Current Landscape of Methods to Evaluate Antimicrobial Activity of Natural Extracts
Abstract
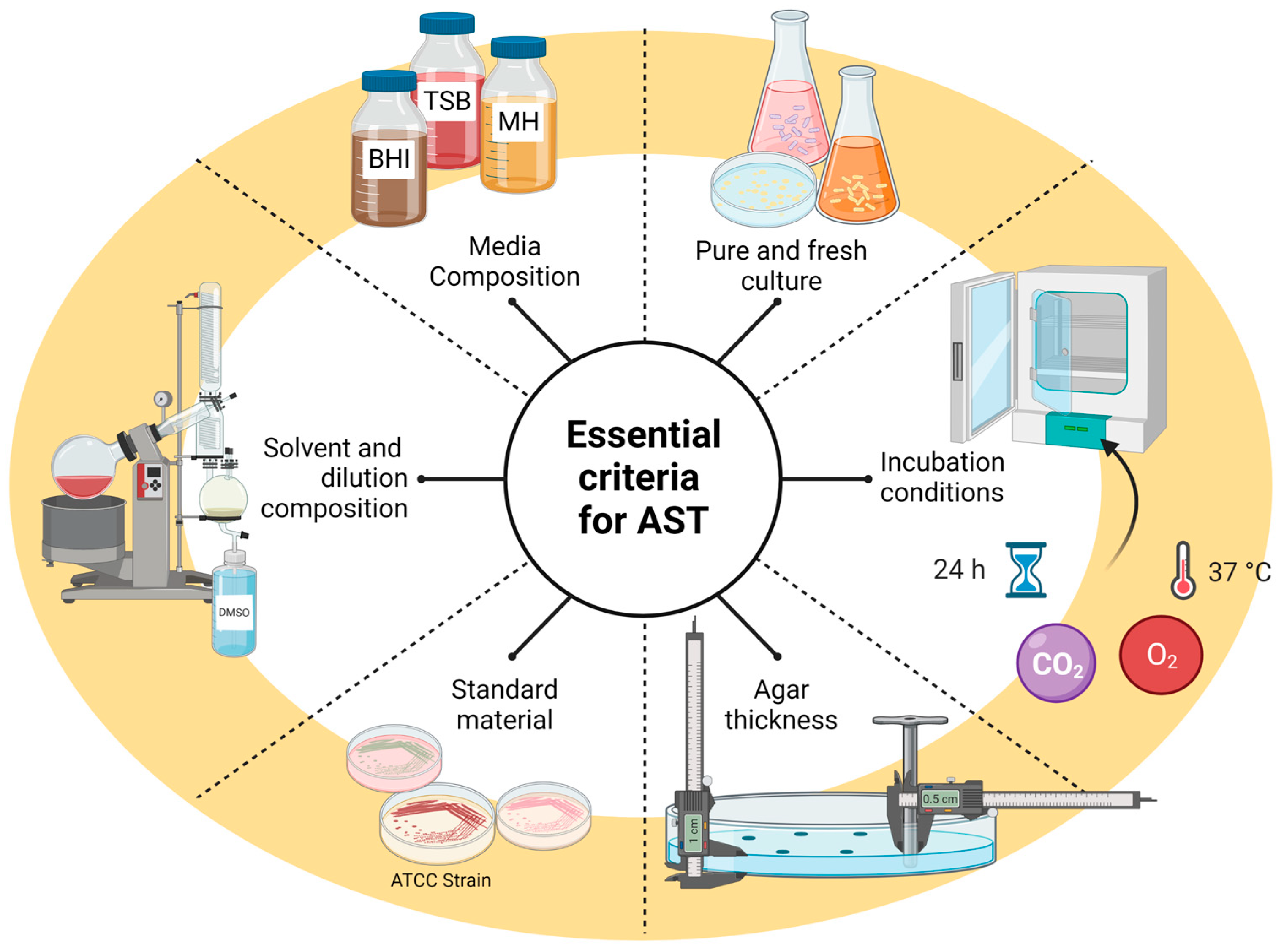
1. Introduction
2. Antibacterial Activity
2.1. Disk Well Diffusion
2.2. Agar Dilution Method
2.3. Broth Dilution Method
2.4. Thin-Layer Chromatography-Bioautography
2.5. Molecular Methods
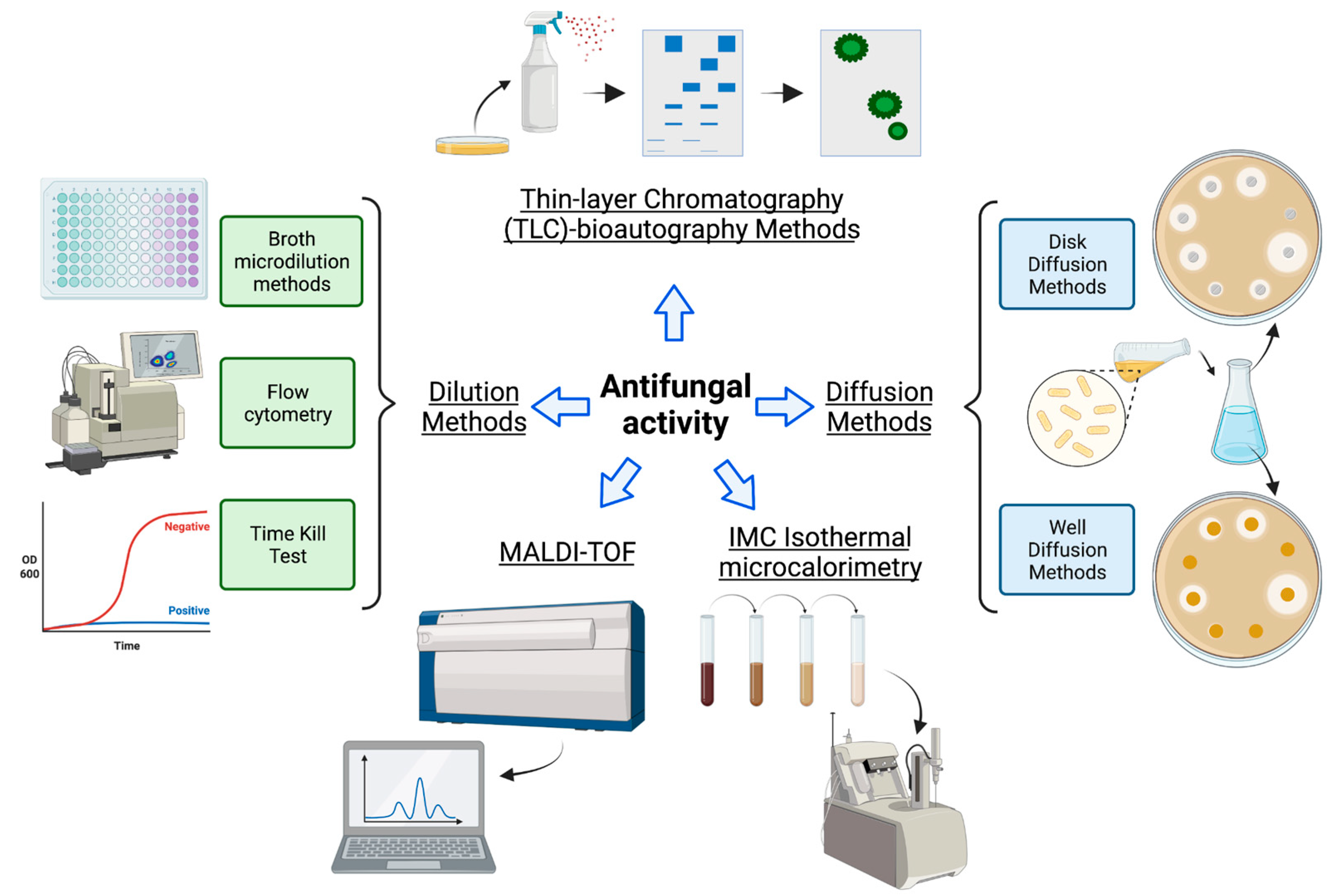
3. Antifungal Activity
| Method/Type of Assay | Description | Media and Microorganism | Advantages | Disadvantages | Ref. | |
|---|---|---|---|---|---|---|
| Diffusion methods | Disk/well diffusion |
|
|
|
| [74,75] |
| Dilution methods | Broth microdilution |
|
|
|
| [76,77,78] |
| Time-kill test |
|
|
|
| [73,79,80] | |
| Flow cytometry, or fluorescence-activated cell sorting (FACS) |
|
|
|
| [68,73,81,82] | |
| Calorimetry | Isothermal microcalorimetry (IMC) |
|
|
|
| [83] |
| Mass Spectrometry | MALDI-TOF |
|
|
|
| [84,85] |
| Thin-layer chromatography (TLC) | Agar overlay bioautography |
|
|
|
| [53,86,87] |
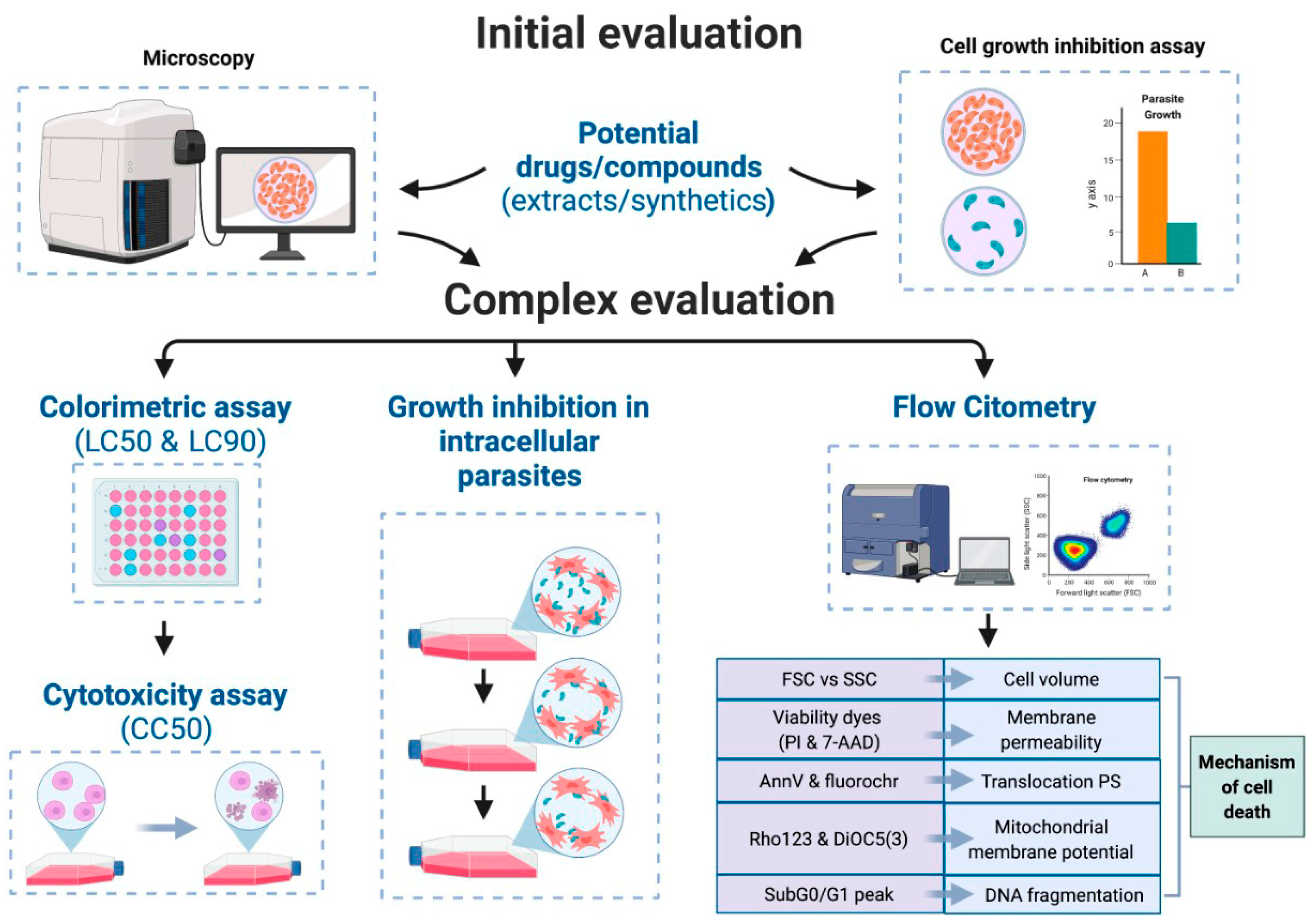
4. Antiparasitic Activity
4.1. Antiprotozoal Activity
4.2. Anthelmintic Activity
5. Antiviral Activity
6. Conclusions and Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Larsson, D.G.J.; Flach, C.-F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Laxminarayan, R. The overlooked pandemic of antimicrobial resistance. Lancet 2022, 399, 606–607. [Google Scholar] [CrossRef] [PubMed]
- Nadimpalli, M.L.; Chan, C.W.; Doron, S. Antibiotic resistance: A call to action to prevent the next epidemic of inequality. Nat. Med. 2021, 27, 187–188. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global Antimicrobial Resistance Surveillance System (GLASS) Report: Early Implementation 2020; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Cohen, T. The Next Pandemic: A Pragmatic and Ethical Discussion About the Looming Threat of Antibiotic Resistance. Voices Bioethics 2022, 8. [Google Scholar] [CrossRef]
- Vaughn, V.M.; Gandhi, T.N.; Petty, L.A.; Patel, P.K.; Prescott, H.C.; Malani, A.N.; Ratz, D.; McLaughlin, E.; Chopra, V.; Flanders, S.A. Empiric Antibacterial Therapy and Community-onset Bacterial Coinfection in Patients Hospitalized With Coronavirus Disease 2019 (COVID-19): A Multi-hospital Cohort Study. Clin. Infect. Dis. 2021, 72, e533–e541. [Google Scholar] [CrossRef]
- Ghimpețeanu, O.M.; Pogurschi, E.N.; Popa, D.C.; Dragomir, N.; Drăgotoiu, T.; Mihai, O.D.; Petcu, C.D. Antibiotic Use in Livestock and Residues in Food-A Public Health Threat: A Review. Foods 2022, 11, 1430. [Google Scholar] [CrossRef]
- Plackett, B. Why big pharma has abandoned antibiotics. Nature 2020, 586, S50–S52. [Google Scholar] [CrossRef]
- Klug, D.M.; Idiris, F.I.M.; Blaskovich, M.A.T.; von Delft, F.; Dowson, C.G.; Kirchhelle, C.; Roberts, A.P.; Singer, A.C.; Todd, M.H. There is no market for new antibiotics: This allows an open approach to research and development. Wellcome Open Res. 2021, 6, 146. [Google Scholar] [CrossRef]
- Baker, S.J.; Payne, D.J.; Rappuoli, R.; De Gregorio, E. Technologies to address antimicrobial resistance. Proc. Natl. Acad. Sci. USA 2018, 115, 12887–12895. [Google Scholar] [CrossRef]
- Thomford, N.E.; Senthebane, D.A.; Rowe, A.; Munro, D.; Seele, P.; Maroyi, A.; Dzobo, K. Natural products for drug discovery in the 21st century: Innovations for novel drug discovery. Int. J. Mol. Sci. 2018, 19, 1578. [Google Scholar] [CrossRef]
- Lautié, E.; Russo, O.; Ducrot, P.; Boutin, J.A. Unraveling plant natural chemical diversity for drug discovery purposes. Front. Pharmacol. 2020, 11, 397. [Google Scholar] [CrossRef] [PubMed]
- Omokhefe Bruce, S. Secondary Metabolites from Natural Products. In Secondary Metabolites [Working Title]; IntechOpen: Rijeka, Croatia, 2022. [Google Scholar]
- Stan, D.; Enciu, A.-M.; Mateescu, A.L.; Ion, A.C.; Brezeanu, A.C.; Stan, D.; Tanase, C. Natural compounds with antimicrobial and antiviral effect and nanocarriers used for their transportation. Front. Pharmacol. 2021, 12, 723233. [Google Scholar] [CrossRef] [PubMed]
