Antibacterial Activity and Untargeted Metabolomics Profiling of Acalypha arvensis Poepp
Abstract
:1. Introduction
2. Results
2.1. Antibacterial Screening of Acalypha arvensis Poepp.
2.2. Phytochemical Characteristics of A. arvensis
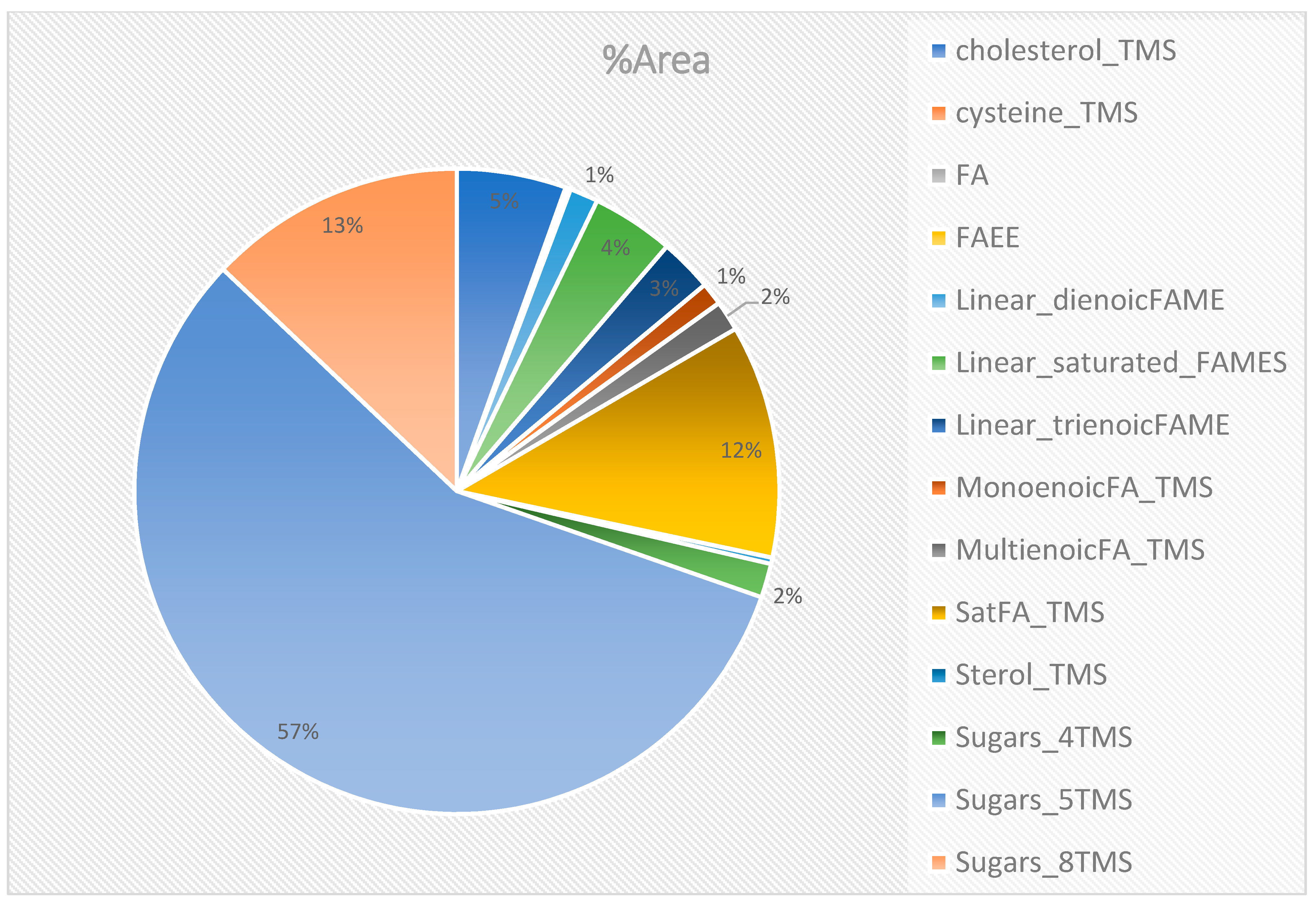
2.3. Untargeted Analysis of GC×GC-TOFMS Metabolite Profiles of the Methanol: Chloroform Extract (50:50), F6
3. Discussion
4. Materials and Methods
4.1. Plant Material, Extraction, and Chemicals
4.2. Bacterial Strains
4.2.1. Performance of the Antibiogram
4.2.2. TLC Agar Overlay Bioautography
4.3. Phytochemical Screening
- To test for flavonoids, a Neu’s reagent or NP/PEG method was used. Two solutions were prepared for the test. Solution A, which was made up of 1 g of 2-aminoethyldiphenylboric acid and 100 mL of methanol, and solution B, which was made up of 5 g of PEG 4000 and 100 mL of ethanol. Then, a mixture of 10 mL of solution A and 8 mL of solution B was sprayed onto a TLC plate. The TLC plate was heated at 110 °C for approximately 2 min. The flavonoids were then observed under UV light at 366 nm and appeared as yellow, green, or orange fluorescent spots.
- A Dragendorff reagent test was used to detect the presence of alkaloids. To perform the test, we first prepared two solutions. Solution A, which was made up of 0.85 g of basic bismuth nitrate and 10 g of tartaric acid dissolved in 40 mL of water, and solution B, which was made up of 16 g of KI dissolved in 40 mL of water. The two solutions were mixed extemporaneously with 5 mL of solution A, 5 mL of solution B, 100 mL of water, and 20 g of tartaric acid. The mixture was then sprayed onto a TLC plate. Alkaloids appeared as orange spots on the plate.
- To detect the presence of sugars, a sulfuric thymol reagent test was used. A solution was prepared by dissolving 0.5 g of thymol in 95 mL of ethanol, and then 5 mL of concentrated sulfuric acid was added. The mixture was sprayed onto a TLC plate and heated at 110 °C for about 15 min. The sugars appeared as pink spots on the plate.
