Glucuronidation Pathways of 5- and 7-Hydroxypropranolol: Determination of Glucuronide Structures and Enzyme Selectivity
Abstract
:1. Introduction
2. Results
2.1. Identification of 4, 5- and 7-OHPGs
2.2. Glucuronidation of 5- and 7-OHP by 19 UGT Enzyme Bags
2.3. Determination of the Regioselectivity of 5- and 7-OHP Glucuronidation
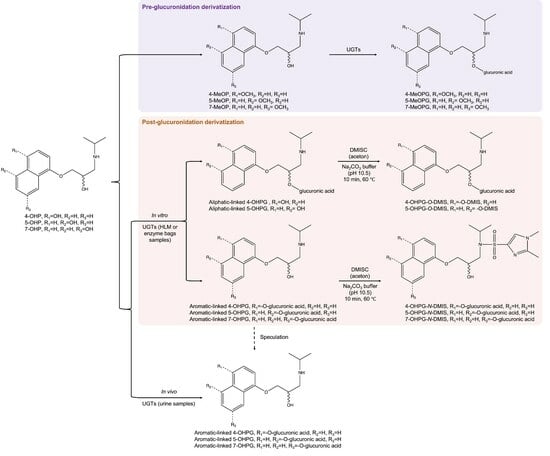
2.3.1. Pre-Glucuronidation Derivatization
2.3.2. Post-Glucuronidation Derivatization
DMIS Derivatives of Propranolol and Hydroxypropranolols
DMIS Derivatives of Hydroxypropranolol Glucuronides
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Preparation of Compound Solutions
4.3. Biotransformation with 19 Human Recombinant UGTs by Enzyme Bags Method
4.4. Incubation with Human Liver Microsomes
4.5. Urine Collection
4.6. Derivatization with DMISC
4.7. LC-MS/MS Analysis
4.7.1. LC-QQQ-MS/MS
4.7.2. LC-QTOF-MS/MS
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tukey, R.H.; Strassburg, C.P. Human UDP-glucuronosyltransferases: Metabolism, expression, and disease. Annu. Rev. Pharmacol. Toxicol. 2000, 40, 581–616. [Google Scholar] [CrossRef] [PubMed]
- Rowland, A.; Miners, J.O.; Mackenzie, P.I. The UDP-glucuronosyltransferases: Their role in drug metabolism and detoxification. Int. J. Biochem. Cell Biol. 2013, 45, 1121–1132. [Google Scholar] [CrossRef] [PubMed]
- Guillemette, C.; Levesque, E.; Rouleau, M. Pharmacogenomics of human uridine diphospho-glucuronosyltransferases and clinical implications. Clin. Pharmacol. Ther. 2014, 96, 324–339. [Google Scholar] [CrossRef] [PubMed]
- Miners, J.O.; Mackenzie, P.I. Drug glucuronidation in humans. Pharmacol. Ther. 1991, 51, 347–369. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, P.I.; Bock, K.W.; Burchell, B.; Guillemette, C.; Ikushiro, S.; Iyanagi, T.; Miners, J.O.; Owens, I.S.; Nebert, D.W. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet. Genom. 2005, 15, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, P.I.; Rogers, A.; Treloar, J.; Jorgensen, B.R.; Miners, J.O.; Meech, R. Identification of UDP Glycosyltransferase 3A1 as a UDP N-Acetylglucosaminyltransferase. J. Biol. Chem. 2008, 283, 36205–36210. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, P.I.; Rogers, A.; Elliot, D.J.; Chau, N.; Hulin, J.A.; Miners, J.O.; Meech, R. The novel UDP glycosyltransferase 3A2: Cloning, catalytic properties, and tissue distribution. Mol. Pharmacol. 2011, 79, 472–478. [Google Scholar] [CrossRef]
- Meech, R.; Mubarokah, N.; Shivasami, A.; Rogers, A.; Nair, P.C.; Hu, D.G.; McKinnon, R.A.; Mackenzie, P.I. A novel function for UDP glycosyltransferase 8: Galactosidation of bile acids. Mol. Pharmacol. 2015, 87, 442. [Google Scholar] [CrossRef]
- Al-Majed, A.A.; Bakheit, A.H.H.; Abdel Aziz, H.A.; Alajmi, F.M.; AlRabiah, H. Chapter Six—Propranolol. In Profiles of Drug Substances, Excipients and Related Methodology; Brittain, H.G., Ed.; Academic Press: Cambridge, MA, USA, 2017; Volume 42, pp. 287–338. [Google Scholar]
- Shand, D.G. Propranolol. N. Engl. J. Med. 1975, 293, 280–285. [Google Scholar] [CrossRef]
- Routledge, P.A.; Shand, D.G. Clinical pharmacokinetics of propranolol. Clin. Pharmacokinet. 1979, 4, 73–90. [Google Scholar] [CrossRef]