- Dejani, N.N.; Elshabrawy, H.A.; Bezerra Filho, C.d.S.M.; de Sousa, D.P. Anticoronavirus and immunomodulatory phenolic compounds: Opportunities and pharmacotherapeutic perspectives. Biomolecules 2021, 11, 1254. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-W.; Lin, L.-G.; Ye, W.-C. Techniques for extraction and isolation of natural products: A comprehensive review. Chin. Med. 2018, 13, 20. [Google Scholar] [CrossRef] [PubMed]
- Bucar, F.; Wube, A.; Schmid, M. Natural product isolation--how to get from biological material to pure compounds. Nat. Prod. Rep. 2013, 30, 525–545. [Google Scholar] [CrossRef]
- Long, F.; Yang, H.; Xu, Y.; Hao, H.; Li, P. A strategy for the identification of combinatorial bioactive compounds contributing to the holistic effect of herbal medicines. Sci. Rep. 2015, 5, 12361. [Google Scholar] [CrossRef] [PubMed]
- Wagner, H.; Ulrich-Merzenich, G. Synergy research: Approaching a new generation of phytopharmaceuticals. Phytomedicine 2009, 16, 97–110. [Google Scholar] [CrossRef]
- Borges, A.; José, H.; Homem, V.; Simões, M. Comparison of Techniques and Solvents on the Antimicrobial and Antioxidant Potential of Extracts from Acacia dealbata and Olea europaea. Antibiotics 2020, 9, 48. [Google Scholar] [CrossRef]
- Sosa-Hernández, J.E.; Escobedo-Avellaneda, Z.; Iqbal, H.M.N.; Welti-Chanes, J. State-of-the-Art Extraction Methodologies for Bioactive Compounds from Algal Biome to Meet Bio-Economy Challenges and Opportunities. Molecules 2018, 23, 2953. [Google Scholar] [CrossRef]
- Gullón, P.; Gullón, B.; Romaní, A.; Rocchetti, G.; Lorenzo, J.M. Smart advanced solvents for bioactive compounds recovery from agri-food by-products: A review. Trends Food Sci. Technol. 2020, 101, 182–197. [Google Scholar] [CrossRef]
- Ngo, T.V.; Scarlett, C.J.; Bowyer, M.C.; Ngo, P.D.; Vuong, Q.V. Impact of Different Extraction Solvents on Bioactive Compounds and Antioxidant Capacity from the Root of Salacia chinensis L. J. Food Qual. 2017, 2017, 1–8. [Google Scholar] [CrossRef]
- Caesar, L.K.; Cech, N.B. Synergy and antagonism in natural product extracts: When 1 + 1 does not equal 2. Nat. Prod. Rep. 2019, 36, 869–888. [Google Scholar] [CrossRef] [PubMed]
- Egan, J.M.; van Santen, J.A.; Liu, D.Y.; Linington, R.G. Development of an NMR-Based Platform for the Direct Structural Annotation of Complex Natural Products Mixtures. J. Nat. Prod. 2021, 84, 1044–1055. [Google Scholar] [CrossRef] [PubMed]
- Mickymaray, S. Efficacy and Mechanism of Traditional Medicinal Plants and Bioactive Compounds against Clinically Important Pathogens. Antibiotics 2019, 8, 257. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xie, S.; Ahmed, S.; Wang, F.; Gu, Y.; Zhang, C.; Chai, X.; Wu, Y.; Cai, J.; Cheng, G. Antimicrobial activity and resistance: Influencing factors. Front. Pharmacol. 2017, 8, 364. [Google Scholar] [CrossRef]
- de Melo, A.L.F.; Rossato, L.; Barbosa, M.D.S.; Palozi, R.A.C.; Alfredo, T.M.; Antunes, K.A.; Eduvirgem, J.; Ribeiro, S.M.; Simionatto, S. From the environment to the hospital: How plants can help to fight bacteria biofilm. Microbiol. Res. 2022, 261, 127074. [Google Scholar] [CrossRef]
- Moloney, M.G. Natural products as a source for novel antibiotics. Trends Pharmacol. Sci. 2016, 37, 689–701. [Google Scholar] [CrossRef]
- Sharma, K.; Guleria, S.; Razdan, V.K.; Babu, V. Synergistic antioxidant and antimicrobial activities of essential oils of some selected medicinal plants in combination and with synthetic compounds. Ind. Crops Prod. 2020, 154, 112569. [Google Scholar] [CrossRef]
- Chusri, S.; Siriyong, T.; Na-Phatthalung, P.; Voravuthikunchai, S.P. Synergistic effects of ethnomedicinal plants of Apocynaceae family and antibiotics against clinical isolates of Acinetobacter baumannii. Asian Pac. J. Trop. Med. 2014, 7, 456–461. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Masota, N.E.; Vogg, G.; Ohlsen, K.; Holzgrabe, U. Reproducibility challenges in the search for antibacterial compounds from nature. PLoS ONE 2021, 16, e0255437. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.A.; Siddiqui, M.F.; Park, S. Current and emerging methods of antibiotic susceptibility testing. Diagnostics 2019, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- Temmerman, R.; Goethals, K.; Garmyn, A.; Vanantwerpen, G.; Vanrobaeys, M.; Haesebrouck, F.; Antonissen, G.; Devreese, M. Agreement of Quantitative and Qualitative Antimicrobial Susceptibility Testing Methodologies: The Case of Enrofloxacin and Avian Pathogenic Escherichia coli. Front. Microbiol. 2020, 11, 570975. [Google Scholar] [CrossRef] [PubMed]