- To identify the presence of sterols, steroids, and triterpenes, a Liebermann and Burchard reagent test was performed. A solution was prepared by mixing 5 mL of acetic anhydride, 5 mL of concentrated sulfuric acid, and 50 mL of 95% ethanol at low temperature just before use, then sprayed onto a TLC plate. The plate was then heated at 110 °C for 10 min. The compounds appeared as fluorescence at 366 nm under UV light.
- A test was performed to detect the presence of tannins. A volume of 5 mL of extract was introduced into a test tube, and 0.5 mL of a 1% aqueous solution of FeCl3 was added. The presence of tannins was indicated by a greenish or blue-blackish color change in the solution.
- A test was performed to identify the presence of coumarins. A volume of 5 mL of extract was placed in a test tube, to which 0.5 mL of a 10% solution of NH4OH was added. The mixture was then observed under UV light at 366 nm. An intense fluorescence indicated the presence of coumarins.
- A test was conducted to detect the presence of terpenoids. A volume of 5 mL of extract was added to a mixture of 2 mL of chloroform and 3 mL of concentrated sulfuric acid. The presence of terpenoids was indicated by the formation of two layers and a brown color at the interface.
- A foam test was performed to identify the presence of saponins. A volume of 10 mL of the extract was added to a test tube and shaken for a few seconds, then left to rest for 15 min. The presence of saponins was indicated by a persistent height of foam on top of the liquid.
- To visualize the compounds using the sulfuric vanillin polyvalent developer, a solution was prepared by combining 1 g of vanillin, 2 mL of sulfuric acid, and 95% ethanol up to a total volume of 100 mL. After spraying the TLC plate with this solution, it was heated at 110 °C for around 5 min. The compounds present on the plate exhibited various colors depending on their chemical nature.