- Elman, M.J.; Sugar, J.; Fiscella, R.; Deutsch, T.A.; Noth, J.; Nyberg, M.; Packo, K.; Anderson, R.J. The effect of propranolol versus placebo on resident surgical performance. Trans. Am. Ophthalmol. Soc. 1998, 96, 283. [Google Scholar] [PubMed]
- Steenen, S.A.; Van Wijk, A.J.; Van Der Heijden, G.J.; van Westrhenen, R.; de Lange, J.; de Jongh, A. Propranolol for the treatment of anxiety disorders: Systematic review and meta-analysis. J. Psychopharmacol. 2016, 30, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, D.S.; Anderson, I.M.; Nutt, D.J.; Allgulander, C.; Bandelow, B.; den Boer, J.A.; Christmas, D.M.; Davies, S.; Fineberg, N.; Lidbetter, N. Evidence-based pharmacological treatment of anxiety disorders, post-traumatic stress disorder and obsessive-compulsive disorder: A revision of the 2005 guidelines from the British Association for Psychopharmacology. J. Psychopharmacol. 2014, 28, 403–439. [Google Scholar] [CrossRef] [PubMed]
- Léauté-Labrèze, C.; De La Roque, E.D.; Hubiche, T.; Boralevi, F.; Thambo, J.-B.; Taïeb, A. Propranolol for severe hemangiomas of infancy. N. Engl. J. Med. 2008, 358, 2649–2651. [Google Scholar] [CrossRef] [PubMed]
- McDonald, J.; Bayrak-Toydemir, P.; Pyeritz, R.E. Hereditary hemorrhagic telangiectasia: An overview of diagnosis, management, and pathogenesis. Genet. Med. 2011, 13, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Horne, M.A.; Flemming, K.D.; Su, I.-C.; Stapf, C.; Jeon, J.P.; Li, D.; Maxwell, S.S.; White, P.; Christianson, T.J.; Agid, R. Clinical course of untreated cerebral cavernous malformations: A meta-analysis of individual patient data. Lancet Neurol. 2016, 15, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Bichara, N.; Ching, M.S.; Blake, C.L.; Ghabrial, H.; Smallwood, R.A. Propranolol hydroxylation and N-desisopropylation by cytochrome P4502D6: Studies using the yeast-expressed enzyme and NADPH/O2 and cumene hydroperoxide-supported reactions. Drug Metab. Dispos. 1996, 24, 112–118. [Google Scholar]
- Silber, B.; Holford, N.H.; Riegelman, S. Stereoselective disposition and glucuronidation of propranolol in humans. J. Pharm. Sci. 1982, 71, 699–704. [Google Scholar] [CrossRef]
- Masubuchi, Y.; Kagimoto, N.; Narimatsu, S.; Fujita, S.; Suzuki, T. Regioselective contribution of the cytochrome P-450 2D subfamily to propranolol metabolism in rat liver microsomes. Drug Metab. Dispos. 1993, 21, 1012–1016. [Google Scholar]
- Walle, T.; Oatis, J.E.; Walle, U.K.; Knapp, D.R. New ring-hydroxylated metabolites of propranolol: Species differences and stereospecific 7-hydroxylation. Drug Metab. Dispos. 1982, 10, 122–127. [Google Scholar]
- Harps, L.C.; Schipperges, S.; Bredendiek, F.; Wuest, B.; Borowiak, A.; Parr, M.K. Two dimensional chromatography mass spectrometry: Quantitation of chiral shifts in metabolism of propranolol in bioanalysis. J. Chromatogr. A 2020, 1617, 460828. [Google Scholar] [CrossRef] [PubMed]
- Vashistha, V.K.; Kumar, A. Stereochemical facets of clinical β-blockers: An overview. Chirality 2020, 32, 722–735. [Google Scholar] [CrossRef] [PubMed]
- Walle, T.; Gaffney, T.E. N-dealkylation and oxidative deamination of propranolol in the cardio-pulmonary circuit of dogs. In Proceedings of the Volunteer Presentations, Fifth International Congress on Pharmacology, San Francisco, CA, USA, 23–28 July 1972. [Google Scholar]