- Humphries, R.M.; Ambler, J.; Mitchell, S.L.; Castanheira, M.; Dingle, T.; Hindler, J.A.; Koeth, L.; Sei, K. CLSI Methods Development and Standardization Working Group of the Subcommittee on Antimicrobial Susceptibility Testing CLSI methods development and standardization working group best practices for evaluation of antimicrobial susceptibility tests. J. Clin. Microbiol. 2018, 56, e01934-17. [Google Scholar] [CrossRef]
- CLSI Dilution AST for Aerobically Grown Bacteria—CLSI. Available online: https://clsi.org/standards/products/microbiology/documents/m07/ (accessed on 26 May 2022).
- Bubonja-Šonje, M.; Knežević, S.; Abram, M. Challenges to antimicrobial susceptibility testing of plant-derived polyphenolic compounds. Arh. Hig. Rada Toksikol. 2020, 71, 300–311. [Google Scholar] [CrossRef]
- Tan, J.B.L.; Lim, Y.Y. Critical analysis of current methods for assessing the in vitro antioxidant and antibacterial activity of plant extracts. Food Chem. 2015, 172, 814–822. [Google Scholar] [CrossRef]
- Silva, A.C.O.; Santana, E.F.; Saraiva, A.M.; Coutinho, F.N.; Castro, R.H.A.; Pisciottano, M.N.C.; Amorim, E.L.C.; Albuquerque, U.P. Which approach is more effective in the selection of plants with antimicrobial activity? Evid. Based Complement. Alternat. Med. 2013, 2013, 308980. [Google Scholar] [CrossRef]
- Jorgensen, J.H.; Turnidge, J.D. Susceptibility test methods: Dilution and disk diffusion methods*. In Manual of Clinical Microbiology, 11th ed.; Pfaller, M.A., Richter, S.S., Funke, G., Jorgensen, J.H., Landry, M.L., Carroll, K.C., Warnock, D.W., Eds.; American Society of Microbiology: Washington, DC, USA, 2015; pp. 1253–1273. ISBN 9781555817374. [Google Scholar]
- Vaou, N.; Stavropoulou, E.; Voidarou, C.; Tsigalou, C.; Bezirtzoglou, E. Towards advances in medicinal plant antimicrobial activity: A review study on challenges and future perspectives. Microorganisms 2021, 9, 41. [Google Scholar] [CrossRef]
- Mamedov, N. Medicinal plants studies: History, challenges and prospective. Med. Aromat. Plants 2012, 1, 133. [Google Scholar] [CrossRef]
- Guimarães, A.C.; Meireles, L.M.; Lemos, M.F.; Guimarães, M.C.C.; Endringer, D.C.; Fronza, M.; Scherer, R. Antibacterial activity of terpenes and terpenoids present in essential oils. Molecules 2019, 24, 2471. [Google Scholar] [CrossRef]
- Horváth, G.; Bencsik, T.; Ács, K.; Kocsis, B. Sensitivity of ESBL-Producing Gram-Negative Bacteria to Essential Oils, Plant Extracts, and Their Isolated Compounds. In Antibiotic Resistance; Elsevier: Amsterdam, The Netherlands, 2016; pp. 239–269. ISBN 9780128036426. [Google Scholar]
- Sadd, M.H. Encyclopedia of Microbiology, 4th ed.; Elsevier S & T: Amsterdam, The Netherlands, 2019; p. 1. ISBN 978-0-12-811737-8. [Google Scholar]
- Massoud, R.; Saffari, H.; Massoud, A.; Moteian, M.Y. Screening methods for assessment of antibacterial activity in nature. In Screening Methods for Assessment of Antibacterial Activity in Nature; University of Brussel: Brussel, Belgium, 2020. [Google Scholar]
- Christenson, J.C.; Korgenski, E.K.; Relich, R.F. Laboratory diagnosis of infection due to bacteria, fungi, parasites, and rickettsiae. In Principles and Practice of Pediatric Infectious Diseases; Elsevier: Amsterdam, The Netherlands, 2018; pp. 1422–1434.e3. ISBN 9780323401814. [Google Scholar]
- 20776–1; Susceptibility Testing of Infectious Agents and Evaluation of Performance of Antimicrobial Susceptibility Test Devices–Part 1: Broth Micro-Dilution Reference Method for Testing the In Vitro Activity of Antimicrobial Agents against Rapidly Growing Aerobic Bacteria Involved in Infectious Diseases. International Organization for Standardization: Geneva, Switzerland, 2019.