4.4. Isolation of Compounds from A. arvensis F6 Fraction Using SPME and Derivatization and GC×GC-TOFMS
4.4.1. SPME Analysis
4.4.2. Derivatization Analysis
4.4.3. GC×GC-TOFMS Method
4.4.4. Data Processing and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gross, M. Antibiotics in Crisis. Curr. Biol. 2013, 23, R1063–R1065. [Google Scholar] [CrossRef] [PubMed]
- Golkar, Z.; Bagasra, O.; Pace, D.G. Bacteriophage Therapy: A Potential Solution for the Antibiotic Resistance Crisis. J. Infect. Dev. Ctries. 2014, 8, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Lushniak, B.D. Surgeon General’s Perspectives. Public Health Rep. 2014, 129, 314–316. [Google Scholar] [CrossRef] [PubMed]
- Abraham, E.P. Chapter XI—The Antibiotics. In Comprehensive Biochemistry; Florkin, M., Stotz, E.H., Eds.; Water-Soluble Vitamins, Hormones, Antibiotics; Elsevier: Amsterdam, The Netherlands, 1963; Volume 11, pp. 181–224. [Google Scholar] [CrossRef]
- Projan, S.J. Why Is Big Pharma Getting out of Antibacterial Drug Discovery? Curr. Opin. Microbiol. 2003, 6, 427–430. [Google Scholar] [CrossRef]
- Power, E. Impact of Antibiotic Restrictions: The Pharmaceutical Perspective. Clin. Microbiol. Infect. 2006, 12, 25–34. [Google Scholar] [CrossRef]
- Gray, D.A.; Wenzel, M. More Than a Pore: A Current Perspective on the In Vivo Mode of Action of the Lipopeptide Antibiotic Daptomycin. Antibiotics 2020, 9, 17. [Google Scholar] [CrossRef]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Report; Government of the United Kingdom: London, UK, 2016; Available online: https://apo.org.au/node/63983 (accessed on 21 May 2023).
- Sermakkani, M.; Thangapandian, V. GC-MS Analysis of Cassia Italica Leaf Methanol Extract; Asian Journal of Pharmaceutical and Clinical Research. Available online: https://asset-pdf.scinapse.io/prod/2186275073/2186275073.pdf (accessed on 3 January 2023).
- McGaw, L.J.; Lall, N.; Meyer, J.J.M.; Eloff, J.N. The Potential of South African Plants against Mycobacterium Infections. J. Ethnopharmacol. 2008, 119, 482–500. [Google Scholar] [CrossRef]
- Courric, E.; Brinvilier, D.; Couderc, P.; Ponce-Mora, A.; Méril-Mamert, V.; Sylvestre, M.; Pelage, J.H.; Vaillant, J.; Rousteau, A.; Bejarano, E.; et al. Medicinal Plants and Plant-Based Remedies in Grande-Terre: An Ethnopharmacological Approach. Plants 2023, 12, 654. [Google Scholar] [CrossRef]
- Turi, C.E.; Murch, S.J. Spiritual and Ceremonial Plants in North America: An Assessment of Moerman’s Ethnobotanical Database Comparing Residual, Binomial, Bayesian and Imprecise Dirichlet Model (IDM) Analysis. J. Ethnopharmacol. 2013, 148, 386–394. [Google Scholar] [CrossRef]
- Volpato, G.; Godínez, D.; Beyra, A.; Barreto, A. Uses of Medicinal Plants by Haitian Immigrants and Their Descendants in the Province of Camagüey, Cuba. J. Ethnobiol. Ethnomed. 2009, 5, 16. [Google Scholar] [CrossRef]
- Boulogne, I.; Germosén-Robineau, L.; Ozier-Lafontaine, H.; Fleury, M.; Loranger-Merciris, G. TRAMIL Ethnopharmalogical Survey in Les Saintes (Guadeloupe, French West Indies): A Comparative Study. J. Ethnopharmacol. 2011, 133, 1039–1050. [Google Scholar] [CrossRef] [PubMed]
- Salla, B. Prise en charge des symptômes douloureux par la médecine traditionnelle haïtienne: Résultats d’une enquête réalisée dans le quartier de Martissant à Port-au-Prince. Douleurs Éval.-Diagn.-Trait. 2017, 18, 223–233. [Google Scholar] [CrossRef]
- TRAMIL. Program of Applied Research to Popular Medicine in the Caribbean. Available online: https://www.tramil.net/fr (accessed on 2 January 2023).
