Nondestructive Metabolomic Fingerprinting: FTIR, NIR and Raman Spectroscopy in Food Screening
Abstract
:1. Introduction
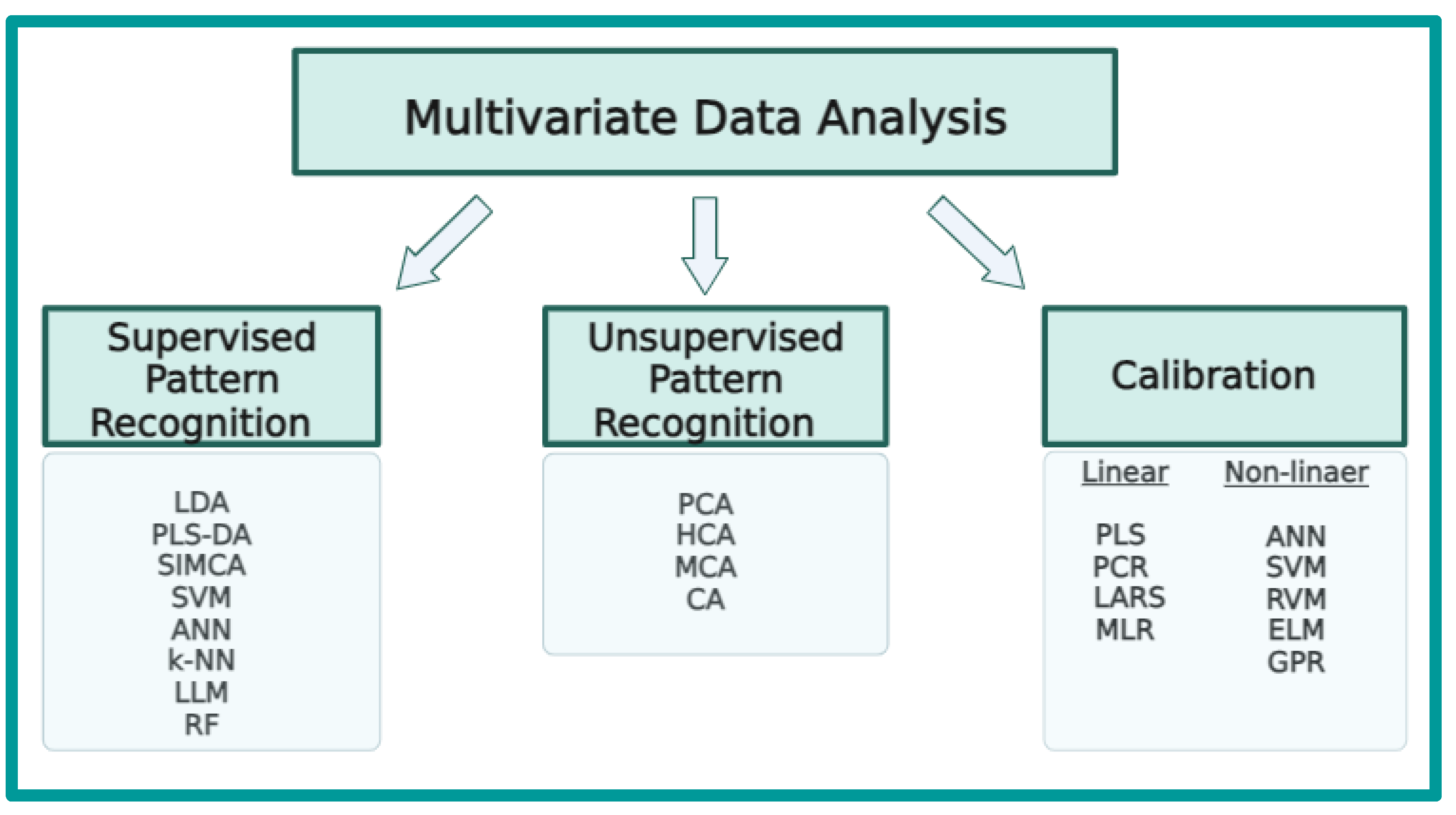
2. Multivariate Data Analysis—Chemometrics
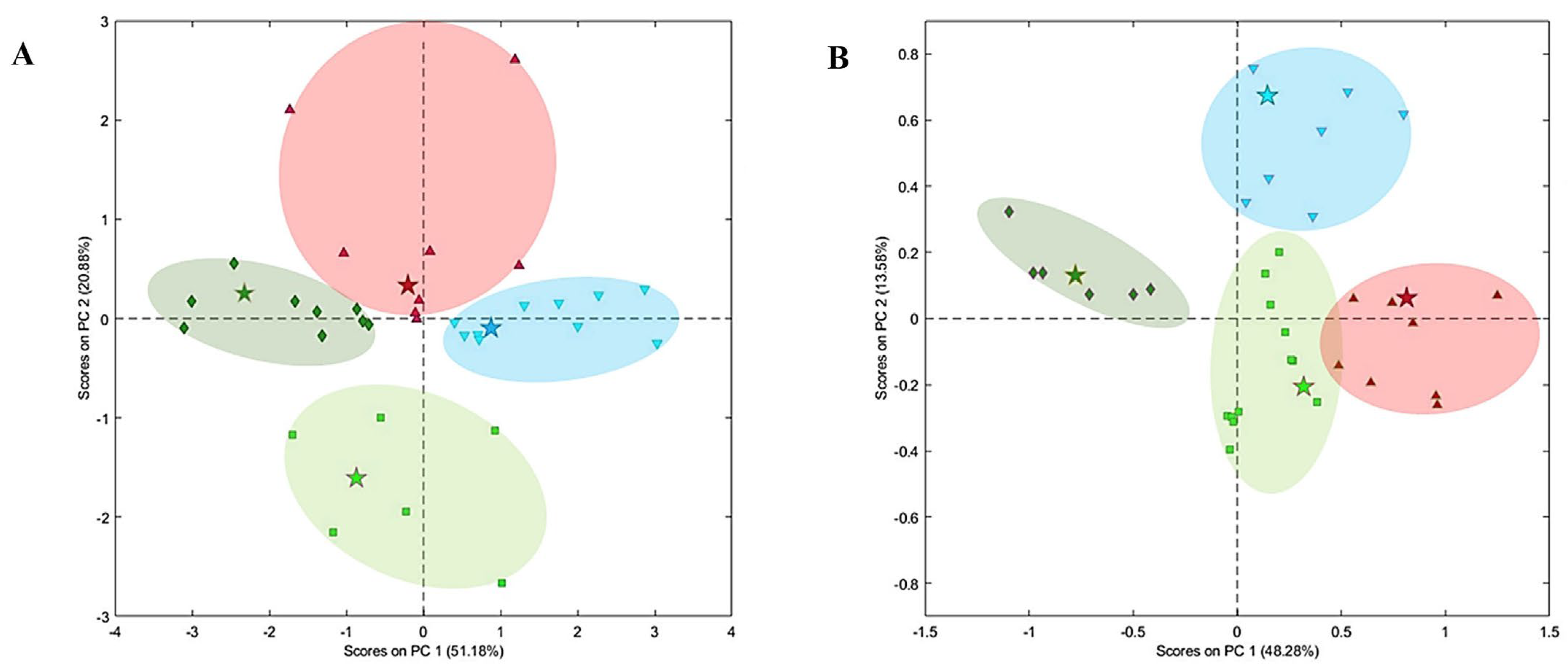
2.1. Unsupervised Pattern Recognition
2.2. Supervised Pattern Recognition
2.3. Multivariate Calibration
2.4. Advantages of Pattern Recognition Techniques in Food Analysis
3. IR Spectroscopy Applications
3.1. MIR (Mid-Infrared Spectroscopy) Applications
3.2. Raman and FT-Raman Applications
3.3. NIR Applications
NIR (Near-Infrared Spectroscopy)
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fraga-Corral, M.; Carpena, M.; Garcia-Oliveira, P.; Pereira, A.G.; Prieto, M.A.; Simal-Gandara, J. Analytical Metabolomics and Applications in Health, Environmental and Food Science. Crit. Rev. Anal. Chem. 2022, 52, 712–734. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Wang, P.; Han, Y.; Wang, X. Modern analytical techniques in metabolomics analysis. Analyst 2012, 137, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Bailey, N.J.C.; Johnson, H.E. Measuring the metabolome: Current analytical technologies. Analyst 2005, 130, 606–625. [Google Scholar] [CrossRef] [PubMed]
- Boccard, J.; Rudaz, S. Harnessing the complexity of metabolomic data with chemometrics. J. Chemom. 2014, 28, 1–9. [Google Scholar] [CrossRef]
- Granato, D.; Putnik, P.; Kovačević, D.B.; Santos, J.S.; Calado, V.; Rocha, R.S.; Da Cruz, A.G.; Jarvis, B.; Rodionova, O.Y.; Pomerantsev, A. Trends in Chemometrics: Food Authentication, Microbiology, and Effects of Processing. Compr. Rev. Food Sci. Food Saf. 2018, 17, 663–677. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Sharma, V. Chemometrics in forensic science. TrAC—Trends Anal. Chem. 2018, 105, 191–201. [Google Scholar] [CrossRef]
- Khakimov, B.; Gürdeniz, G.; Engelse, S.B. Trends in the application of chemometrics to foodomics studies. Acta Aliment. 2015, 44, 4–31. [Google Scholar] [CrossRef]
- Rodriguez-Saona, L.E.; Pujolras, M.P.; Giusti, M.M. Targeted and Non-Targeted Analysis. In Analytical Separation Science; Wiley-VCH Verlag GmbH & Co. KGaA: Hoboken, NJ, USA, 2015; pp. 1401–1436. [Google Scholar]
- Mafata, M.; Brand, J.; Medvedovici, A.; Buica, A. Chemometric and sensometric techniques in enological data analysis. Crit. Rev. Food Sci. Nutr. 2022, 1–15. [Google Scholar] [CrossRef]
- Mariey, L.; Signolle, J.P.; Amiel, C.; Travert, J. Discrimination, classification, identification of microorganisms using FTIR spectroscopy and chemometrics. Vib. Spectrosc. 2001, 26, 151–159. [Google Scholar] [CrossRef]
- González-Domínguez, R.; Sayago, A.; Fernández-Recamales, Á. An Overview on the Application of Chemometrics Tools in Food Authenticity and Traceability. Foods 2022, 11, 3940. [Google Scholar] [CrossRef]
- Wang, H.P.; Chen, P.; Dai, J.W.; Liu, D.; Li, J.Y.; Xu, Y.P.; Chu, X.L. Recent advances of chemometric calibration methods in modern spectroscopy: Algorithms, strategy, and related issues. TrAC—Trends Anal. Chem. 2022, 153, 116648. [Google Scholar] [CrossRef]
- Brereton, R.G. Chemometrics: Data Analysis for the Laboratory and Chemical Plant; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2003; Volume 8, ISBN 0471489778. [Google Scholar]