- Foerster, S.; Desilvestro, V.; Hathaway, L.J.; Althaus, C.L.; Unemo, M. A new rapid resazurin-based microdilution assay for antimicrobial susceptibility testing of Neisseria gonorrhoeae. J. Antimicrob. Chemother. 2017, 72, 1961–1968. [Google Scholar] [CrossRef] [PubMed]
- Kowalska-Krochmal, B.; Dudek-Wicher, R. The minimum inhibitory concentration of antibiotics: Methods, interpretation, clinical relevance. Pathogens 2021, 10, 165. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, A.; Vranken, T.; Malhotra, A.; Arts, J.J.C.; Habibovic, P. In vitro antimicrobial susceptibility testing methods: Agar dilution to 3D tissue-engineered models. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 187–208. [Google Scholar] [CrossRef] [PubMed]
- Dewanjee, S.; Gangopadhyay, M.; Bhattacharya, N.; Khanra, R.; Dua, T.K. Bioautography and its scope in the field of natural product chemistry. J. Pharm. Anal. 2015, 5, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, Y.; Wang, R.; Wang, Z.; Yang, B.; Kuang, H. An Evolving Technology That Integrates Classical Methods with Continuous Technological Developments: Thin-Layer Chromatography Bioautography. Molecules 2021, 26, 647. [Google Scholar] [CrossRef]
- Suleimana, M.M.; McGaw, L.J.; Naidoo, V.; Eloff, J.N. Detection of antimicrobial compounds by bioautography of different extracts of leaves of selected South African tree species. Afr. J. Tradit. Complement. Altern. Med. 2009, 7, 64–78. [Google Scholar] [CrossRef]
- Shakeri, A.; Sharifi, M.J.; Fazly Bazzaz, B.S.; Emami, A.; Soheili, V.; Sahebkar, A.; Asili, J. Bioautography Detection of Antimicrobial Compounds from the Essential Oil of Salvia Pachystachys. Curr. Bioact. Compd. 2018, 14, 80–85. [Google Scholar] [CrossRef]
- Berney, M.; Hammes, F.; Bosshard, F.; Weilenmann, H.-U.; Egli, T. Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight Kit in combination with flow cytometry. Appl. Environ. Microbiol. 2007, 73, 3283–3290. [Google Scholar] [CrossRef]
- Bankier, C.; Cheong, Y.; Mahalingam, S.; Edirisinghe, M.; Ren, G.; Cloutman-Green, E.; Ciric, L. A comparison of methods to assess the antimicrobial activity of nanoparticle combinations on bacterial cells. PLoS ONE 2018, 13, e0192093. [Google Scholar] [CrossRef]
- Nieto-Velázquez, N.G.; Gomez-Valdez, A.A.; González-Ávila, M.; Sánchez-Navarrete, J.; Toscano-Garibay, J.D.; Ruiz-Pérez, N.J. Preliminary Study on Citrus Oils Antibacterial Activity Measured by Flow Cytometry: A Step-by-Step Development. Antibiotics 2021, 10, 218. [Google Scholar] [CrossRef]
- Lagier, J.-C.; Hugon, P.; Khelaifia, S.; Fournier, P.-E.; La Scola, B.; Raoult, D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin. Microbiol. Rev. 2015, 28, 237–264. [Google Scholar] [CrossRef] [PubMed]
- Diakite, A.; Dubourg, G.; Dione, N.; Afouda, P.; Bellali, S.; Ngom, I.I.; Valles, C.; Tall, M.L.; Lagier, J.-C.; Raoult, D. Optimization and standardization of the culturomics technique for human microbiome exploration. Sci. Rep. 2020, 10, 9674. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, M.; Lagier, J.C.; Raoult, D.; Khelaifia, S. Bacterial culture through selective and non-selective conditions: The evolution of culture media in clinical microbiology. New Microbes New Infect. 2020, 34, 100622. [Google Scholar] [CrossRef] [PubMed]
- Aloni-Grinstein, R.; Shifman, O.; Lazar, S.; Steinberger-Levy, I.; Maoz, S.; Ber, R. A rapid real-time quantitative PCR assay to determine the minimal inhibitory extracellular concentration of antibiotics against an intracellular Francisella tularensis Live Vaccine Strain. Front. Microbiol. 2015, 6, 1213. [Google Scholar] [CrossRef] [PubMed]
- Kainz, K.; Bauer, M.A.; Madeo, F.; Carmona-Gutierrez, D. Fungal infections in humans: The silent crisis. Microb. Cell 2020, 7, 143–145. [Google Scholar] [CrossRef]
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and Multi-National Prevalence of Fungal Diseases-Estimate Precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef]
- Robbins, N.; Caplan, T.; Cowen, L.E. Molecular evolution of antifungal drug resistance. Annu. Rev. Microbiol. 2017, 71, 753–775. [Google Scholar] [CrossRef]
- Aldholmi, M.; Marchand, P.; Ourliac-Garnier, I.; Le Pape, P.; Ganesan, A. A Decade of Antifungal Leads from Natural Products: 2010-2019. Pharmaceuticals 2019, 12, 182. [Google Scholar] [CrossRef]
- Berkow, E.L.; Lockhart, S.R.; Ostrosky-Zeichner, L. Antifungal susceptibility testing: Current approaches. Clin. Microbiol. Rev. 2020, 33, e00069-19. [Google Scholar] [CrossRef]
- Fisher, M.C.; Alastruey-Izquierdo, A.; Berman, J.; Bicanic, T.; Bignell, E.M.; Bowyer, P.; Bromley, M.; Brüggemann, R.; Garber, G.; Cornely, O.A.; et al. Tackling the emerging threat of antifungal resistance to human health. Nat. Rev. Microbiol. 2022, 20, 557–571. [Google Scholar] [CrossRef]
- Perlin, D.S.; Rautemaa-Richardson, R.; Alastruey-Izquierdo, A. The global problem of antifungal resistance: Prevalence, mechanisms, and management. Lancet Infect. Dis. 2017, 17, e383–e392. [Google Scholar] [CrossRef] [PubMed]
- Posteraro, B.; De Carolis, E.; Vella, A.; Sanguinetti, M. MALDI-TOF mass spectrometry in the clinical mycology laboratory: Identification of fungi and beyond. Expert Rev. Proteom. 2013, 10, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef]
- Sanchez Armengol, E.; Harmanci, M.; Laffleur, F. Current strategies to determine antifungal and antimicrobial activity of natural compounds. Microbiol. Res. 2021, 252, 126867. [Google Scholar] [CrossRef] [PubMed]
- Scorzoni, L.; Benaducci, T.; Almeida, A.M.F.; Silva, D.H.S.; Bolzani, V.d.S.; Gianinni, M.J.S.M. The use of standard methodology for determination of antifungal activity of natural products against medical yeasts Candida sp and Cryptococcus sp. Braz. J. Microbiol. 2007, 38, 391–397. [Google Scholar] [CrossRef]
- Hadacek, F.; Greger, H. Testing of antifungal natural products: Methodologies, comparability of results and assay choice. Phytochem. Anal. 2000, 11, 137–147. [Google Scholar] [CrossRef]
- CLSI M27Ed4: Broth Dilution Antifungal Susceptibility, Yeasts. Available online: https://clsi.org/standards/products/microbiology/documents/m27/ (accessed on 30 May 2022).
- Rodriguez-Tudela, J.L.; Arendrup, M.C.; Barchiesi, F.; Bille, J.; Chryssanthou, E.; Cuenca-Estrella, M.; Dannaoui, E.; Denning, D.W.; Donnelly, J.P.; Dromer, F.; et al. Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID European Committee for Antimicrobial Susceptibility Testing (EUCAST) EUCAST definitive document EDef 7.1: Method for the determination of broth dilution MICs of antifungal agents for fermentative yeasts. Clin. Microbiol. Infect. 2008, 14, 398–405. [Google Scholar] [CrossRef]
- Alexander, B.D. Reference Method For Broth Dilution Antifungal Susceptibility Testing Of Filamentous Fungi; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017; p. 50. ISBN 1-56238-831-2. [Google Scholar]
- Arikan, S. Current status of antifungal susceptibility testing methods. Med. Mycol. 2007, 45, 569–587. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Sheehan, D.J.; Rex, J.H. Determination of fungicidal activities against yeasts and molds: Lessons learned from bactericidal testing and the need for standardization. Clin. Microbiol. Rev. 2004, 17, 268–280. [Google Scholar] [CrossRef]
- Vale-Silva, L.A.; Buchta, V. Antifungal susceptibility testing by flow cytometry: Is it the future? Mycoses 2006, 49, 261–273. [Google Scholar] [CrossRef]
- Bleichrodt, R.-J.; Read, N.D. Flow cytometry and FACS applied to filamentous fungi. Fungal Biol. Rev. 2019, 33, 1–15. [Google Scholar] [CrossRef]
- Covarrubias-Rivera, L.; López-Cruz, R.; Ragazzo-Sánchez, J.A.; Iñiguez-Moreno, M.; Calderón-Santoyo, M. Determination by isothermal microcalorimetry of the sensitivity of phytopathogenic fungi of tropical fruits against an ethanolic extract of jackfruit leaf (Artocarpus heterophyllus Lam.). J. Microbiol. Methods 2022, 195, 106457. [Google Scholar] [CrossRef] [PubMed]