- Fournet, J. Flore Illustrée des Phanérogames de Guadeloupe et de Martinique; CIRAD: Montpellier, France, 2002. [Google Scholar]
- Caceres, A.; Cano, O.; Samayoa, B.; Aguilar, L. Plants Used in Guatemala for the Treatment of Gastrointestinal Disorders. 1. Screening of 84 Plants against Enterobacteria. J. Ethnopharmacol. 1990, 30, 55–73. [Google Scholar] [CrossRef] [PubMed]
- Ble-González, E.A.; Gómez-Rivera, A.; Zamilpa, A.; López-Rodríguez, R.; Lobato-García, C.E.; Álvarez-Fitz, P.; Gutierrez-Roman, A.S.; Perez-García, M.D.; Bugarin, A.; González-Cortazar, M. Ellagitannin, Phenols, and Flavonoids as Antibacterials from Acalypha arvensis (Euphorbiaceae). Plants 2022, 11, 300. [Google Scholar] [CrossRef] [PubMed]
- Caceres, A.; Alvarez, A.V.; Ovando, A.E.; Samayoa, B.E. Plants Used in Guatemala for the Treatment of Respiratory Diseases. 1. Screening of 68 Plants against Gram-Positive Bacteria. J. Ethnopharmacol. 1991, 31, 193–208. [Google Scholar] [CrossRef]
- Wang, M.; Lamers, R.-J.A.N.; Korthout, H.A.A.J.; van Nesselrooij, J.H.J.; Witkamp, R.F.; van der Heijden, R.; Voshol, P.J.; Havekes, L.M.; Verpoorte, R.; van der Greef, J. Metabolomics in the Context of Systems Biology: Bridging Traditional Chinese Medicine and Molecular Pharmacology. Phytother. Res. 2005, 19, 173–182. [Google Scholar] [CrossRef]
- Satheeshkumar, N.; Nisha, N.; Sonali, N.; Nirmal, J.; Jain, G.K.; Spandana, V. Analytical Profiling of Bioactive Constituents from Herbal Products, Using Metabolomics—A Review. Nat. Prod. Commun. 2012, 7, 1934578X1200700837. [Google Scholar] [CrossRef]
- Giebelhaus, R.T.; Erland, L.A.E.; Murch, S.J. HormonomicsDB: A Novel Workflow for the Untargeted Analysis of Plant Growth Regulators and Hormones. F1000Research 2022, 11, 1191. [Google Scholar] [CrossRef]
- Erland, L.A.E.; Giebelhaus, R.T.; Victor, J.M.R.; Murch, S.J.; Saxena, P.K. The Morphoregulatory Role of Thidiazuron: Metabolomics-Guided Hypothesis Generation for Mechanisms of Activity. Biomolecules 2020, 10, 1253. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Harwansh, R.K.; Bahadur, S.; Biswas, S.; Kuchibhatla, L.N.; Tetali, S.D.; Raghavendra, A.S. Metabolomics of Medicinal Plants—A Versatile Tool for Standardization of Herbal Products and Quality Evaluation of Ayurvedic Formulations. Curr. Sci. 2016, 111, 1624–1630. [Google Scholar] [CrossRef]
- Cai, Y.; Weng, K.; Guo, Y.; Peng, J.; Zhu, Z.-J. An Integrated Targeted Metabolomic Platform for High-Throughput Metabolite Profiling and Automated Data Processing. Metabolomics 2015, 11, 1575–1586. [Google Scholar] [CrossRef]