- Zhou, L.; Zhang, C.; Liu, F.; Qiu, Z.; He, Y. Application of Deep Learning in Food: A Review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1793–1811. [Google Scholar] [CrossRef] [PubMed]
- Berrueta, L.A.; Alonso-Salces, R.M.; Héberger, K. Supervised pattern recognition in food analysis. J. Chromatogr. A 2007, 1158, 196–214. [Google Scholar] [CrossRef]
- Cebi, N.; Bekiroglu, H.; Erarslan, A.; Rodriguez-saona, L. Rapid Sensing: Hand-Held and Portable FTIR Applications for On-Site Food Quality Control from Farm to Fork. Molecules 2023, 28, 3727. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro Da Cunha, B.; Fonseca, L.P.; Calado, C.R.C. Metabolic fingerprinting with fourier-transform infrared (Ftir) spectroscopy: Towards a high-throughput screening assay for antibiotic discovery and mechanism-of-action elucidation. Metabolites 2020, 10, 145. [Google Scholar] [CrossRef] [PubMed]
- Umar, A.H.; Syahruni, R.; Ranteta’dung, I.; Rafi, M. FTIR-based fingerprinting combined with chemometrics method for rapid discrimination of Jatropha spp. (Euphorbiaceae) from different regions in South Sulawesi. J. Appl. Pharm. Sci. 2023, 13, 139–149. [Google Scholar] [CrossRef]
- Borges, C.V.; Amorim, V.B.D.O.; Ramlov, F.; Ledo, C.A.D.S.; Donato, M.; Maraschin, M.; Amorim, E.P. Characterisation of metabolic profile of banana genotypes, aiming at biofortified Musa spp. cultivars. Food Chem. 2014, 145, 496–504. [Google Scholar] [CrossRef]
- Easmin, S.; Zaidul, I.S.M.; Ghafoor, K.; Ferdosh, S.; Jaffri, J.; Ali, M.E.; Mirhosseini, H.; Al-Juhaimi, F.Y.; Perumal, V.; Khatib, A. Rapid investigation of α-glucosidase inhibitory activity of Phaleria macrocarpa extracts using FTIR-ATR based fingerprinting. J. Food Drug Anal. 2017, 25, 306–315. [Google Scholar] [CrossRef]
- Sahoo, M.R.; Umashankara, M.S. FTIR Based Metabolomics Profiling and Fingerprinting of Some Medicinal Plants: An Attempt to Develop an Approach for Quality Control and Standardization of Herbal Materials. Pharmacogn. Res. 2022, 15, 163–167. [Google Scholar] [CrossRef]
- Kwon, Y.K.; Ahn, M.S.; Park, J.S.; Liu, J.R.; In, D.S.; Min, B.W.; Kim, S.W. Discrimination of cultivation ages and cultivars of ginseng leaves using Fourier transform infrared spectroscopy combined with multivariate analysis. J. Ginseng Res. 2014, 38, 52–58. [Google Scholar] [CrossRef]
- Osman, S.O.M.; Saad, A.S.I.; Tadano, S.; Takeda, Y.; Yamasaki, Y.; Tahir, I.S.A.; Tsujimoto, H.; Akashi, K. Probing Differential Metabolome Responses among Wheat Genotypes to Heat Stress Using Fourier Transform Infrared-Based Chemical Fingerprinting. Agriculture 2022, 12, 753. [Google Scholar] [CrossRef]
- Abramovic, B.; Jajic, I.; Abramovic, B.; Cosic, J.; Juric, V. Detection of deoxynivalenol in wheat by Fourier transform infrared spectroscopy. Acta Chim. Slov. 2007, 54, 859. [Google Scholar]
- Kurniawan, M.F.; Andarwulan, N.; Wulandari, N.; Rafi, M. Metabolomic approach for understanding phenolic compounds and melanoidin roles on antioxidant activity of Indonesia robusta and arabica coffee extracts. Food Sci. Biotechnol. 2017, 26, 1475–1480. [Google Scholar] [CrossRef] [PubMed]