- Marinach, C.; Alanio, A.; Palous, M.; Kwasek, S.; Fekkar, A.; Brossas, J.-Y.; Brun, S.; Snounou, G.; Hennequin, C.; Sanglard, D.; et al. MALDI-TOF MS-based drug susceptibility testing of pathogens: The example of Candida albicans and fluconazole. Proteomics 2009, 9, 4627–4631. [Google Scholar] [CrossRef] [PubMed]
- Durand, C.; Maubon, D.; Cornet, M.; Wang, Y.; Aldebert, D.; Garnaud, C. Can we improve antifungal susceptibility testing? Front. Cell. Infect. Microbiol. 2021, 11, 720609. [Google Scholar] [CrossRef]
- Nuthan, B.R.; Rakshith, D.; Marulasiddaswamy, K.M.; Rao, H.C.Y.; Ramesha, K.P.; Mohana, N.C.; Siddappa, S.; Darshan, D.; Kumara, K.K.S.; Satish, S. Application of Optimized and Validated Agar Overlay TLC-Bioautography Assay for Detecting the Antimicrobial Metabolites of Pharmaceutical Interest. J. Chromatogr. Sci. 2020, 58, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Marston, A. Thin-layer chromatography with biological detection in phytochemistry. J. Chromatogr. A 2011, 1218, 2676–2683. [Google Scholar] [CrossRef]
- Pisarski, K. The global burden of disease of zoonotic parasitic diseases: Top 5 contenders for priority consideration. Trop. Med. Infect. Dis. 2019, 4, 44. [Google Scholar] [CrossRef] [PubMed]
- Goupil, L.S.; McKerrow, J.H. Introduction: Drug discovery and development for neglected diseases. Chem. Rev. 2014, 114, 11131–11137. [Google Scholar] [CrossRef]
- Campbell, S.; Soman-Faulkner, K. Antiparasitic Drugs. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Wink, M. Medicinal plants: A source of anti-parasitic secondary metabolites. Molecules 2012, 17, 12771–12791. [Google Scholar] [CrossRef]
- Tagboto, S.; Townson, S. Antiparasitic properties of medicinal plants and other naturally occurring products. Adv. Parasitol. 2001, 50, 199–295. [Google Scholar] [CrossRef]
- Das, K.; Tiwari, R.K.; Shrivastava, D.K. Techniques for evaluation of medicinal plant products as antimicrobial agents: Current methods and future trends. J. Med. Plants Res. 2010, 4, 104–111. [Google Scholar]
- McHardy, I.H.; Wu, M.; Shimizu-Cohen, R.; Couturier, M.R.; Humphries, R.M. Detection of intestinal protozoa in the clinical laboratory. J. Clin. Microbiol. 2014, 52, 712–720. [Google Scholar] [CrossRef] [PubMed]
- Doonan, F.; Cotter, T.G. Morphological assessment of apoptosis. Methods 2008, 44, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Henry, C.M.; Hollville, E.; Martin, S.J. Measuring apoptosis by microscopy and flow cytometry. Methods 2013, 61, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Riss, T.L.; Moravec, R.A.; Niles, A.L.; Benink, H.A.; Worzella, T.J.; Minor, L. Cell Viability Assays. In Assay Guidance Manual; Sittampalam, G.S., Coussens, N.P., Nelson, H., Arkin, M., Auld, D., Austin, C., Bejcek, B., Glicksman, M., Inglese, J., Iversen, P.W., et al., Eds.; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2004. [Google Scholar]
- Adan, A.; Alizada, G.; Kiraz, Y.; Baran, Y.; Nalbant, A. Flow cytometry: Basic principles and applications. Crit. Rev. Biotechnol. 2017, 37, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Soeiro, M.N.C.; Werbovetz, K.; Boykin, D.W.; Wilson, W.D.; Wang, M.Z.; Hemphill, A. Novel amidines and analogues as promising agents against intracellular parasites: A systematic review. Parasitology 2013, 140, 929–951. [Google Scholar] [CrossRef]
- Aslantürk, Ö.S. In vitro cytotoxicity and cell viability assays: Principles, advantages, and disadvantages. In Genotoxicity—A Predictable Risk to Our Actual World; Larramendy, M.L., Soloneski, S., Eds.; InTech: London, UK, 2018; ISBN 978-1-78923-418-3. [Google Scholar]
- Präbst, K.; Engelhardt, H.; Ringgeler, S.; Hübner, H. Basic colorimetric proliferation assays: MTT, WST, and resazurin. Methods Mol. Biol. 2017, 1601, 1–17. [Google Scholar] [CrossRef]
- García Díaz, J.; Tuenter, E.; Escalona Arranz, J.C.; Llauradó Maury, G.; Cos, P.; Pieters, L. Antimicrobial activity of leaf extracts and isolated constituents of Croton linearis. J. Ethnopharmacol. 2019, 236, 250–257. [Google Scholar] [CrossRef]
- Henriques, C.; Moreira, T.L.B.; Maia-Brigagão, C.; Henriques-Pons, A.; Carvalho, T.M.U.; de Souza, W. Tetrazolium salt based methods for high-throughput evaluation of anti-parasite chemotherapy. Anal. Methods 2011, 3, 2148. [Google Scholar] [CrossRef]
- Ilaghi, M.; Sharifi, I.; Sharififar, F.; Sharifi, F.; Oliaee, R.T.; Babaei, Z.; Meimamandi, M.S.; Keyhani, A.; Bamorovat, M. The potential role and apoptotic profile of three medicinal plant extracts on Leishmania tropica by MTT assay, macrophage model and flow cytometry analysis. Parasite Epidemiol. Control 2021, 12, e00201. [Google Scholar] [CrossRef]
- Barrio, G.; Grueiro, M.; Montero, D.; Nogal, J.J.; Escario, J.A.; Muelas, S.; Fernández, C.; Vega, C.; Rolón, M.; Fernández, M.; et al. In Vitro Antiparasitic Activity of Plant Extracts from Panama. Pharm. Biol. 2004, 42, 332–337. [Google Scholar] [CrossRef]
- Jiménez-Ruiz, A.; Alzate, J.F.; Macleod, E.T.; Lüder, C.G.K.; Fasel, N.; Hurd, H. Apoptotic markers in protozoan parasites. Parasit. Vectors 2010, 3, 104. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Godínez, C.; Ontiveros-Rodríguez, J.C.; Ríos-Valencia, D.G.; Herbert-Pucheta, J.E.; Zepeda-Vallejo, L.G.; Carrero, J.C. Anti-amoebic Activity of Leaf Extracts and Aporphine Alkaloids Obtained from Annona purpurea. Planta Med. 2020, 86, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Sederstrom, J.M. Assaying cell cycle status using flow cytometry. Curr. Protoc. Mol. Biol. 2015, 111, 28.6.1–28.6.11. [Google Scholar] [CrossRef]
- Mukherjee, D.; Singh, C.B.; Dey, S.; Mandal, S.; Ghosh, J.; Mallick, S.; Hussain, A.; Swapana, N.; Ross, S.A.; Pal, C. Induction of apoptosis by zerumbone isolated from Zingiber zerumbet (L.) Smith in protozoan parasite Leishmania donovani due to oxidative stress. Braz. J. Infect. Dis. 2016, 20, 48–55. [Google Scholar] [CrossRef]
- Souza, R.O.d.S.; Sousa, P.L.; de Menezes, R.R.P.P.B.; Sampaio, T.L.; Tessarolo, L.D.; Silva, F.C.O.; Pereira, M.G.; Martins, A.M.C. Trypanocidal activity of polysaccharide extract from Genipa americana leaves. J. Ethnopharmacol. 2018, 210, 311–317. [Google Scholar] [CrossRef]
- Vermes, I.; Haanen, C.; Steffens-Nakken, H.; Reutelingsperger, C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J. Immunol. Methods 1995, 184, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Crowley, L.C.; Marfell, B.J.; Scott, A.P.; Waterhouse, N.J. Quantitation of apoptosis and necrosis by annexin V binding, propidium iodide uptake, and flow cytometry. Cold Spring Harb. Protoc. 2016, 2016, pdb-prot087288. [Google Scholar] [CrossRef]
- Khademvatan, S.; Saki, J.; Gharavi, M.J.; Rahim, F. Allium sativum extract induces apoptosis in Leishmania major (MRHO/IR/75/ER) promastigotes. J. Med. Plants Res. 2011, 5, 3725–3732. [Google Scholar]
- Proto, W.R.; Coombs, G.H.; Mottram, J.C. Cell death in parasitic protozoa: Regulated or incidental? Nat. Rev. Microbiol. 2013, 11, 58–66. [Google Scholar] [CrossRef]
- Moreira, A.L.; Scariot, D.B.; Pelegrini, B.L.; Pessini, G.L.; Ueda-Nakamura, T.; Nakamura, C.V.; Ferreira, I.C.P. Acyclic Sesquiterpenes from the Fruit Pericarp of Sapindus saponaria Induce Ultrastructural Alterations and Cell Death in Leishmania amazonensis. Evid. Based Complement. Alternat. Med. 2017, 2017, 5620693. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, R.D.D.G.; Oliveira, A.P.; Ferreira, C.; Passos, C.L.A.; Fialho, E.; Soares, D.C.; Amaral, V.F.; Bezerra, G.B.; Esteves, R.S.; Santos, M.G.; et al. Anti-Leishmania amazonensis activity of the terpenoid fraction from Eugenia pruniformis leaves. An. Acad. Bras. Cienc. 2020, 92, e20201181. [Google Scholar] [CrossRef] [PubMed]