- Giebelhaus, R.T.; Tarazona Carrillo, K.; Nam, S.L.; de la Mata, A.P.; Araneda, J.F.; Hui, P.; Ma, J.; Harynuk, J.J. Detection of Common Adulterants in Olive Oils by Bench Top 60 MHz 1H NMR with Partial Least Squares Regression. J. Food Compos. Anal. 2023, 122, 105465. [Google Scholar] [CrossRef]
- Seebaluck, R.; Gurib-Fakim, A.; Mahomoodally, F. Medicinal Plants from the Genus Acalypha (Euphorbiaceae)—A Review of Their Ethnopharmacology and Phytochemistry. J. Ethnopharmacol. 2015, 159, 137–157. [Google Scholar] [CrossRef] [PubMed]
- Bussmann, R.W.; Ashley, G.; Sharon, D.; Chait, G.; Diaz, D.; Pourmand, K.; Jonat, B.; Somogy, S.; Guardado, G.; Aguirre, C.; et al. Proving That Traditional Knowledge Works: The Antibacterial Activity of Northern Peruvian Medicinal Plants. Ethnobot. Res. Appl. 2011, 9, 067–096. [Google Scholar] [CrossRef]
- Barbieri, R.; Coppo, E.; Marchese, A.; Daglia, M.; Sobarzo-Sánchez, E.; Nabavi, S.F.; Nabavi, S.M. Phytochemicals for Human Disease: An Update on Plant-Derived Compounds Antibacterial Activity. Microbiol. Res. 2017, 196, 44–68. [Google Scholar] [CrossRef] [PubMed]
- Cushnie, T.P.T.; Lamb, A.J. Antimicrobial Activity of Flavonoids. Int. J. Antimicrob. Agents 2005, 26, 343–356. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Tyagi, A.; Bhansali, P.; Pareek, S.; Singh, V.; Ilyas, A.; Mishra, R.; Poddar, N.K. Saponins: Extraction, Bio-Medicinal Properties and Way Forward to Anti-Viral Representatives. Food Chem. Toxicol. 2021, 150, 112075. [Google Scholar] [CrossRef]
- Boakye, Y.; Agyare, C.; Hensel, A. Anti-Infective Properties and Time-Kill Kinetics of Phyllanthus Muellerianus and Its Major Constituent, Geraniin. Med. Chem. Curr. Res. 2016, 6, 95–105. [Google Scholar] [CrossRef]
- Delaquis, P.J.; Stanich, K.; Girard, B.; Mazza, G. Antimicrobial Activity of Individual and Mixed Fractions of Dill, Cilantro, Coriander and Eucalyptus Essential Oils. Int. J. Food Microbiol. 2002, 74, 101–109. [Google Scholar] [CrossRef]
- Abdelhafez, O.H.; Othman, E.M.; Fahim, J.R.; Desoukey, S.Y.; Pimentel-Elardo, S.M.; Nodwell, J.R.; Schirmeister, T.; Tawfike, A.; Abdelmohsen, U.R. Metabolomics Analysis and Biological Investigation of Three Malvaceae Plants. Phytochem. Anal. 2020, 31, 204–214. [Google Scholar] [CrossRef]
- Wishart, D.S. Metabolomics: Applications to Food Science and Nutrition Research. Trends Food Sci. Technol. 2008, 19, 482–493. [Google Scholar] [CrossRef]
- Xiao, D.; Ye, R.; Davidson, P.M.; Hayes, D.G.; Golden, D.A.; Zhong, Q. Sucrose Monolaurate Improves the Efficacy of Sodium Hypochlorite against Escherichia Coli O157:H7 on Spinach. Int. J. Food Microbiol. 2011, 145, 64–68. [Google Scholar] [CrossRef]
