. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Chhikara, B.S.; Parang, K. Global Cancer Statistics 2022: The trends projection analysis. Chem. Biol. Lett. 2023, 10, 451. [Google Scholar]
- Hadjiyianni, A.; Kalali, D. The potential anticancer activities of telmisartan– a literature review. Ann. Acad. Med. Silesiensis 2023, 77, 176–181. [Google Scholar] [CrossRef]
- Thiel, O. Heterocyclic Chemistry in Drug Discovery; Li, J.J., Ed.; Wiley: Hoboken, NJ, USA, 2013; pp. 1–16. ISBN 978-1118148907. [Google Scholar]
- Zhang, J.; Duan, D.; Song, Z.; Liu, T.; Hou, Y.; Fan, J. Small molecules regulating reactive oxygenspecies homeostasis for cancer therapy. Med. Res. Rev. 2021, 41, 342–394. [Google Scholar] [CrossRef]
- Fallahi, P.; Ferrari, S.M.; Elia, G.; Ragusa, F.; Patrizio, A.; Paparo, S.R.; Marone, G.; Galdiero, M.R.; Guglielmi, G.; Foddis, R.; et al. Primary Cell Cultures for the Personalized Therapy in Aggressive Thyroid Cancer of Follicular Origin. Semin. Cancer Biol. 2022, 79, 203–216. [Google Scholar] [CrossRef]
- Jakobsen, G.; Sjue, K.; Paulsen, Ø.; Kaasa, S.; Hjermstad, M.J.; Klepstad, P. Zopiclone versus placebo for short-term treatment of insomnia in patients with advanced cancer—A double-blind, randomized placebo-controlled clinical multicenter phase IV trial. Support Care Cancer 2022, 31, 60. [Google Scholar] [CrossRef]
- Assefa, D.G.; Zeleke, E.D.; Bekele, D.; Ejigu, D.A.; Molla, W.; Woldesenbet, T.T.; Aynalem, A.; Abebe, M.; Mebratu, A.; Manyazewal, T. Isoniazid Preventive Therapy for Prevention of Tuberculosis among People Living with HIV in Ethiopia: A Systematic Review of Implementation and Impacts. Int. J. Environ. Res. Public Health 2023, 20, 621. [Google Scholar] [CrossRef]
- Serafini, G.; De Biase, A.; Lamazza, L.; Mazzucchi, G.; Lollobrigida, M. Efficacy of Topical Treatments for the Management of Symptomatic Oral Lichen Planus: A Systematic Review. Int. J. Environ. Res. Public Health 2023, 20, 1202. [Google Scholar] [CrossRef]
- Sobańska, A.W.; Żydek, G.; Włodno, P.; Brzezińska, E. Comparative (Q)SAR Analysis of Benzodiazepine Derivatives with Different Biological Activity. Eur. J. Med. Chem. 2015, 89, 147–155. [Google Scholar] [CrossRef]
- Fagerström, K. Nicotine: Pharmacology, Toxicity and Therapeutic use. J. Smok. Cessat. 2014, 9, 53–59. [Google Scholar] [CrossRef]
- Perez-Castro, L.; Garcia, R.; Venkateswaran, N.; Barnes, S.; Conacci-Sorrell, M. Tryptophan and its metabolites in normal physiology and cancer etiology. FEBS J. 2023, 290, 7–27. [Google Scholar] [CrossRef] [PubMed]
- Stipanuk, M.H.; Caudill, M.A. Biochemical, Physiological, and Molecular Aspects of Human Nutrition—E-Book; Elsevier Health Sciences: Philadelphia, PA, USA, 2013; p. 541. ISBN 9780323266956. [Google Scholar]
- Mosnaim, A.D.; Ranade, V.V.; Wolf, M.E.; Puente, J.; Valenzuela, M.A. Phenothiazine molecule provides the basic chemical structure for various classes of pharmacotherapeutic agents. Am. J. Ther. 2006, 13, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhao, D.; Liu, Z.; Liu, F. Repurposing psychiatric drugs as anticancer agents. Cancer Lett. 2018, 419, 257–265. [Google Scholar] [CrossRef]
- Otręba, M.; Stojko, J.; Rzepecka-Stojko, A. Phenothiazine derivatives and their impact on the necroptosis and necrosis processes. A review. Toxicology 2023, 492, 153528. [Google Scholar] [CrossRef]
- Ohlow, M.; Moosmann, B. Phenothiazine: The seven lives of pharmacology’s first lead structure. Drug Discov. Today 2011, 16, 119–131. [Google Scholar] [CrossRef]