- Dayakar, A.; Chandrasekaran, S.; Veronica, J.; Sundar, S.; Maurya, R. In vitro and in vivo evaluation of anti-leishmanial and immunomodulatory activity of Neem leaf extract in Leishmania donovani infection. Exp. Parasitol. 2015, 153, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Shinjyo, N.; Nakayama, H.; Ishimaru, K.; Hikosaka, K.; Mi-Ichi, F.; Norose, K.; Yoshida, H. Hypericum erectum alcoholic extract inhibits Toxoplasma growth and Entamoeba encystation: An exploratory study on the anti-protozoan potential. J. Nat. Med. 2020, 74, 294–305. [Google Scholar] [CrossRef]
- Hendrickx, S.; Caljon, G.; Maes, L. In Vitro Growth Inhibition Assays of Leishmania spp. Methods Mol. Biol. 2020, 2116, 791–800. [Google Scholar] [CrossRef]
- García, M.; Monzote, L.; Montalvo, A.M.; Scull, R. Screening of medicinal plants against Leishmania amazonensis. Pharm. Biol. 2010, 48, 1053–1058. [Google Scholar] [CrossRef]
- Kashif, M.; Hira, S.K.; Upadhyaya, A.; Gupta, U.; Singh, R.; Paladhi, A.; Khan, F.I.; Rub, A.; Manna, P.P. In silico studies and evaluation of antiparasitic role of a novel pyruvate phosphate dikinase inhibitor in Leishmania donovani infected macrophages. Int. J. Antimicrob. Agents 2019, 53, 508–514. [Google Scholar] [CrossRef]
- Wolf, K.; Dormeyer, M. Information-based methods in the development of antiparasitic drugs. Parasitol. Res. 2003, 90 (Suppl. 2), S91–S96. [Google Scholar] [CrossRef]
- Ogungbe, I.V.; Setzer, W.N. In-silico Leishmania target selectivity of antiparasitic terpenoids. Molecules 2013, 18, 7761–7847. [Google Scholar] [CrossRef]
- Johnson, R.M.; Rawson, S.; McPhillie, M.J.; Fishwick, C.W.G.; Muench, S.P. The Growing Role of Electron Microscopy in Anti-parasitic Drug Discovery. Curr. Med. Chem. 2018, 25, 5279–5290. [Google Scholar] [CrossRef]
- Vannier-Santos, M.A.; De Castro, S.L. Electron microscopy in antiparasitic chemotherapy: A (close) view to a kill. Curr. Drug Targets 2009, 10, 246–260. [Google Scholar] [CrossRef] [PubMed]
- Hendrickx, S.; Caljon, G.; Maes, L. Need for sustainable approaches in antileishmanial drug discovery. Parasitol. Res. 2019, 118, 2743–2752. [Google Scholar] [CrossRef] [PubMed]
- Finger, S.; Wiegand, C.; Buschmann, H.-J.; Hipler, U.-C. Antibacterial properties of cyclodextrin-antiseptics-complexes determined by microplate laser nephelometry and ATP bioluminescence assay. Int. J. Pharm. 2013, 452, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, R.; Yadav, J.S. Development of a rapid ATP bioluminescence assay for biocidal susceptibility testing of rapidly growing mycobacteria. J. Clin. Microbiol. 2010, 48, 3725–3728. [Google Scholar] [CrossRef]
- Villalta, F.; Rachakonda, G. Advances in preclinical approaches to Chagas disease drug discovery. Expert Opin. Drug Discov. 2019, 14, 1161–1174. [Google Scholar] [CrossRef] [PubMed]
- Weng, H.-B.; Chen, H.-X.; Wang, M.-W. Innovation in neglected tropical disease drug discovery and development. Infect. Dis. Poverty 2018, 7, 67. [Google Scholar] [CrossRef]
- Cowell, A.N.; Winzeler, E.A. Advances in omics-based methods to identify novel targets for malaria and other parasitic protozoan infections. Genome Med. 2019, 11, 63. [Google Scholar] [CrossRef]
- Munguía, B.; Saldaña, J.; Nieves, M.; Melian, M.E.; Ferrer, M.; Teixeira, R.; Porcal, W.; Manta, E.; Domínguez, L. Sensitivity of Haemonchus contortus to anthelmintics using different in vitro screening assays: A comparative study. Parasit. Vectors 2022, 15, 129. [Google Scholar] [CrossRef]
- Zenebe, S.; Feyera, T.; Assefa, S. In Vitro Anthelmintic Activity of Crude Extracts of Aerial Parts of Cissus quadrangularis L. and Leaves of Schinus molle L. against Haemonchus contortus. Biomed Res. Int. 2017, 2017, 1905987. [Google Scholar] [CrossRef]
- Garbin, V.P.; Munguía, B.; Saldaña, J.C.; Deschamps, C.; Cipriano, R.R.; Molento, M.B. Chemical characterization and in vitro anthelmintic activity of Citrus bergamia Risso and Citrus X paradisii Macfad essential oil against Haemonchus contortus Kirby isolate. Acta Trop. 2021, 217, 105869. [Google Scholar] [CrossRef]
- Jayawardene, K.L.T.D.; Palombo, E.A.; Boag, P.R. Natural products are a promising source for anthelmintic drug discovery. Biomolecules 2021, 11, 457. [Google Scholar] [CrossRef] [PubMed]
- Vijayakumar, S.; Malaikozhundan, B.; Saravanakumar, K.; Durán-Lara, E.F.; Wang, M.-H.; Vaseeharan, B. Garlic clove extract assisted silver nanoparticle—Antibacterial, antibiofilm, antihelminthic, anti-inflammatory, anticancer and ecotoxicity assessment. J. Photochem. Photobiol. B Biol. 2019, 198, 111558. [Google Scholar] [CrossRef] [PubMed]
- Athanasiadou, S.; Almvik, M.; Hellström, J.; Madland, E.; Simic, N.; Steinshamn, H. Chemical Analysis and Anthelmintic Activity Against Teladorsagia Circumcincta of Nordic Bark Extracts In vitro. Front. Vet. Sci. 2021, 8, 666924. [Google Scholar] [CrossRef]
- Eguale, T.; Tadesse, D.; Giday, M. In vitro anthelmintic activity of crude extracts of five medicinal plants against egg-hatching and larval development of Haemonchus contortus. J. Ethnopharmacol. 2011, 137, 108–113. [Google Scholar] [CrossRef]
- Giovanelli, F.; Mattellini, M.; Fichi, G.; Flamini, G.; Perrucci, S. In Vitro Anthelmintic Activity of Four Plant-Derived Compounds against Sheep Gastrointestinal Nematodes. Vet. Sci. 2018, 5, 78. [Google Scholar] [CrossRef]
- Carvalho, C.O.; Chagas, A.C.S.; Cotinguiba, F.; Furlan, M.; Brito, L.G.; Chaves, F.C.M.; Stephan, M.P.; Bizzo, H.R.; Amarante, A.F.T. The anthelmintic effect of plant extracts on Haemonchus contortus and Strongyloides venezuelensis. Vet. Parasitol. 2012, 183, 260–268. [Google Scholar] [CrossRef]
- Ferreira, L.E.; Benincasa, B.I.; Fachin, A.L.; Contini, S.H.T.; França, S.C.; Chagas, A.C.S.; Beleboni, R.O. Essential oils of Citrus aurantifolia, Anthemis nobile and Lavandula officinalis: In vitro anthelmintic activities against Haemonchus contortus. Parasit. Vectors 2018, 11, 269. [Google Scholar] [CrossRef] [PubMed]
- Maestrini, M.; Tava, A.; Mancini, S.; Tedesco, D.; Perrucci, S. In Vitro Anthelmintic Activity of Saponins from Medicago spp. Against Sheep Gastrointestinal Nematodes. Molecules 2020, 25, 242. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Tak, H.; Nazir, R.; Lone, B.A. In vitro and in vivo anthelmintic activities of Iris kashmiriana Linn. J. Saudi Soc. Agric. Sci. 2016, 17, 235–240. [Google Scholar] [CrossRef]
- Mathew, M.D.; Mathew, N.D.; Miller, A.; Simpson, M.; Au, V.; Garland, S.; Gestin, M.; Edgley, M.L.; Flibotte, S.; Balgi, A.; et al. Elegans Forward and Reverse Genetics to Identify New Compounds with Anthelmintic Activity. PLoS Negl. Trop. Dis. 2016, 10, e0005058. [Google Scholar] [CrossRef]
- Corsi, A.K.; Wightman, B.; Chalfie, M. A Transparent Window into Biology: A Primer on Caenorhabditis elegans. Genetics 2015, 200, 387–407. [Google Scholar] [CrossRef] [PubMed]
- Zamanian, M.; Chan, J.D. High-content approaches to anthelmintic drug screening. Trends Parasitol. 2021, 37, 780–789. [Google Scholar] [CrossRef] [PubMed]
- Adamson, C.S.; Chibale, K.; Goss, R.J.M.; Jaspars, M.; Newman, D.J.; Dorrington, R.A. Antiviral drug discovery: Preparing for the next pandemic. Chem. Soc. Rev. 2021, 50, 3647–3655. [Google Scholar] [CrossRef] [PubMed]
- Irwin, K.K.; Renzette, N.; Kowalik, T.F.; Jensen, J.D. Antiviral drug resistance as an adaptive process. Virus Evol. 2016, 2, vew014. [Google Scholar] [CrossRef]
- De Clercq, E.; Li, G. Approved Antiviral Drugs over the Past 50 Years. Clin. Microbiol. Rev. 2016, 29, 695–747. [Google Scholar] [CrossRef]
- Li, G.; De Clercq, E. Chapter 1. overview of antiviral drug discovery and development: Viral versus host targets. In Antiviral Discovery for Highly Pathogenic Emerging Viruses; Muñoz-Fontela, C., Delgado, R., Eds.; Drug Discovery; Royal Society of Chemistry: Cambridge, UK, 2021; pp. 1–27. ISBN 978-1-78801-564-6. [Google Scholar]