- Saelens, D.A.; Walle, T.; Gaffney, T.E.; Privitera, P.J. Studies on the contribution of active metabolites to the anticonvulsant effects of propranolol. Eur. J. Pharmacol. 1977, 42, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, J.; O’donnell, S.R. Pharmacology of 4-hydroxypropranolol, a metabolite of propranolol. Br. J. Pharmacol. 1971, 43, 222–235. [Google Scholar] [CrossRef] [PubMed]
- Ram, N.; Bauer, E.W.; Hesse, U.C.; Heilman, R.D. Cardiovascular effects of 5-hydroxypropranolol (ORF 12592) in dogs. Arch. Int. de Pharmacodyn. et de Ther. 1977, 228, 118–125. [Google Scholar]
- Yang, F.; Liu, S.; Wolber, G.; Bureik, M.; Parr, M.K. Complete reaction phenotyping of propranolol and 4-hydroxypropranolol with the 19 enzymes of the human UGT1 and UGT2 families. Int. J. Mol. Sci. 2022, 23, 7476. [Google Scholar] [CrossRef] [PubMed]
- Dorp, E.L.v.; Morariu, A.; Dahan, A. Morphine-6-glucuronide: Potency and safety compared with morphine. Expert Opin. Pharmacother. 2008, 9, 1955–1961. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.A.; Hull, J.E.; Norris, K.J. Glucuronidation of propranolol and 4′-hydroxypropranolol. Substrate specificity and stereoselectivity of rat liver microsomal glucuronyltransferases. Drug Metab. Dispos. 1981, 9, 466–471. [Google Scholar]
- Salomonsson, M.L.; Bondesson, U.; Hedeland, M. In vitro formation of phase I and II metabolites of propranolol and determination of their structures using chemical derivatization and liquid chromatography–tandem mass spectrometry. J. Mass Spectrom. 2009, 44, 742–754. [Google Scholar] [CrossRef]
- Salomonsson, M.L.; Bondesson, U.; Hedeland, M. Structural evaluation of the glucuronides of morphine and formoterol using chemical derivatization with 1, 2-dimethylimidazole-4-sulfonyl chloride and liquid chromatography/ion trap mass spectrometry. Rapid Commun. Mass Spectrom. Int. J. Devoted Rapid Dissem. Up-Minute Res. Mass Spectrom. 2008, 22, 2685–2697. [Google Scholar] [CrossRef]
- Xu, L.; Spink, D.C. 1, 2-Dimethylimidazole-4-sulfonyl chloride, a novel derivatization reagent for the analysis of phenolic compounds by liquid chromatography electrospray tandem mass spectrometry: Application to 1-hydroxypyrene in human urine. J. Chromatogr. B 2007, 855, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Walle, T.; Walle, U.K.; Olanoff, L.S. Quantitative account of propranolol metabolism in urine of normal man. Drug Metab. Dispos. Biol. Fate Chem. 1985, 13, 204–209. [Google Scholar] [PubMed]
- Cleaveland, C.R.; Shand, D.G. Effect of route of administration on the relationship between β-adrenergic blockade and plasma propranolol level. Clin. Pharmacol. Ther. 1972, 13, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Otton, S.; Gillam, E.; Lennard, M.; Tucker, G.; Woods, H. Propranolol oxidation by human liver microsomes-the use of cumene hydroperoxide to probe isoenzyme specificity and regio-and stereoselectivity. Br. J. Clin. Pharmacol. 1990, 30, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.K.; Wong, C.S. Simultaneous quantification of propranolol and sulfamethoxazole and major human metabolite conjugates 4-hydroxy-propranolol sulfate and sulfamethoxazole-β-glucuronide in municipal wastewater—A framework for multiple classes of drugs and conjugates. J. Chromatogr. A 2016, 1471, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Proctor, L.D.; Warr, A.J. Development of a continuous process for the industrial generation of diazomethane1. Org. Process Res. Dev. 2002, 6, 884–892. [Google Scholar] [CrossRef]