- Jaszczyszyn, A.; Gąsiorowski, K.; Świątek, P.; Malinka, W.; Cieślik-Boczula, K.; Petrus, J.; Czarnik-Matusewicz, B. Chemical structure of phenothiazines and their biological activity. Pharmacol. Rep. 2012, 64, 16–23. [Google Scholar] [CrossRef]
- Pluta, K.; Morak-Młodawska, B.; Jeleń, M. Synthesis and properties of diaza-, triaza- and tetraazaphenothiazines. J. Heterocycl. Chem. 2009, 46, 355–391. [Google Scholar] [CrossRef]
- Morak-Młodawska, B.; Pluta, K.; Jeleń, M. Phenothiazines Modified with the Pyridine Ring as Promising Anticancer Agents. Life 2021, 11, 206. [Google Scholar] [CrossRef]
- Zhang, J.; Ming, C.; Zhang, W.; Okechukwu, P.N.; Morak-Mlodawska, B.; Pluta, K.; Jeleń, M.; Akim, A.M.; Ang, K.-P.; Ooi, K.K. 10H-3,6-Diazaphenothiazines Induce G2/M Phase Cell Cycle Arrest, Caspase-dependent Apoptosis and Inhibits Cell Invasion of A2780 Ovarian Carcinoma Cells through Regulation on NF-κB and (BIRC6-XIAP) Complexes. Drug Des. Dev. Ther. 2017, 11, 3045–3063. [Google Scholar] [CrossRef] [PubMed]
- Pluta, K.; Jeleń, M.; Morak-Młodawska, B.; Zimecki, M.; Artym, J.; Kocięba, M. Anticancer activity of newly synthesized azaphenothiazines in NCI’s anticancer screening. Pharmacol. Rep. 2010, 62, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Morak-Młodawska, B.; Pluta, K.; Matralis, A.N.; Kourounakis, A.P. Antioxidant Activity of Newly Synthesized 2,7-Diazaphenothiazines. Arch. Pharm. Chem. Life Sci. 2010, 343, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.F.; Cao, H.; Liu, H.; Li, B.Q.; Ma, Y.M. Synthesis and bioactivity of novel bis-heterocyclic compounds containing pyrazole and oxadiazoline. J. Chin. Chem. Soc. 2011, 58, 369–375. [Google Scholar] [CrossRef]
- May, B.C.; Fafarman, A.T.; Hong, S.B.; Rogers, M.; Deady, L.W.; Prusiner, S.B.; Cohen, F.E. Potent inhibition of scrapie prion replication in cultured cells by bis-acridines. Proc. Natl. Acad. Sci. USA 2003, 100, 3416–3421. [Google Scholar] [CrossRef]
- Kushwaha, K.; Sakhuja, R.; Jain, S.C. Synthesis and antimicrobial activity of novel bis-azaphenothiazines. Med. Chem. Res. 2013, 22, 4459–4467. [Google Scholar] [CrossRef]
- Madrid, P.B.; Polgar, W.E.; Tolla, L.; Tanga, M.J. Synthesis and antitubercular activity of phenothiazines withreduced binding to dopamine and serotonin receptors. Bioorg. Med. Chem. Lett. 2007, 17, 3014–3017. [Google Scholar] [CrossRef]
- Morak-Młodawska, B.; Pluta, K.; Latocha, M.; Jeleń, M. Synthesis, spectroscopic characterization, and anticancer activity of new 10-substituted 1,6-diazaphenothiazines. Med. Chem. Res. 2016, 25, 2425–2433. [Google Scholar] [CrossRef]
- Morak-Młodawska, B.; Pluta, K.; Zimecki, M.; Jeleń, M.; Artym, J.; Kocięba, M. Synthesis and selected immunological properties of 10-substituted 1,8-diazaphenothiazines. Med. Chem. Res. 2015, 24, 1408–1418. [Google Scholar] [CrossRef]
- Morak-Młodawska, B.; Pluta, K. Synthesis of novel dipyrido-1,4-thiazines. Heterocycles 2007, 71, 1347–1361. [Google Scholar] [CrossRef]
- Morak-Młodawska, B.; Pluta, K.; Latocha, M.; Suwińska, K.; Jeleń, M.; Kuśmierz, D. 3,6-Diazaphenothiazines as potential lead molecules—Synthesis, characterization and anticancer activity. J. Enzym. Inhib. Med. Chem. 2016, 31, 1512–1519. [Google Scholar] [CrossRef] [PubMed]
- Way2Drug, Understanding Chemical-Biological Interactions, Predictive Services. Available online: www.way2drug.com/passonline/index.php (accessed on 20 October 2022).