- Owen, L.; Laird, K.; Shivkumar, M. Antiviral plant-derived natural products to combat RNA viruses: Targets throughout the viral life cycle. Lett. Appl. Microbiol. 2021, 75, 476–499. [Google Scholar] [CrossRef]
- Musarra-Pizzo, M.; Pennisi, R.; Ben-Amor, I.; Mandalari, G.; Sciortino, M.T. Antiviral Activity Exerted by Natural Products against Human Viruses. Viruses 2021, 13, 828. [Google Scholar] [CrossRef]
- Prichard, M.N.; Turk, S.R.; Coleman, L.A.; Engelhardt, S.L.; Shipman, C.; Drach, J.C. A microtiter virus yield reduction assay for the evaluation of antiviral compounds against human cytomegalovirus and herpes simplex virus. J. Virol. Methods 1990, 28, 101–106. [Google Scholar] [CrossRef]
- Cirne-Santos, C.C.; Barros, C.d.S.; Gomes, M.W.L.; Gomes, R.; Cavalcanti, D.N.; Obando, J.M.C.; Ramos, C.J.B.; Villaça, R.C.; Teixeira, V.L.; Paixão, I.C.N.d.P. In Vitro Antiviral Activity Against Zika Virus From a Natural Product of the Brazilian Brown Seaweed Dictyota menstrualis. Nat. Prod. Commun. 2019, 14, 1934578X1985912. [Google Scholar] [CrossRef]
- Gu, L.; Schneller, S.W.; Li, Q. Assays for the identification of novel antivirals against bluetongue virus. J. Vis. Exp. 2013, 80, e50820. [Google Scholar] [CrossRef]
- Chiamenti, L.; da Silva, F.P.; Schallemberger, K.; Demoliner, M.; Rigotto, C.; Fleck, J.D. Cytotoxicity and antiviral activity evaluation of Cymbopogon spp hydroethanolic extracts. Braz. J. Pharm. Sci. 2019, 55, e18063. [Google Scholar] [CrossRef]
- Smither, S.J.; Lear-Rooney, C.; Biggins, J.; Pettitt, J.; Lever, M.S.; Olinger, G.G. Comparison of the plaque assay and 50% tissue culture infectious dose assay as methods for measuring filovirus infectivity. J. Virol. Methods 2013, 193, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Baer, A.; Kehn-Hall, K. Viral concentration determination through plaque assays: Using traditional and novel overlay systems. J. Vis. Exp. 2014, 93, e52065. [Google Scholar] [CrossRef] [PubMed]
- Visintini Jaime, M.F.; Redko, F.; Muschietti, L.V.; Campos, R.H.; Martino, V.S.; Cavallaro, L.V. In vitro antiviral activity of plant extracts from Asteraceae medicinal plants. Virol. J. 2013, 10, 245. [Google Scholar] [CrossRef]
- Mendoza, E.J.; Manguiat, K.; Wood, H.; Drebot, M. Two Detailed Plaque Assay Protocols for the Quantification of Infectious SARS-CoV-2. Curr. Protoc. Microbiol. 2020, 57, ecpmc105. [Google Scholar] [CrossRef]
- Cresta, D.; Warren, D.C.; Quirouette, C.; Smith, A.P.; Lane, L.C.; Smith, A.M.; Beauchemin, C.A.A. Time to revisit the endpoint dilution assay and to replace the TCID50 as a measure of a virus sample’s infection concentration. PLoS Comput. Biol. 2021, 17, e1009480. [Google Scholar] [CrossRef]
- Bullen, C.K.; Davis, S.L.; Looney, M.M. Quantification of Infectious SARS-CoV-2 by the 50% Tissue Culture Infectious Dose Endpoint Dilution Assay. Methods Mol. Biol. 2022, 2452, 131–146. [Google Scholar] [CrossRef]
- Stewart, H.; Bartlett, C.; Ross-Thriepland, D.; Shaw, J.; Griffin, S.; Harris, M. A novel method for the measurement of hepatitis C virus infectious titres using the IncuCyte ZOOM and its application to antiviral screening. J. Virol. Methods 2015, 218, 59–65. [Google Scholar] [CrossRef]
- Singh, P.; Singh, G.; Karsky, J.; Nelson, E.; Ramamoorthy, S. A convenient colorimetric assay for the quantification of porcine epidemic diarrhea virus and neutralizing antibodies. J. Virol. Methods 2018, 262, 32–37. [Google Scholar] [CrossRef]
- Bolívar-Marin, S.; Bosch, I.; Narváez, C.F. Combination of the Focus-Forming Assay and Digital Automated Imaging Analysis for the Detection of Dengue and Zika Viral Loads in Cultures and Acute Disease. J. Trop. Med. 2022, 2022, 2177183. [Google Scholar] [CrossRef]
- Coimbra, L.D.; Borin, A.; Fontoura, M.; Gravina, H.D.; Nagai, A.; Shimizu, J.F.; Bispo-dos-Santos, K.; Granja, F.; Oliveira, P.S.L.; Franchini, K.G.; et al. Identification of Compounds With Antiviral Activity Against SARS-CoV-2 in the MMV Pathogen Box Using a Phenotypic High-Throughput Screening Assay. Front.Virol. 2022, 2, 854363. [Google Scholar] [CrossRef]
- Koishi, A.C.; Zanello, P.R.; Bianco, É.M.; Bordignon, J.; Nunes Duarte dos Santos, C. Screening of Dengue virus antiviral activity of marine seaweeds by an in situ enzyme-linked immunosorbent assay. PLoS ONE 2012, 7, e51089. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, V.; Brognaro, H.; Prabhu, P.R.; de Souza, E.E.; Günther, S.; Reinke, P.Y.A.; Lane, T.J.; Ginn, H.; Han, H.; Ewert, W.; et al. Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease. Commun. Biol. 2022, 5, 805. [Google Scholar] [CrossRef] [PubMed]
- Sachse, M.; Tenorio, R.; Fernández de Castro, I.; Muñoz-Basagoiti, J.; Perez-Zsolt, D.; Raïch-Regué, D.; Rodon, J.; Losada, A.; Avilés, P.; Cuevas, C.; et al. Unraveling the antiviral activity of plitidepsin against SARS-CoV-2 by subcellular and morphological analysis. Antivir. Res. 2022, 200, 105270. [Google Scholar] [CrossRef] [PubMed]
- Dolskiy, A.A.; Grishchenko, I.V.; Yudkin, D.V. Cell cultures for virology: Usability, advantages, and prospects. Int. J. Mol. Sci. 2020, 21, 978. [Google Scholar] [CrossRef]
- Rumlová, M.; Ruml, T. In vitro methods for testing antiviral drugs. Biotechnol. Adv. 2018, 36, 557–576. [Google Scholar] [CrossRef]
- Aliabadi, N.; Jamalidoust, M.; Pouladfar, G.; Ziyaeyan, A.; Ziyaeyan, M. Antiviral activity of triptolide on herpes simplex virus in vitro. Immun. Inflamm. Dis. 2022, 10, e667. [Google Scholar] [CrossRef] [PubMed]
- Sureram, S.; Arduino, I.; Ueoka, R.; Rittà, M.; Francese, R.; Srivibool, R.; Darshana, D.; Piel, J.; Ruchirawat, S.; Muratori, L.; et al. The Peptide A-3302-B Isolated from a Marine Bacterium Micromonospora sp. Inhibits HSV-2 Infection by Preventing the Viral Egress from Host Cells. Int. J. Mol. Sci. 2022, 23, 947. [Google Scholar] [CrossRef]
- Marin, M.; Du, Y.; Giroud, C.; Kim, J.H.; Qui, M.; Fu, H.; Melikyan, G.B. High-Throughput HIV-Cell Fusion Assay for Discovery of Virus Entry Inhibitors. Assay Drug Dev. Technol. 2015, 13, 155–166. [Google Scholar] [CrossRef]
- Ho, H.P.T.; Vo, D.N.K.; Lin, T.-Y.; Hung, J.-N.; Chiu, Y.-H.; Tsai, M.-H. Ganoderma microsporum immunomodulatory protein acts as a multifunctional broad-spectrum antiviral against SARS-CoV-2 by interfering virus binding to the host cells and spike-mediated cell fusion. Biomed. Pharmacother. 2022, 155, 113766. [Google Scholar] [CrossRef]
- Meunier, T.; Desmarets, L.; Bordage, S.; Bamba, M.; Hervouet, K.; Rouillé, Y.; François, N.; Decossas, M.; Sencio, V.; Trottein, F.; et al. A Photoactivable Natural Product with Broad Antiviral Activity against Enveloped Viruses, Including Highly Pathogenic Coronaviruses. Antimicrob. Agents Chemother. 2022, 66, e0158121. [Google Scholar] [CrossRef] [PubMed]
- Kausar, S.; Said Khan, F.; Ishaq Mujeeb Ur Rehman, M.; Akram, M.; Riaz, M.; Rasool, G.; Hamid Khan, A.; Saleem, I.; Shamim, S.; Malik, A. A review: Mechanism of action of antiviral drugs. Int. J. Immunopathol. Pharmacol. 2021, 35, 20587384211002620. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Wang, H.; Wang, Z.; Wang, Q. Progression of antiviral agents targeting viral polymerases. Molecules 2022, 27, 370. [Google Scholar] [CrossRef] [PubMed]
- Gabaglio, S.; Alvarenga, N.; Cantero-González, G.; Degen, R.; Ferro, E.A.; Langjahr, P.; Chnaiderman, J.; Sotelo, P.H. A quantitative PCR assay for antiviral activity screening of medicinal plants against Herpes simplex 1. Nat. Prod. Res. 2021, 35, 2926–2930. [Google Scholar] [CrossRef] [PubMed]
- Sáez-Álvarez, Y.; Arias, A.; Del Águila, C.; Agudo, R. Development of a fluorescence-based method for the rapid determination of Zika virus polymerase activity and the screening of antiviral drugs. Sci. Rep. 2019, 9, 5397. [Google Scholar] [CrossRef] [PubMed]