- Maas, A.; Maier, C.; Michel-Lauter, B.; Madea, B.; Hess, C. 1,2-Dimethylimidazole-4-sulfonyl chloride (DMISC), a novel derivatization strategy for the analysis of propofol by LC-ESI-MS/MS. Anal. Bioanal. Chem. 2017, 409, 1547–1554. [Google Scholar] [CrossRef] [PubMed]
- Oatis, J.E., Jr.; Russell, M.P.; Knapp, D.R.; Walle, T. Ring-hydroxylated propranolol: Synthesis and beta-receptor antagonist and vasodilating activities of the seven isomers. J. Med. Chem. 1981, 24, 309–314. [Google Scholar] [CrossRef]
- Yang, F.; Machalz, D.; Wang, S.; Li, Z.; Wolber, G.; Bureik, M. A common polymorphic variant of UGT1A5 displays increased activity due to optimized cofactor binding. FEBS Lett. 2018, 592, 1837–1846. [Google Scholar] [CrossRef]
- Alfa, C.; Fantes, P.; Hyams, J.; McLeod, M.; Warbrick, E. Experiments with Fission Yeast. A Laboratory Course Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1993. [Google Scholar]
- Yang, F.; Sharma, S.S.; Bureik, M.; Parr, M.K. Mutual modulation of the activities of human CYP2D6 and four UGTs during the metabolism of propranolol. Curr. Issues Mol. Biol. 2023, 45, 7130–7146. [Google Scholar] [CrossRef]







| UGTs | (S)-5-OHPG | (R)-5-OHPG | (S)-7-OHPG | (R)-7-OHPG |
|---|---|---|---|---|
| UGT1A1 | + | + | + | + |
| UGT1A3 | + | + | + | + |
| UGT1A4 | - | - | - | - |
| UGT1A5 | - | - | - | - |
| UGT1A6 | - | - | + | + |
| UGT1A7 | + + + + | + + + + | + + + + + | + + + + |
| UGT1A8 | + + | + | + | + |
| UGT1A9 | + + + + | + + + | + + + + + | + + + + |
| UGT1A10 | + | + | + | + |
| UGT2A1 | + + + | + + + | + + + + | + + |
| UGT2A2 | + | + | + | + |
| UGT2A3 | - | - | - | - |
| UGT2B4 | - | - | - | - |
| UGT2B7 | - | - | - | - |
| UGT2B10 | - | - | - | - |
| UGT2B11 | - | - | - | - |
| UGT2B15 | - | - | - | - |
| UGT2B17 | - | - | - | - |
| UGT2B28 | - | - | - | - |
| UGTs | 5-MeOPG I | 5-MeOPG II | 7-MeOPG I | 7-MeOPG II |
|---|---|---|---|---|
| UGT1A1 | + + | + + | + | + + |
| UGT1A3 | + + | + + | + + | + + |
| UGT1A6 | ** | ** | - | - |
| UGT1A7 | + + + | + + + + | + + + | + + + + |
| UGT1A8 | - | - | - | - |
| UGT1A9 | + + + + | + + + + | + + + + | + + + + |
| UGT1A10 | + + + | + | + + | + |
| UGT2A1 | + + + + | + + + + | + + + + | + + + + |
| UGT2A2 | - | - | - | - |
| Postulated Fragment | Elementary Composition | Theoretical Mass (m/z) | Experimental Mass (m/z) | Mass Error (ppm) |
|---|---|---|---|---|
| [M+H]+ | [C21H28N3O4S]+ | 418.1795 | 418.1797 | 0.48 |
| [M+H–C3H6]+ | [C18H22N3O4S]+ | 376.1326 | 376.1332 | 1.60 |
| [M+H–C3H6–H2O]+ | [C18H20N3O3S]+ | 358.1220 | 358.1212 | –2.23 |
| [M+H–C10H8O]+ | [C11H20N3O3S]+ | 274.1220 | 274.1214 | –2.19 |
| [M+H–C10H8O–C3H6]+ | [C8H14N3O3S]+ | 232.0750 | 232.0752 | 0.86 |
| [M+H–C10H8O–C3H6–H2O]+ | [C8H12N3O2S]+ | 214.0645 | 214.0642 | –1.40 |
| [DMIS-NCH2+H]+ | [C6H10N3O2S]+ | 188.0488 | 188.0487 | –0.53 |
| [DMIS-NH2+H]+ | [C5H10N3O2S]+ | 176.0488 | 176.0488 | 0.00 |
| Postulated Fragment | Elementary Composition | Theoretical Mass (m/z) | Experimental Mass (m/z) | Mass Error (ppm) |
|---|---|---|---|---|
| [M+H]+ | [C21H28N3O5S]+ | 434.1744 | 434.1750 | 1.38 |