- ATTC Company. Available online: https://www.atcc.org/support/contact-us (accessed on 30 July 2023).


| 1H NMR (ppm) | NOESY | COSY | 13C NMR (ppm) | HSQC | HMBC |
|---|---|---|---|---|---|
| CH2 4.99 | 4.99–7.48 | 6.95–8.05 | CH2 51.89. | 6.30–123.208 | Hγ 7.84–120.44/154.90 |
| H9 6.02 | 4.99–6.02 | 6.02–7.51 | C9 109.53 | H9 6.02–109.53 | H1 7.48–133.16 |
| H4 6.95 | 4.99–7.34 | 7.34–7.84 | C4a 115.73 | H4 6.95–121.55 | H9 6.02–137.82 |
| Hβ 7.34 | 6.02–7.51 | Cβ 120.44 | Hβ 7.34–120.44 | H4 6.95–115.73 | |
| H1 7.48 | 6.95–8.05 | C4 121.55 | H1 7.48–135.25 | H6 7.91–150.24 | |
| H8 7.51 | C10a 133.16 | H8 7.51–148.38 | |||
| Hγ 7.84 | C1 135.25 | Hγ 7.84–138.97 | |||
| H6 7.91 | C9a 137.82 | H6 7.91–145.30 | |||
| H3 8.05 | Cγ 138.97 | H3 8.05–144.52 | |||
| C3 144.52 | |||||
| C6 145.30 | |||||
| C8 148.38 | |||||
| C5a 150.24 | |||||
| Cα 154.90 |
| Probability of Activity Spectrum (Pa %) | |||||
|---|---|---|---|---|---|
| 1a | (36%) Apoptosis antagonist | (27%) Histone deacetylase stimulant | (35%) Mitochondrial processing peptidase inhibitor | (79%) Glycosyl-phosphatidylinositol phospholipase D inhibitor | (56%) Chenodeoxy-choloyltaurine hydrolase inhibitor |
| 1b | (29%) Apoptosis antagonist | (30%) Histone deacetylase stimulant | (34%) Alzheimer’s disease treatment | (80%) Glycosyl-phosphatidylinositol phospholipase D inhibitor | (58%)(S)-6-hydroxy-nicotine oxidase inhibitor |
| 1c | (27%) Apoptosis antagonist | (39%) Histone deacetylase stimulant | (36%) Alzheimer’s disease treatment | (77%) Glycosyl-phosphatidylinositol phospholipase D inhibitor | (50%) Antiallergic |
| 2a | (32%) Cytochrome P450 inhibitor | (63%) Anaphylatoxin receptor antagonist | (27%) p21-activated kinase 1 inhibitor | (85%) Glycosyl-phosphatidylinositol phospholipase D inhibitor | (54%) Treatment of neurodegenerative diseases |
| 2b | (18%) Apoptosis antagonist | (13%) Protein kinase B alpha inhibitor | (33%) Mitochondrial processing peptidase inhibitor | (85%) Glycosyl-phosphatidylinositol phospholipase D inhibitor | (55%) Treatment of neurodegenerative diseases |
| 2c | (22%) Cytochrome P450 inhibitor | (9%) Histone deacetylase stimulant | (61%) Anaphylatoxin receptor antagonist | (83%) Glycosyl-phosphatidylinositol phospholipase D inhibitor | (53%) Treatment of neurodegenerative diseases |
| 3a | (43%) Cytochrome P450 inhibitor | (65%) Histone deacetylase stimulant | (30%) Mitochondrial processing peptidase inhibitor | (85%) Glycosyl-phosphatidylinositol phospholipase D inhibitor | (76%) Treatment of neurodegenerative diseases |
| 3b | (40%) Cytochrome P450 inhibitor | (67%) Histone deacetylase stimulant | (43%) Alzheimer’s disease treatment | (86%) Glycosyl-phosphatidylinositol phospholipase D inhibitor | (77%) Treatment of neurodegenerative diseases |