- Beadle, J.R.; Valiaeva, N.; Yang, G.; Yu, J.-H.; Broker, T.R.; Aldern, K.A.; Harden, E.A.; Keith, K.A.; Prichard, M.N.; Hartman, T.; et al. Synthesis and Antiviral Evaluation of Octadecyloxyethyl Benzyl 9-[(2-Phosphonomethoxy)ethyl]guanine (ODE-Bn-PMEG), a Potent Inhibitor of Transient HPV DNA Amplification. J. Med. Chem. 2016, 59, 10470–10478. [Google Scholar] [CrossRef]
- Vicenti, I.; Dragoni, F.; Giannini, A.; Giammarino, F.; Spinicci, M.; Saladini, F.; Boccuto, A.; Zazzi, M. Development of a Cell-Based Immunodetection Assay for Simultaneous Screening of Antiviral Compounds Inhibiting Zika and Dengue Virus Replication. SLAS Discov. 2020, 25, 506–514. [Google Scholar] [CrossRef]
- Case, J.B.; Bailey, A.L.; Kim, A.S.; Chen, R.E.; Diamond, M.S. Growth, detection, quantification, and inactivation of SARS-CoV-2. Virology 2020, 548, 39–48. [Google Scholar] [CrossRef]
- Liu, T.; Li, Y.; Koydemir, H.C.; Zhang, Y.; Yang, E.; Wang, H.; Li, J.; Bai, B.; Ozcan, A. Stain-free, rapid, and quantitative viral plaque assay using deep learning and holography. arXiv 2022, arXiv:2207.00089. [Google Scholar]
- Theuerkauf, S.A.; Michels, A.; Riechert, V.; Maier, T.J.; Flory, E.; Cichutek, K.; Buchholz, C.J. Quantitative assays reveal cell fusion at minimal levels of SARS-CoV-2 spike protein and fusion from without. iScience 2021, 24, 102170. [Google Scholar] [CrossRef]
- Chan, S.-W. Fusion assays for screening of fusion inhibitors targeting SARS-CoV-2 entry and syncytia formation. Front. Pharmacol. 2022, 13, 1007527. [Google Scholar] [CrossRef]
- Hochdorfer, D.; Businger, R.; Hotter, D.; Seifried, C.; Solzin, J. Automated, label-free TCID50 assay to determine the infectious titer of virus-based therapeutics. J. Virol. Methods 2022, 299, 114318. [Google Scholar] [CrossRef] [PubMed]
- Romano, J.D.; Tatonetti, N.P. Informatics and computational methods in natural product drug discovery: A review and perspectives. Front. Genet. 2019, 10, 368. [Google Scholar] [CrossRef]
- Gomaa, H.; Elshoubaky, G. Antiviral Activity of Sulfated Polysaccharides Carrageenan from Some Marine Seaweeds. Int. J. Curr. Pharm. Rev. Res. 2015, 7, 34–42. [Google Scholar]
- Padmanabhan, P.; Khaleefathullah, S.; Kaveri, K.; Palani, G.; Ramanathan, G.; Thennarasu, S.; Tirichurapalli Sivagnanam, U. Antiviral activity of Thiosemicarbazones derived from α-amino acids against Dengue virus. J. Med. Virol. 2017, 89, 546–552. [Google Scholar] [CrossRef] [PubMed]
- Golus, J.; Sawicki, R.; Widelski, J.; Ginalska, G. The agar microdilution method—A new method for antimicrobial susceptibility testing for essential oils and plant extracts. J. Appl. Microbiol. 2016, 121, 1291–1299. [Google Scholar] [CrossRef]
- Lu, W.; Su, X.; Klein, M.S.; Lewis, I.A.; Fiehn, O.; Rabinowitz, J.D. Metabolite measurement: Pitfalls to avoid and practices to follow. Annu. Rev. Biochem. 2017, 86, 277–304. [Google Scholar] [CrossRef]
- Periwal, V.; Bassler, S.; Andrejev, S.; Gabrielli, N.; Patil, K.R.; Typas, A.; Patil, K.R. Bioactivity assessment of natural compounds using machine learning models trained on target similarity between drugs. PLoS Comput. Biol. 2022, 18, e1010029. [Google Scholar] [CrossRef]
- Zhang, R.; Li, X.; Zhang, X.; Qin, H.; Xiao, W. Machine learning approaches for elucidating the biological effects of natural products. Nat. Prod. Rep. 2021, 38, 346–361. [Google Scholar] [CrossRef]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef]
- Mantravadi, P.K.; Kalesh, K.A.; Dobson, R.C.J.; Hudson, A.O.; Parthasarathy, A. The quest for novel antimicrobial compounds: Emerging trends in research, development, and technologies. Antibiotics 2019, 8, 8. [Google Scholar] [CrossRef] [PubMed]


| Type of Assay | Description Detection/Equipment | Activity Under Evaluation | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|---|
| Dye Exclusion | |||||
| Resazurin | Colorimetric/M Determine the viability of cells by measuring the chromogenic reaction concentration of colored compounds in a solution | Metabolic activity. Mitochondrial dehydrogenase and oxidoreductase activity |
|
| [100,101,102] |
| Tetrazolium salts (MTT, XTT, MTS, WST) |
|
| [103,104,105] | ||
| Flow Cytometry | |||||
| FSC vs SSC | FC Variations in Forward Scattered (FSC) light determine volume changes and variations in Side Scatter (SSC) can determine internal composition | Cell size and volume |
|
| [106,107] |
| Viability dyes PI, 7-AAD | Fluorescent/FC Loss of the integrity of the membrane can be determined by using “viability dies” | Cell cycle arrest Apoptosis/Necrosis Evaluation of membrane integrity |
|
| [108,109,110] |
| Annexin V (AnnV) conjugated to fluorochromes (FITC, PE, APC, etc.) | Fluorescent/FC Apoptotic cells show the migration of PS from the inner layer of the plasma membrane towards the outer layer (become exposed) | Translocation of phosphatidylserine (PS) Evaluation of membrane integrity |
|
| [111,112,113] |
| Permeable cationic lipophilic fluorochrome -(Rho 123) and DiOC5(3) | Fluorescent/FC Apoptotic cells show alterations in the synthesis of ATP and the mitochondrial electron transport chain in the mitochondrial membrane | Mitochondrial membrane Potential |
|
| [114,115] |
| Sub-G0/G1 | Fluorescent/M Percentage quantification of cells with fragmented DNA by analyzing the “sub-G0/G1” peak in a DNA histogram | DNA fragmentation (strand breaks) |
|
| [116,117] |
| In Vitro culture | |||||
| Growth Inhibition Assay | Colorimetric/M The candidate drug is added to the culture, and the parasite grows during a determined time. Variables: drug concentration and time collection points | Cell viability/proliferation |
|
| [118] |
| Intracellular Parasite Growth Inhibition Assay | Colorimetric/M Eukaryotic cells are infected with intracellular parasites in vitro. Then, test compounds are added, and the culture grows during a determined time | Cell viability/proliferation inside host |
|
| [119,120] |
| Assay | Description | Activity Under Evaluation | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|---|
| Adult Motility Assay (AMA) |
| Inhibition of adult worm motility, which can indicate mortality or paralysis |
|
| [133,134,136] |
| Larval development test (LDT) |
| Development of L1 to infective L3 larvae |
|
| [137,138] |
| Larval mortality/paralysis test (LMT) |
| Inhibition of L3 larvae motility |
| Motile L3 larvae have to be grown prior testing | [139] |
| Egg hatch test (EHT) or Egg hatch inhibition assay (EHIA) |
| Inhibition of eggs hatching |
| Eggs have to be isolated from infected samples or adult females | [133,138,140,141,142] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzalez-Pastor, R.; Carrera-Pacheco, S.E.; Zúñiga-Miranda, J.; Rodríguez-Pólit, C.; Mayorga-Ramos, A.; Guamán, L.P.; Barba-Ostria, C. Current Landscape of Methods to Evaluate Antimicrobial Activity of Natural Extracts. Molecules 2023, 28, 1068. https://doi.org/10.3390/molecules28031068
Gonzalez-Pastor R, Carrera-Pacheco SE, Zúñiga-Miranda J, Rodríguez-Pólit C, Mayorga-Ramos A, Guamán LP, Barba-Ostria C. Current Landscape of Methods to Evaluate Antimicrobial Activity of Natural Extracts. Molecules. 2023; 28(3):1068. https://doi.org/10.3390/molecules28031068
Chicago/Turabian StyleGonzalez-Pastor, Rebeca, Saskya E. Carrera-Pacheco, Johana Zúñiga-Miranda, Cristina Rodríguez-Pólit, Arianna Mayorga-Ramos, Linda P. Guamán, and Carlos Barba-Ostria. 2023. "Current Landscape of Methods to Evaluate Antimicrobial Activity of Natural Extracts" Molecules 28, no. 3: 1068. https://doi.org/10.3390/molecules28031068
APA StyleGonzalez-Pastor, R., Carrera-Pacheco, S. E., Zúñiga-Miranda, J., Rodríguez-Pólit, C., Mayorga-Ramos, A., Guamán, L. P., & Barba-Ostria, C. (2023). Current Landscape of Methods to Evaluate Antimicrobial Activity of Natural Extracts. Molecules, 28(3), 1068. https://doi.org/10.3390/molecules28031068