- Nurrulhidayah, A.F.; Che Man, Y.B.; Amin, I.; Arieff Salleh, R.; Farawahidah, M.Y.; Shuhaimi, M.; Khatib, A. FTIR-ATR spectroscopy based metabolite fingerprinting as a direct determination of butter adulterated with lard. Int. J. Food Prop. 2015, 18, 372–379. [Google Scholar] [CrossRef]
- Fu, Y.; Toyoda, K.; Ihara, I. Application of ATR-FTIR spectroscopy and principal component analysis in characterization of 15-acetyldeoxynivalenol in corn oil. Eng. Agric. Environ. Food 2014, 7, 163–168. [Google Scholar] [CrossRef]
- Del Bove, M.; Lattanzi, M.; Rellini, P.; Pelliccia, C.; Fatichenti, F.; Cardinali, G. Comparison of molecular and metabolomic methods as characterization tools of Debaryomyces hansenii cheese isolates. Food Microbiol. 2009, 26, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Skotti, E.; Kountouri, S.; Bouchagier, P.; Tsitsigiannis, D.I.; Polissiou, M.; Tarantilis, P.A. FTIR spectroscopic evaluation of changes in the cellular biochemical composition of the phytopathogenic fungus Alternaria alternata induced by extracts of some Greek medicinal and aromatic plants. Spectrochim. Acta—Part A Mol. Biomol. Spectrosc. 2014, 127, 463–472. [Google Scholar] [CrossRef]
- Estelles-Lopez, L.; Ropodi, A.; Pavlidis, D.; Fotopoulou, J.; Gkousari, C.; Peyrodie, A.; Panagou, E.; Nychas, G.J.; Mohareb, F. An automated ranking platform for machine learning regression models for meat spoilage prediction using multi-spectral imaging and metabolic profiling. Food Res. Int. 2017, 99, 206–215. [Google Scholar] [CrossRef]
- Okere, E.E.; Arendse, E.; Nieuwoudt, H.; Perold, W.J.; Opara, U.L. Non-destructive evaluation of the quality characteristics of pomegranate kernel oil by fourier transform near-infrared and mid-infrared spectroscopy. Front. Plant Sci. 2022, 13, 867555. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, Y.; He, C.; Xie, M.; Luo, H.; Wang, Y.; Zhang, J. Rapid screen of aflatoxin-contaminated peanut oil using Fourier transform infrared spectroscopy combined with multivariate decision tree. Int. J. Food Sci. Technol. 2018, 53, 2386–2393. [Google Scholar] [CrossRef]
- Kos, G.; Sieger, M.; McMullin, D.; Zahradnik, C.; Sulyok, M.; Öner, T.; Mizaikoff, B.; Krska, R. A novel chemometric classification for FTIR spectra of mycotoxin-contaminated maize and peanuts at regulatory limits. Food Addit. Contam. Part A 2016, 33, 1596–1607. [Google Scholar] [CrossRef] [PubMed]
- Debebe, A.; Redi-Abshiro, M.; Chandravanshi, B.S. Non-destructive determination of ethanol levels in fermented alcoholic beverages using Fourier transform mid-infrared spectroscopy. Chem. Cent. J. 2017, 11, 27. [Google Scholar] [CrossRef] [PubMed]
- Krähmer, A.; Böttcher, C.; Rode, A.; Nothnagel, T.; Schulz, H. Quantifying biochemical quality parameters in carrots (Daucus carota L.)—FT-Raman spectroscopy as efficient tool for rapid metabolite profiling. Food Chem. 2016, 212, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Nache, M.; Hinrichs, J.; Scheier, R.; Schmidt, H.; Hitzmann, B. Prediction of the pH as indicator of porcine meat quality using Raman spectroscopy and metaheuristics. Chemom. Intell. Lab. Syst. 2016, 154, 45–51. [Google Scholar] [CrossRef]