- Nobmann, P.; Smith, A.; Dunne, J.; Henehan, G.; Bourke, P. The Antimicrobial Efficacy and Structure Activity Relationship of Novel Carbohydrate Fatty Acid Derivatives against Listeria Spp. and Food Spoilage Microorganisms. Int. J. Food Microbiol. 2009, 128, 440–445. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, H.; Hao, T.; Li, S. In Vitro Antibacterial Activities and Mechanism of Sugar Fatty Acid Esters against Five Food-Related Bacteria. Food Chem. 2015, 187, 370–377. [Google Scholar] [CrossRef]
- Watanabe, T.; Katayama, S.; Matsubara, M.; Honda, Y.; Kuwahara, M. Antibacterial Carbohydrate Monoesters Suppressing Cell Growth of Streptococcus Mutans in the Presence of Sucrose. Curr. Microbiol. 2000, 41, 210–213. [Google Scholar] [CrossRef] [PubMed]
- Burt, S. Essential Oils: Their Antibacterial Properties and Potential Applications in Foods—A Review. Int. J. Food Microbiol. 2004, 94, 223–253. [Google Scholar] [CrossRef] [PubMed]
- Tariq, S.; Wani, S.; Rasool, W.; Shafi, K.; Bhat, M.A.; Prabhakar, A.; Shalla, A.H.; Rather, M.A. A Comprehensive Review of the Antibacterial, Antifungal and Antiviral Potential of Essential Oils and Their Chemical Constituents against Drug-Resistant Microbial Pathogens. Microb. Pathog. 2019, 134, 103580. [Google Scholar] [CrossRef]
- Farha, A.K.; Yang, Q.-Q.; Kim, G.; Li, H.-B.; Zhu, F.; Liu, H.-Y.; Gan, R.-Y.; Corke, H. Tannins as an Alternative to Antibiotics. Food Biosci. 2020, 38, 100751. [Google Scholar] [CrossRef]
- Chung, K.-T.; Stevens, S.E., Jr.; Lin, W.-F.; Wei, C.I. Growth Inhibition of Selected Food-Borne Bacteria by Tannic Acid, Propyl Gallate and Related Compounds. Lett. Appl. Microbiol. 1993, 17, 29–32. [Google Scholar] [CrossRef]
- Lo, M.-M.; Benfodda, Z.; Dunyach-Rémy, C.; Bénimélis, D.; Roulard, R.; Fontaine, J.-X.; Mathiron, D.; Quéro, A.; Molinié, R.; Meffre, P. Isolation and Identification of Flavones Responsible for the Antibacterial Activities of Tillandsia Bergeri Extracts. ACS Omega 2022, 7, 35851–35862. [Google Scholar] [CrossRef]
- Farouil, L.; Dias, R.P.; Popotte-Julisson, G.; Bibian, G.; Adou, A.I.; de la Mata, A.P.; Sylvestre, M.; Harynuk, J.J.; Cebrián-Torrejón, G. The Metabolomic Profile of the Essential Oil from Zanthoxylum Caribaeum (Syn. Chiloperone) Growing in Guadeloupe FWI Using GC × GC-TOFMS. Metabolites 2022, 12, 1293. [Google Scholar] [CrossRef] [PubMed]
- Nam, S.L.; de la Mata, A.P.; Harynuk, J.J. Automated Screening and Filtering Scripts for GC×GC-TOFMS Metabolomics Data. Separations 2021, 8, 84. [Google Scholar] [CrossRef]

| Gram-Positive Bacteria | Gram-Negative Bacteria | |||||
|---|---|---|---|---|---|---|
| Staphylococcus aureus ATCC 25923 | Enterococcus faecalis ATCC 29212 | Staphylococcus aureus 024 | Staphylococcus aureus 003 | Pseudomona aeruginosa ATCC 27853 | Escherichia coli ATCC 25922 | |