| 3c | (33%) Cytochrome P450 inhibitor | (70%) Histone deacetylase stimulant | (20%) Protein kinase inhibitor | (84%) Glycosyl-phosphatidylinositol phospholipase D inhibitor | (75%) Treatment of neurodegenerative diseases |
| 4a | (29%) Cytochrome P450 inhibitor | (76%) Histone deacetylase stimulant | (60%) Alzheimer’s disease treatment | (80%) Glycosyl-phosphatidylinositol phospholipase D inhibitor | (61%) Treatment of neurodegenerative diseases |
| 4b | (26%) Cytochrome P450 inhibitor | (78%) Histone deacetylase stimulant | (60%) Alzheimer’s disease treatment | (80%) Glycosyl-phosphatidylinositol phospholipase D inhibitor | (62%) Treatment of neurodegenerative diseases |
| 4c | (28%) Mitochondrial processing peptidase inhibitor | (79%) Histone deacetylase stimulant | (60%) Alzheimer’s disease treatment | (77%) Glycosyl-phosphatidylinositol phospholipase D inhibitor | (60%) Treatment of neurodegenerative diseases |
| Compounds | Anticancer Activity IC50 (µM) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 24 h Treatment Period | 48 h Treatment Period | 72 h Treatment Period | |||||||
| L6 | MCF-7 | SW480 | L6 | MCF-7 | SW480 | L6 | MCF-7 | SW480 | |
| 1a | 20.63 ± 0.29 | 7.85 ± 1.67 | 21.84 ± 0.36 | 19.13 ± 1.08 | 7.12 ± 0.42 | 20.52 ± 0.79 | 16.14 ± 0.42 | 7.49 ± 1.13 | 17.47 ± 0.5 |
| 1b | 15.98 ± 0.36 | 11.55 ± 0.15 | 28.65 ± 1.28 | 1.92± 1.39 | 15.92 ± 0.83 | 20.73 ± 1.22 | 11.71 ± 0.59 | 9.60 ± 0.46 | 19.76 ± 1.21 |
| 1c | 11.28 ± 0.99 | 5.00 ± 1.29 | 0.67 ± 2.05 | 2.67 ± 2.13 | 0.75 ± 1.85 | 3.97 ± 1.27 | 6.82 ± 1.24 | 1.14 ± 0.51 | 4.27 ± 0.19 |
| 2a | 22.06 ± 1.42 | 6.27 ± 0.95 | 9.68 ± 0.17 | 17.73± 0.27 | 9.65 ± 0.63 | 8.74 ± 0.03 | 13.04 ± 1.32 | 8.16 ± 0.97 | 7.433 ± 0.02 |
| 2b | 25.08 ± 1.12 | 14.74 ± 0.86 | 11.89 ± 0.44 | 17.41 ± 2.4 | 12.18 ± 0.15 | 9.85 ± 0.18 | 11.95 ± 1.68 | 11.14 ± 1.27 | 3.11 ± 0.88 |
| 2c | 17.63 ± 0.85 | 23.17 ± 0.33 | 17.24 ± 1.46 | 19.23± 0.13 | 10.02 ± 0.05 | 21.06 ± 0.99 | 17.96 ± 0.18 | 9.96 ± 0.03 | 15.92 ± 1.48 |
| 3a | 30.82 ± 0.47 | 4.774 ± 0.84 | 6.12 ± 0.21 | 21.73 ± 0.48 | 3.17 ± 0.13 | 4.16 ± 0.08 | 21.41 ± 0.17 | 6.10 ± 1.33 | 4.56 ± 1.18 |
| 3b | 20.83 ± 0.28 | 19.04 ± 1.23 | 24.49 ± 0.26 | 22.67 ± 0.29 | 28.03 ± 0.65 | 21.19 ± 0.14 | 22.15 ± 0.22 | 18.89 ± 0.15 | 19.30 ± 0.12 |
| 3c | 48.98 ± 0.33 | 15.03 ± 0.73 | 22.32 ± 0.93 | 28.39 ± 0.37 | 11.74 ± 0.38 | 28.75 ± 1.55 | 35.53 ± 0.04 | 25.17 ± 1.74 | 25.95 ± 0.84 |
| 4a | 35.90 ± 0.57 | 32.68 ± 2.31 | 22.98 ± 1.22 | 22.95 ± 1.39 | 33.77 ± 1.46 | 24.06 ± 0.66 | 14.94 ± 1.15 | 19.87 ± 2.36 | 24.40 ± 0.83 |