- Jayan, H.; Sun, D.W.; Pu, H.; Wei, Q. Surface-enhanced Raman spectroscopy combined with stable isotope probing to assess the metabolic activity of Escherichia coli cells in chicken carcass wash water. Spectrochim. Acta—Part A Mol. Biomol. Spectrosc. 2022, 280, 121549. [Google Scholar] [CrossRef] [PubMed]
- Magdas, D.A.; Cozar, B.I.; Feher, I.; Guyon, F.; Dehelean, A.; Cinta Pinzaru, S. Testing the limits of FT-Raman spectroscopy for wine authentication: Cultivar, geographical origin, vintage and terroir effect influence. Sci. Rep. 2019, 9, 19954. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Chen, L.; Chou, K.C.; Lu, X. Campylobacter jejuni Antimicrobial Resistance Profiles and Mechanisms Determined Using a Raman Spectroscopy-Based Metabolomic Approach. Appl. Environ. Microbiol. 2021, 87, e00388-21. [Google Scholar] [CrossRef]
- Huayhongthong, S.; Khuntayaporn, P.; Thirapanmethee, K.; Wanapaisan, P.; Chomnawang, M.T. Raman spectroscopic analysis of food-borne microorganisms. LWT 2019, 114, 108419. [Google Scholar] [CrossRef]
- Lafhal, S.; Vanloot, P.; Bombarda, I.; Valls, R.; Kister, J.; Dupuy, N. Raman spectroscopy for identification and quantification analysis of essential oil varieties: A multivariate approach applied to lavender and lavandin essential oils. J. Raman Spectrosc. 2015, 46, 577–585. [Google Scholar] [CrossRef]
- McLeod, G.; Clelland, K.; Tapp, H.; Kemsley, E.K.; Wilson, R.H.; Poulter, G.; Coombs, D.; Hewitt, C.J. A comparison of variate pre-selection methods for use in partial least squares regression: A case study on NIR spectroscopy applied to monitoring beer fermentation. J. Food Eng. 2009, 90, 300–307. [Google Scholar] [CrossRef]
- Debebe, A.; Anberbir, A.; Redi-Abshiro, M.; Chandravanshi, B.S.; Asfaw, A.; Asfaw, N.; Retta, N. Alcohol determination in distilled alcoholic beverages by liquid phase fourier transform mid-infrared and near-infrared spectrophotometries. Food Anal. Methods 2017, 10, 172–179. [Google Scholar] [CrossRef]
- Cozzolino, D.; Flood, L.; Bellon, J.; Gishen, M.; De Barros Lopes, M. Combining near infrared spectroscopy and multivariate analysis as a tool to differentiate different strains of Saccharomyces cerevisiae: A metabolomic study. Yeast 2006, 23, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Lafhal, S.; Vanloot, P.; Bombarda, I.; Kister, J.; Dupuy, N. Chemometric analysis of French lavender and lavandin essential oils by near infrared spectroscopy. Ind. Crop. Prod. 2016, 80, 156–164. [Google Scholar] [CrossRef]
- Falade, T.D.; Sultanbawa, Y.; Fletcher, M.T.; Fox, G. Near infrared spectrometry for rapid non-invasive modelling of Aspergillus-contaminated maturing kernels of maize (Zea mays L.). Agriculture 2017, 7, 77. [Google Scholar] [CrossRef]
- Levasseur-Garcia, C.; Bailly, S.; Kleiber, D.; Bailly, J.D. Assessing risk of fumonisin contamination in maize using near-infrared spectroscopy. J. Chem. 2015, 2015, 485864. [Google Scholar] [CrossRef]
- Tao, F.; Yao, H.; Hruska, Z.; Kincaid, R.; Rajasekaran, K.; Bhatnagar, D. A novel hyperspectral-based approach for identification of maize kernels infected with diverse Aspergillus flavus fungi. Biosyst. Eng. 2020, 200, 415–430. [Google Scholar] [CrossRef]