| F6 | 5.5 mg/mL | |||||
| Diameter (mm) | 13.67 ± 1.15 | 12.5 ± 0.71 | 13 ± 0 | 14 ± 0 | 7 ± 0 | 13 ± 0 |
| Oxa5 | 5 µg | |||||
| Diameter (mm) | 46 ± 1 | N/D | 46 ± 1 | 36 ± 1 | N/D | N/D |
| Gram-Positive Bacteria | Gram-Negative Bacteria | |||||||
|---|---|---|---|---|---|---|---|---|
| Fractions | S. aureus ATCC 29213 | Clinical SARM BA 22038 | E. faecalis ATCC 51299 | K. pneumonia ATCC 700603 | Clinical resistant K. pneumonia BA 34029 | P. aeruginosa ATCC 27853 | Clinical BMR BA 35014 | E. coli ATCC 25922 |
| 2 mg/mL | ||||||||
| F6 | V | V | X | X | X | X | X | X |
| Fraction | 5 mg/mL | |||||||
| F3 | V | V | X | X | X | X | X | X |
| F4 | V | V | X | X | X | X | X | X |
| F6 | V | V | X | X | X | X | X | X |
| Fraction | 10 mg/mL | |||||||
| F3 | V | V | X | X | X | V | X | X |
| F4 | V | V | X | X | X | V | X | X |
| F6 | V | V | V | X | X | V | X | X |
| Fraction | 20 mg/mL | |||||||
| F3 | V | V | X | X | X | V | X | V |
| F4 | V | V | X | X | X | V | X | V |
| F5 | V | V | X | X | X | V | X | X |
| F6 | V | V | V | X | X | V | X | V |
| Flavonoids (Neu’s Reagent) | Alkaloids (Dragendorff Reagent) | Sugars (Sulfuric Thymol Reagent) | Sterols, Steroids, and Triterpenes (Liebermann and Burchard Reagent) | Tannins | Coumarins | Terpenoids | Saponins | |
|---|---|---|---|---|---|---|---|---|
| F1 | − | − | − | + | − | − | + | − |
| F2 | − | − | − | + | − | − | − | − |
| F3 | + | + | + | + | + | − | − | − |
| F4 | + | + | + | + | + | − | − | − |
| F5 | + | − | + | − | + | − | − | + |
| F6 | + | + | + | + | + | − | + | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thesnor, V.; Molinié, R.; Giebelhaus, R.T.; de la Mata Espinosa, A.P.; Harynuk, J.J.; Bénimélis, D.; Vanhoye, B.; Dunyach-Rémy, C.; Sylvestre, M.; Cheremond, Y.; et al. Antibacterial Activity and Untargeted Metabolomics Profiling of Acalypha arvensis Poepp. Molecules 2023, 28, 7882. https://doi.org/10.3390/molecules28237882
Thesnor V, Molinié R, Giebelhaus RT, de la Mata Espinosa AP, Harynuk JJ, Bénimélis D, Vanhoye B, Dunyach-Rémy C, Sylvestre M, Cheremond Y, et al. Antibacterial Activity and Untargeted Metabolomics Profiling of Acalypha arvensis Poepp. Molecules. 2023; 28(23):7882. https://doi.org/10.3390/molecules28237882
Chicago/Turabian StyleThesnor, Valendy, Roland Molinié, Ryland T. Giebelhaus, A. Paulina de la Mata Espinosa, James J. Harynuk, David Bénimélis, Bérénice Vanhoye, Catherine Dunyach-Rémy, Muriel Sylvestre, Yvens Cheremond, and et al. 2023. "Antibacterial Activity and Untargeted Metabolomics Profiling of Acalypha arvensis Poepp" Molecules 28, no. 23: 7882. https://doi.org/10.3390/molecules28237882