| 4b | 36.74 ± 1.03 | 29.73 ± 1.33 | 29.09 ± 0.37 | 30.84 ± 0.93 | 24.24 ± 0.17 | 26.45 ± 0.74 | 11.61 ± 0.47 | 25.59 ± 0.35 | 21.41 ± 0.21 |
| 4c | 27.05 ± 0.74 | 0.12 ± 1.28 | 16.59 ± 0.38 | 20.16 ± 0.75 | 2.112 × 10−5 ± 3.86 | 12.07 ± 0.35 | 13.84 ± 0.02 | NA | 11.59 ± 0.52 |
| Doxorubicin | 11.52 ± 0.68 | 28.40 ± 0.36 | 13.46 ± 0.11 | 6.62 ± 0.18 | 24.62 ± 2.37 | 10.44 ± 0.1 | 3.07 ± 0.38 | NA | 10.77 ± 0.07 |
| Compounds | Selectivity Index (SI) of Anticancer Activity | |||||
|---|---|---|---|---|---|---|
| 24 h Treatment Period | 48 h Treatment Period | 72 h Treatment Period | ||||
| MCF-7 | SW480 | MCF-7 | SW480 | MCF-7 | SW480 | |
| 1a | 2.62 | 0.94 | 2.69 | 0.93 | 2.15 | 0.92 |
| 1b | 1.38 | 0.55 | 0.12 | 0.1 | 1.22 | 0.59 |
| 1c | 2.25 | 16.5 | 3.56 | 0.67 | 5.5 | 1.5 |
| 2a | 3.5 | 2.32 | 1.83 | 2.03 | 1.59 | 1.75 |
| 2b | 1.71 | 2.10 | 1.42 | 1.77 | 1.02 | 3.84 |
| 2c | 0.76 | 1.02 | 1.91 | 0.91 | 1.80 | 1.12 |
| 3a | 6.46 | 5.02 | 6.85 | 5.22 | 3.51 | 4.69 |
| 3b | 1.09 | 0.95 | 0.81 | 1.07 | 1.17 | 1.15 |
| 3c | 3.25 | 2.19 | 2.42 | 0.94 | 1.41 | 1.37 |
| 4a | 1.10 | 1.56 | 0.68 | 0.95 | 0.75 | 0.61 |
| 4b | 1.23 | 1.21 | 1.27 | 1.16 | 0.45 | 0.54 |
| 4c | >100 | 1.63 | >100 | 1.67 | - | 1.34 |
| Doxorubicin | 0.41 | 0.85 | 0.25 | 0.63 | - | 0.28 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martula, E.; Morak-Młodawska, B.; Jeleń, M.; Okechukwu, P.N.; Balachandran, A.; Tehirunavukarasu, P.; Anamalay, K.; Ulaganathan, V. Synthesis and Structural Characterization of Novel Dimers of Dipyridothiazine as Promising Antiproliferative Agents. Molecules 2023, 28, 7662. https://doi.org/10.3390/molecules28227662
Martula E, Morak-Młodawska B, Jeleń M, Okechukwu PN, Balachandran A, Tehirunavukarasu P, Anamalay K, Ulaganathan V. Synthesis and Structural Characterization of Novel Dimers of Dipyridothiazine as Promising Antiproliferative Agents. Molecules. 2023; 28(22):7662. https://doi.org/10.3390/molecules28227662
Chicago/Turabian StyleMartula, Emilia, Beata Morak-Młodawska, Małgorzata Jeleń, Patrick N. Okechukwu, Abbirami Balachandran, Prethika Tehirunavukarasu, Kirthani Anamalay, and Vaidehi Ulaganathan. 2023. "Synthesis and Structural Characterization of Novel Dimers of Dipyridothiazine as Promising Antiproliferative Agents" Molecules 28, no. 22: 7662. https://doi.org/10.3390/molecules28227662
APA StyleMartula, E., Morak-Młodawska, B., Jeleń, M., Okechukwu, P. N., Balachandran, A., Tehirunavukarasu, P., Anamalay, K., & Ulaganathan, V. (2023). Synthesis and Structural Characterization of Novel Dimers of Dipyridothiazine as Promising Antiproliferative Agents. Molecules, 28(22), 7662. https://doi.org/10.3390/molecules28227662