- Carames, E.T.D.S.; Piacentini, K.C.; Alves, L.T.; Pallone, J.A.L.; Rocha, L.D.O. NIR spectroscopy and chemometric tools to identify high content of deoxynivalenol in barley. Food Addit. Contam. Part A 2020, 37, 1542–1552. [Google Scholar] [CrossRef]
- Lim, J.; Kim, G.; Mo, C.; Oh, K.; Yoo, H.; Ham, H.; Kim, M.S. Classification of Fusarium-infected Korean hulled barley using near-infrared reflectance spectroscopy and partial least squares discriminant analysis. Sensors 2017, 17, 2258. [Google Scholar] [CrossRef]
- Lim, J.; Kim, G.; Mo, C.; Oh, K.; Kim, G.; Ham, H.; Kim, S.; Kim, M.S. Application of near infrared reflectance spectroscopy for rapid and non-destructive discrimination of hulled barley, naked barley, and wheat contaminated with Fusarium. Sensors 2018, 18, 113. [Google Scholar] [CrossRef]
- Pettersson, H.; Åberg, L. Near infrared spectroscopy for determination of mycotoxins in cereals. Food Control 2003, 14, 229–232. [Google Scholar] [CrossRef]
- Sirisomboon, C.D.; Putthang, R.; Sirisomboon, P. Application of near infrared spectroscopy to detect aflatoxigenic fungal contamination in rice. Food Control 2013, 33, 207–214. [Google Scholar] [CrossRef]
- Shawky, E.; Selim, D.A. NIR spectroscopy-multivariate analysis for discrimination and bioactive compounds prediction of different Citrus species peels. Spectrochim. Acta—Part A Mol. Biomol. Spectrosc. 2019, 219, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Taradolsirithitikul, P.; Sirisomboon, P.; Dachoupakan Sirisomboon, C. Qualitative and quantitative analysis of ochratoxin A contamination in green coffee beans using Fourier transform near infrared spectroscopy. J. Sci. Food Agric. 2017, 97, 1260–1266. [Google Scholar] [CrossRef] [PubMed]
- Krause, D.; Holtz, C.; Gastl, M.; Hussein, M.A.; Becker, T. NIR and PLS discriminant analysis for predicting the processability of malt during lautering. Eur. Food Res. Technol. 2015, 240, 831–846. [Google Scholar] [CrossRef]
- Dvořáček, V.; Prohasková, A.; Chrpová, J.; Štočková, L. Near infrared spectroscopy for deoxynivalenol content estimation in intact wheat grain. Plant Soil Environ. 2012, 58, 196–203. [Google Scholar] [CrossRef]
- De Girolamo, A.; Cervellieri, S.; Visconti, A.; Pascale, M. Rapid analysis of deoxynivalenol in durum wheat by FT-NIR spectroscopy. Toxins 2014, 6, 3129–3143. [Google Scholar] [CrossRef] [PubMed]
- Peiris, K.H.; Dong, Y.; Davis, M.A.; Bockus, W.W.; Dowell, F.E. Estimation of the deoxynivalenol and moisture contents of bulk wheat grain samples by FT-NIR spectroscopy. Cereal Chem. 2017, 94, 677–682. [Google Scholar] [CrossRef]
- Li, Z.; Wang, P.P.; Huang, C.C.; Shang, H.; Pan, S.Y.; Li, X.J. Application of Vis/NIR Spectroscopy for Chinese Liquor Discrimination. Food Anal. Methods 2014, 7, 1337–1344. [Google Scholar] [CrossRef]
- Li, X.; He, Y.; Wu, C.; Sun, D.W. Nondestructive measurement and fingerprint analysis of soluble solid content of tea soft drink based on Vis/NIR spectroscopy. J. Food Eng. 2007, 82, 316–323. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, X.; Xiaoling, D.; Ouyang, A.; Gao, R.; Zhang, H.; Pan, Y. Non-Destructive Measurement of Soluble Solids Content and Vitamin C in Gannan Navel Oranges by Vis-NIR Spectroscopy. Sens. Lett. 2011, 9, 1133–1139. [Google Scholar] [CrossRef]