| [M+H–C3H6]+ | [C18H22N3O5S]+ | 392.1275 | 392.1274 | −0.26 |
| [M+H–C3H9N–H2O]+ | [C18H17N2O4S]+ | 357.0907 | 357.0897 | −2.8 |
| [M+H–DMIS]+ | [C16H20NO3]+ | 274.1438 | 274.1447 | 3.28 |
| [M+H–DMIS–C3H6]+ | [C13H14NO3]+ | 232.0968 | 232.0966 | −0.86 |
| [M+H–DMIS–C3H9N]+ | [C13H11O3]+ | 215.0703 | 215.0703 | 0.00 |
| [M+H–DMIS–C10H8O]+ | [C6H14NO]+ | 116.1072 | 116.1072 | 0.00 |
| Postulated Fragment | Elementary Composition | Theoretical Mass (m/z) | Experimental Mass (m/z) | Mass Error (ppm) |
|---|---|---|---|---|
| [M+H]+ | [C26H34N5O7S2]+ | 592.1894 | 592.1892 | −0.33 |
| [M+H–C3H6]+ | [C23H28N5O7S2]+ | 550.1425 | 550.1429 | 0.73 |
| [M+H–DMIS]+ | [C21H26N3O5S]+ | 432.1588 | 432.1575 | −3.00 |
| [M+H–C3H6–DMIS]+ | [C18H20N3O5S]+ | 390.1118 | 390.1118 | 0.00 |
| [M+H–DMIS–C3H9N–H2O]+ | [C18H17N2O4S]+ | 357.0904 | 357.0894 | −2.80 |
| [M+H–DMIS–C10H8O]+ | [C11H20N3O3S]+ | 274.1220 | 274.1217 | −1.09 |
| [M+H–DMIS–C10H8O–C3H6]+ | [C8H14N3O3S]+ | 232.0750 | 232.0752 | 0.86 |
| [NH2-(CH3)2-DMIS]+ | [C8H16N3O2S]+ | 218.0958 | 218.0952 | −2.75 |
| [M+H–2DMIS–C3H9N–H2O]+ | [C13H11O2]+ | 199.0754 | 199.0749 | −2.51 |
| [DMIS-NCH2+H]+ | [C6H10N3O2S]+ | 188.0488 | 188.0486 | −1.06 |
| [DMIS-NH2]+ | [C5H10N3O2S]+ | 176.0488 | 176.0483 | −2.84 |
| Analytes | Retention Time (min) | Precursor Ions (m/z) | Product Ions (m/z) | CE (eV) | ESI |
|---|---|---|---|---|---|
| 4-Methoxypropranolol | 27.6 | 290 | 187 | 12 | + |
| 5-Methoxypropranolol | 25.1 | 116 | 16 | ||
| 7-Methoxypropranolol | 27.6 | 72 | 44 | ||
| (R)-4-Hydroxypropranolol glucuronide | 8.7 | 452 | 276 | 12 | + |
| (S)-4-Hydroxypropranolol glucuronide | 8.5 | 199 | 20 | ||
| (R)-5-Hydroxypropranolol glucuronide | 6.4 | 173 | 24 | ||
| (S)-5-Hydroxypropranolol glucuronide | 6.7 | 116 | 28 | ||
| (R)-7-Hydroxypropranolol glucuronide | 17.3 | 98 | 40 | ||
| (S)-7-Hydroxypropranolol glucuronide | 16.6 | 72 | 44 | ||
| 4-Methoxypropranolol glucuronide I | 13.3 | 466 | 290 | 25 | + |
| 4-Methoxypropranolol glucuronide II | 13.6 | 213 | 30 | ||
| 5-Methoxypropranolol glucuronide I | 24.4 | 187 | 30 | ||
| 5-Methoxypropranolol glucuronide II | 26.1 | 116 | 28 | ||
| 7-Methoxypropranolol glucuronide I | 25.7 | 72 | 44 | ||
| 7-Methoxypropranolol glucuronide II | 27.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, F.; Wenzel, M.; Bureik, M.; Parr, M.K. Glucuronidation Pathways of 5- and 7-Hydroxypropranolol: Determination of Glucuronide Structures and Enzyme Selectivity. Molecules 2023, 28, 7783. https://doi.org/10.3390/molecules28237783
Yang F, Wenzel M, Bureik M, Parr MK. Glucuronidation Pathways of 5- and 7-Hydroxypropranolol: Determination of Glucuronide Structures and Enzyme Selectivity. Molecules. 2023; 28(23):7783. https://doi.org/10.3390/molecules28237783
Chicago/Turabian StyleYang, Fan, Maxi Wenzel, Matthias Bureik, and Maria Kristina Parr. 2023. "Glucuronidation Pathways of 5- and 7-Hydroxypropranolol: Determination of Glucuronide Structures and Enzyme Selectivity" Molecules 28, no. 23: 7783. https://doi.org/10.3390/molecules28237783
APA StyleYang, F., Wenzel, M., Bureik, M., & Parr, M. K. (2023). Glucuronidation Pathways of 5- and 7-Hydroxypropranolol: Determination of Glucuronide Structures and Enzyme Selectivity. Molecules, 28(23), 7783. https://doi.org/10.3390/molecules28237783