- Shen, F.; Zhao, T.; Jiang, X.; Liu, X.; Fang, Y.; Liu, Q.; Hu, Q.; Liu, X. On-line detection of toxigenic fungal infection in wheat by visible/near infrared spectroscopy. LWT 2019, 109, 216–224. [Google Scholar] [CrossRef]
- Tao, F.; Yao, H.; Zhu, F.; Hruska, Z.; Liu, Y.; Rajasekaran, K.; Bhatnagar, D. A rapid and nondestructive method for simultaneous determination of aflatoxigenic fungus and aflatoxin contamination on corn kernels. J. Agric. Food Chem. 2019, 67, 5230–5239. [Google Scholar] [CrossRef] [PubMed]
- Tao, F.; Yao, H.; Hruska, Z.; Liu, Y.; Rajasekaran, K.; Bhatnagar, D. Use of visible–near-infrared (Vis-NIR) spectroscopy to detect aflatoxin B1 on peanut kernels. Appl. Spectrosc. 2019, 73, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Filho, E.A.; Silva, L.M.; Lima, Y.; Ribeiro, P.; Silva, E.; Zocolo, G.; Canuto, K.; Morais, S.; Castro, A.C.; de Brito, E. Metabolomic variability of different genotypes of cashew by lc-ms and correlation with near-infrared spectroscopy as a tool for fast phenotyping. Metabolites 2019, 9, 121. [Google Scholar]
- Das, R.S.; Agrawal, Y.K. Raman spectroscopy: Recent advancements, techniques and applications. Vib. Spectrosc. 2011, 57, 163–176. [Google Scholar] [CrossRef]
- Boyacı, İ.H.; Temiz, H.T.; Geniş, H.E.; Soykut, E.A.; Yazgan, N.N.; Güven, B.; Uysal, R.S.; Bozkurt, A.G.; İlaslan, K.; Torun, Ö.; et al. Dispersive and FT-Raman spectroscopic methods in food analysis. R. Soc. Chem. 2015, 5, 56606–56624. [Google Scholar]
- Ellis, D.I.; Brewster, V.L.; Dunn, W.B.; Allwood, J.W.; Golovanov, A.P.; Goodacre, R. Fingerprinting food: Current technologies for the detection of food adulteration and contamination. Chem. Soc. Rev. 2012, 41, 5706–5727. [Google Scholar] [CrossRef]
- Chen, A.J.; Li, J.; Jannasch, A.; Mutlu, A.S.; Wang, M.C.; Cheng, J.X. Fingerprint Stimulated Raman Scattering Imaging Reveals Retinoid Coupling Lipid Metabolism and Survival. ChemPhysChem 2018, 19, 2500–2506. [Google Scholar] [CrossRef]
- Cozzolino, D. Near infrared spectroscopy in natural products analysis. Planta Med. 2009, 75, 746–756. [Google Scholar] [CrossRef]
- Teixeira Dos Santos, C.A.; Lopo, M.; Páscoa, R.N.M.J.; Lopes, J.A. A review on the applications of portable near-infrared spectrometers in the agro-food industry. Appl. Spectrosc. 2013, 67, 1215–1233. [Google Scholar] [CrossRef]
- Osborne, B.G. Near-Infrared Spectroscopy in Food Analysis. In Encyclopedia of Analytical Chemistry; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2000; pp. 1–14. [Google Scholar]

| Detection Technology | Commodity | Type of Food Product | Parameters Measured | Data Acquisition | Data Treatment | References |
|---|---|---|---|---|---|---|
| FTIR | Fruit | Banana | Carbohydrates, proteins, and lipids | 4000–500 cm−1 | PCA | [19] |
| Herbal | Phaleria macrocarpa (Mahkota Dewa) | Functional groups (CHO, -COOH, -NO2, -NH, and -OH) | 4000–400 cm−1 | SIMCA | [20] | |
| Herbal | Some Medicinal Plants (Glycyrrhiza glabra root aqueous, Terminalia chebula aqueous, Zingiber officinale, Ocimum sanctum leaf aqueous, Piper longum, Curcuma longa | Functional groups (such as phenolics (-OH), carbonyl (C=O), aldehyde (CH=O), ether (C-O-C), aromatic (C=C), and alkyl groups –CH) | 4000 to 500 cm−1 | - | [21] | |
| Herbal | Ginseng leaves | Active ingredients (ginsenosides, polysaccharides, triterpenoids, flavonoids, volatile oils, polyacetylenic alcohols, peptides, amino acids, and fatty acids) | 4000 to 400 cm−1 | PCA, HCA, PLS-DA | [22] | |
| Cereal | Wheat Genotypes | FTIR-based biomarkers (Fm482, Fm576, Fm1251, Fm1465, Fm1502, and Fm1729) | 4000 to 400 cm−1 | PCA, LDA | [23] | |
| Wheat | Mycotoxin deoxynivalenol (DON) | 650 and 4000 cm | PLS1, MLR | [24] | ||
| Coffee | Indonesia robusta and arabica coffee | Functional groups (ester/lactone, aldehyde, ketone, aromatic acids, and aliphatic) | 4000–400 cm−1 | PCA, PLS | [25] | |
| Dairy | Butter | Functional groups (fatty acids, methylene groups, aliphatic groups, CH3 groups) | 4000–650 cm−1 | PLS | [26] | |
| Oil | Corn oil | Aflatoxins 15-acetyldeoxynivalenol | 4000–600 cm−1 | PCA, MSC | [27] | |
| Bacteria | Debaryomyces hanseni (cheese) | Origin of isolation | 4000 and 400 cm−1 | RAPD | [28] | |
| Fungus | Phytopathogenic fungus A. alternata (Greek medicinal and aromatic plants) | Biochemical composition (lipids, proteins and a ratios of lipids/amide II, amide I/total amides and amide II/total amides) | 4000–400 cm−1 | PCA | [29] | |
| Meat | Minced meat | Temperature of storage, the initial contamination, pH | 4000 to 400 cm−1 | PCA | [30] | |
| Oil | Pomegranate kernel oil | Quality parameters according to their respective cultivars (refractive index, peroxide value, total phenolic content, refractive index, total carotenoid content) | 4000–400 cm−1 | PCA and OPLS-DA | [31] | |
| Peanut oil | Aflatoxin B1 (AFB1) and aflatoxin (AFT) | 4000 to 650 cm−1 | MDT | [32] | ||
| Peanut | Aflatoxin B1 | 4000 to 575 cm−1 | PCA | [33] | ||
| Beverage | Fermented alcoholic beverages | Ethanol | 1200–850 cm−1 | PLS | [34] | |
| FT-RAMAN | Vegetables | Carrot | Carbohydrates, carotenoids, and polyacetylenes | 4000 to 100 cm−1 | PCA, PLS | [35] |
| RAMAN | Meat | Porcine meat | pH | 2105 to 323 cm−1 | MSC | [36] |
| Meat | Chicken carcass | Escherichia coli cells | 500–3500 cm−1 | PCA, LDA | [37] | |
| Beverage | Wine | Ethanol | 1000–0 cm−1, 3600–0 cm−1 | SLDA | [38] | |
| Drug | Antibiotics (Campylobacter jejuni) | AMR (antimicrobial resistance) profile | 400 to 1800 cm−1 | HCA, PCA | [39] | |
| Bacteria strains | foodborne microorganisms (E. coli ATCC 25922, B. cereus ATCC 11778, S. aureus ATCC 13565 and Salmonella typhimurium ATCC 13311) | Acids and proteins | 500–1600 cm−1 | PCA | [40] | |
| Essential oils | Lavender (Lavandula angustifolia) and lavandin essential oils | Major terpenoid composition, eucalyptol, camphor, β-Caryophyllene, eucalyptol, linalyl acetate, inalool, β-caryophyllene | 90–4000 cm−1 | PCA, PLS-DA | [41] | |
| NIR | Beverage | Beer | Fermentation parameters (ethanol concentration, specific gravity (SG), optical density, and dry cell weight | 10,000 to 4000 cm−1 | PLS-R | [42] |
| Distilled Alcoholic Beverages | Methanol and ethanol | 1720–1660 nm | [43] | |||
| Yeast | Saccharomyces cerevisiae | O–H second overtone of water and ethanol; C–H3 stretch first overtone or with compounds containing C–H aromatic groups | 400–2500 nm | LDA, PCA | [44] | |
| Essential oils | Lavender (Lavandula angustifolia) and lavandin essential oils | Linalool and eucalyptol content | 4500–9000 cm−1 | PCA, PLS- DA | [45] | |
| Cereal | Maize | Aflatoxigenic Aspergillus spp. contamination | 800–2600 nm | PCA | [46] | |
| Maize | Ergosterol and fumonisins content | 400–2498 nm | PCA | [47] | ||
| Maize | Aspergillus flavus fungi | 900–2500 nm | PLS-DA | [48] | ||
| Barley | Deoxynivalenol (DON) | 10,000 to 4000 cm−1 | PLS-DA, PLS-R | [49] | ||
| Hulled barley | Fusarium | 1175 to 2170 nm | PLS-DA | [50] | ||
| Hulled Barley, Naked Barley, and Wheat | Fusarium | 360–2500 nm | PLS-DA | [51] | ||
| Wheat | Deoxynivalenol (DON) | 570–1100 nm | PLS | [52] | ||
| Rice | Aflatoxigenicfungal contamination | 950–1650 nm | PLSR | [53] | ||
| FT-NIR | Fruit | Citrus species peels | Bioflavonoids (diosmin and hesperidin) | 12,000–4000 cm−1 | HCA, PCA, PLSR | [54] |
| Coffee | Green coffee beans | Ochratoxin A (OTA) | 800–2500 nm | PLS-DA | [55] | |
| Cereal | Malt | Lautering | 800–2500 nm | PLS-DA, PCA | [56] | |
| Wheat | Deoxynivalenol (DON) | 10,000–4000 cm−1 | PLS, DA | [57] | ||
| Durum wheat | Deoxynivalenol (DON) | 10,000 to 4000 cm−1 | LDA, PLS | [58] | ||
| Bulk wheat | Deoxynivalenol (DON), moisture content (MC) | 10,000 to 4000 cm−1 | PLS | [59] | ||
| Vis/NIR | Beverage | Chinese liquor | Alcohol degree, age, flavor | 570–1848 nm | PCA, LDA, SIMCA | [60] |
| Beverage | Tea soft drink | Soluble solids content | 425–1000 nm | PLS, MLR | [61] | |
| Fruit | Gannan navel oranges | Soluble solids content and vitamin C | 350–1800 nm | PLSR | [62] | |
| Cereal | Wheat | Toxigenic fungal infection | 600 to 1600 nm | PCA, LDA, PLSR | [63] | |
| Corn | Aflatoxigenic fungus and aflatoxin | 400–2500 nm | PLS-DA | [64] | ||
| Nut | Peanut | Aflatoxin B1 (AFB1) | 400–2500 nm | PLS-DA | [65] | |
| MicroNIR | Nut | Cashew apple | The °Brix, total acidity, and concentration of ascorbic acid (vitamin C) | 1150–2170 nm | PCA, HCA | [66] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cebi, N.; Bekiroglu, H.; Erarslan, A. Nondestructive Metabolomic Fingerprinting: FTIR, NIR and Raman Spectroscopy in Food Screening. Molecules 2023, 28, 7933. https://doi.org/10.3390/molecules28237933
Cebi N, Bekiroglu H, Erarslan A. Nondestructive Metabolomic Fingerprinting: FTIR, NIR and Raman Spectroscopy in Food Screening. Molecules. 2023; 28(23):7933. https://doi.org/10.3390/molecules28237933
Chicago/Turabian StyleCebi, Nur, Hatice Bekiroglu, and Azime Erarslan. 2023. "Nondestructive Metabolomic Fingerprinting: FTIR, NIR and Raman Spectroscopy in Food Screening" Molecules 28, no. 23: 7933. https://doi.org/10.3390/molecules28237933
APA StyleCebi, N., Bekiroglu, H., & Erarslan, A. (2023). Nondestructive Metabolomic Fingerprinting: FTIR, NIR and Raman Spectroscopy in Food Screening. Molecules, 28(23), 7933. https://doi.org/10.3390/molecules28237933






