Identification of Unstable Ellagitannin Metabolites in the Leaves of Quercus dentata by Chemical Derivatization
Abstract
:1. Introduction
2. Results and Discussion
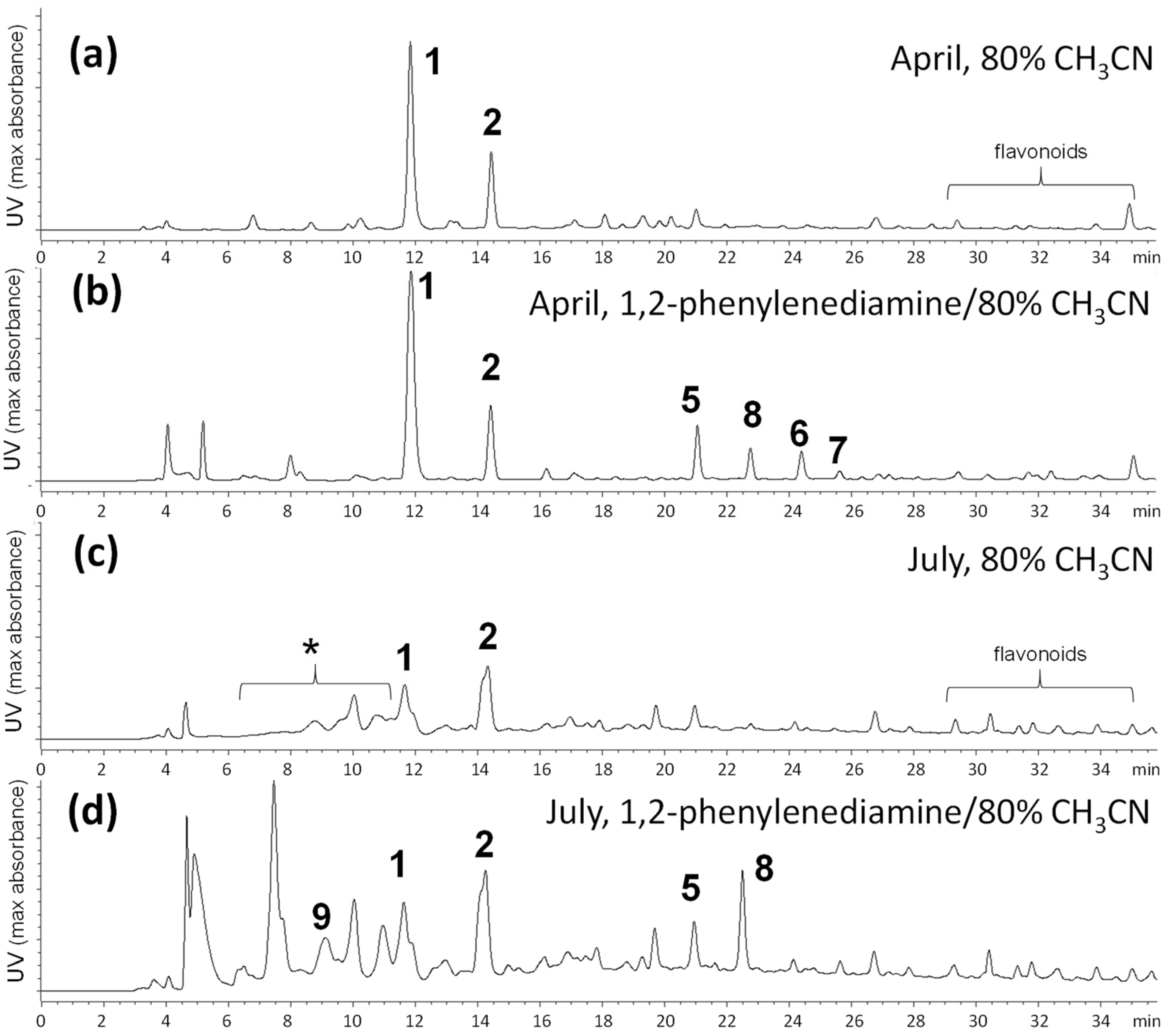
2.1. Comparison of Tannin Compositions in Spring and Summer Leaves
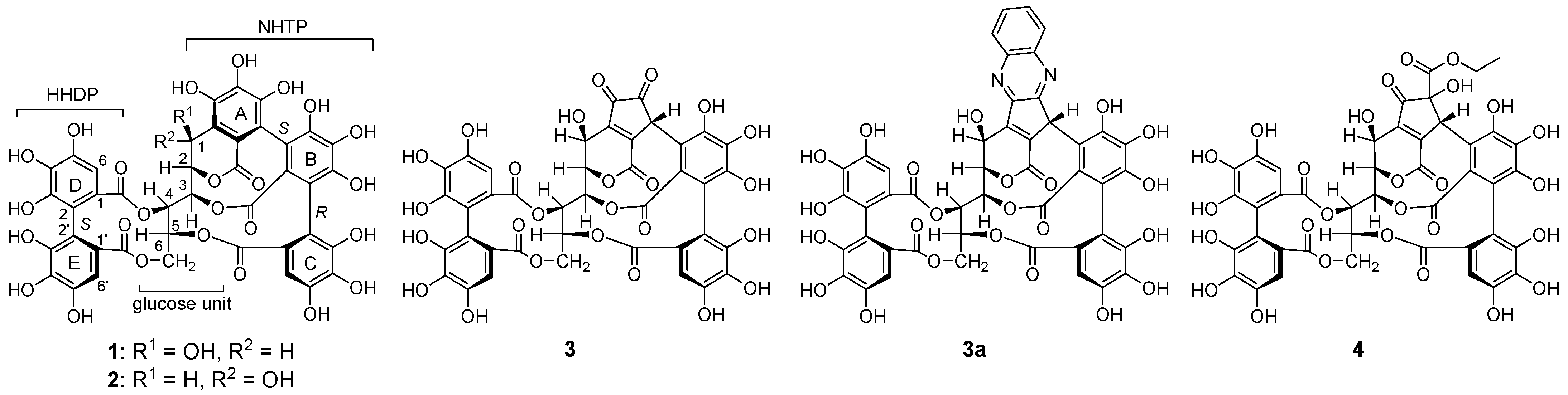
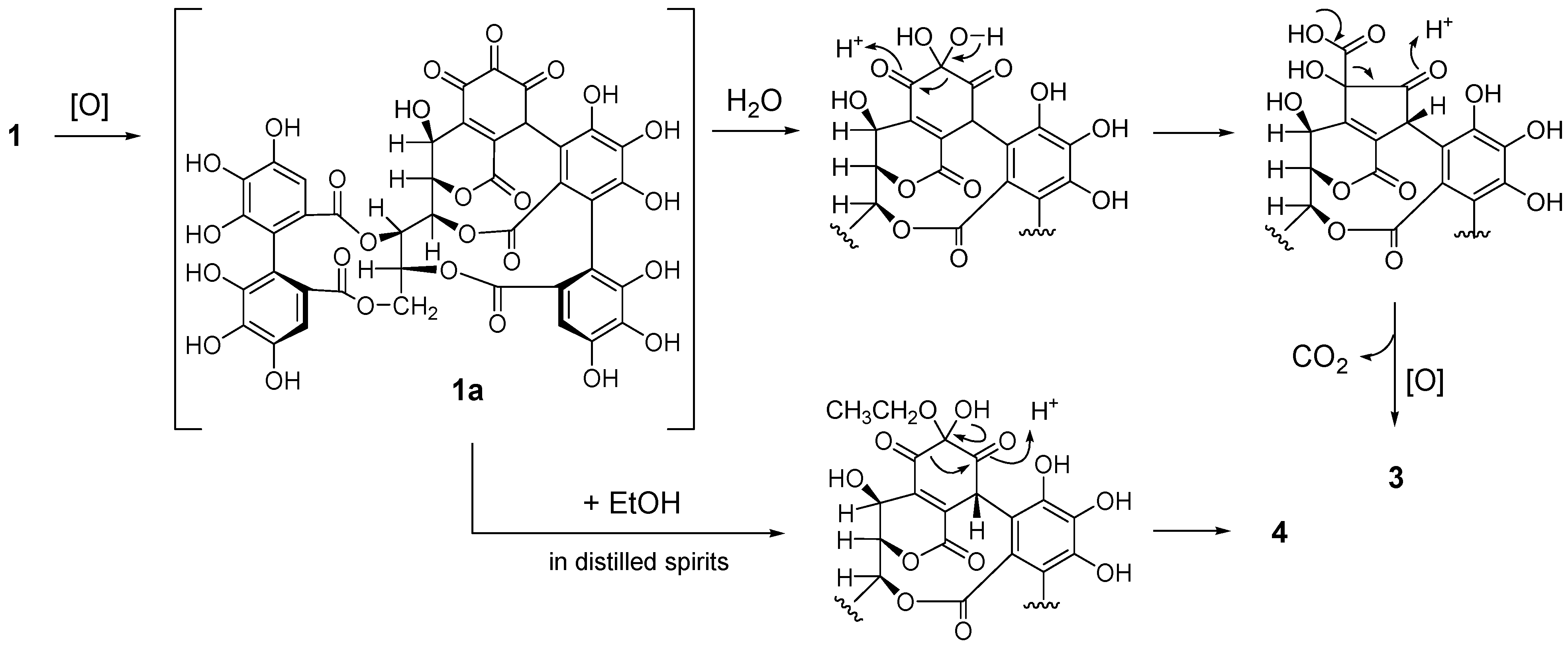
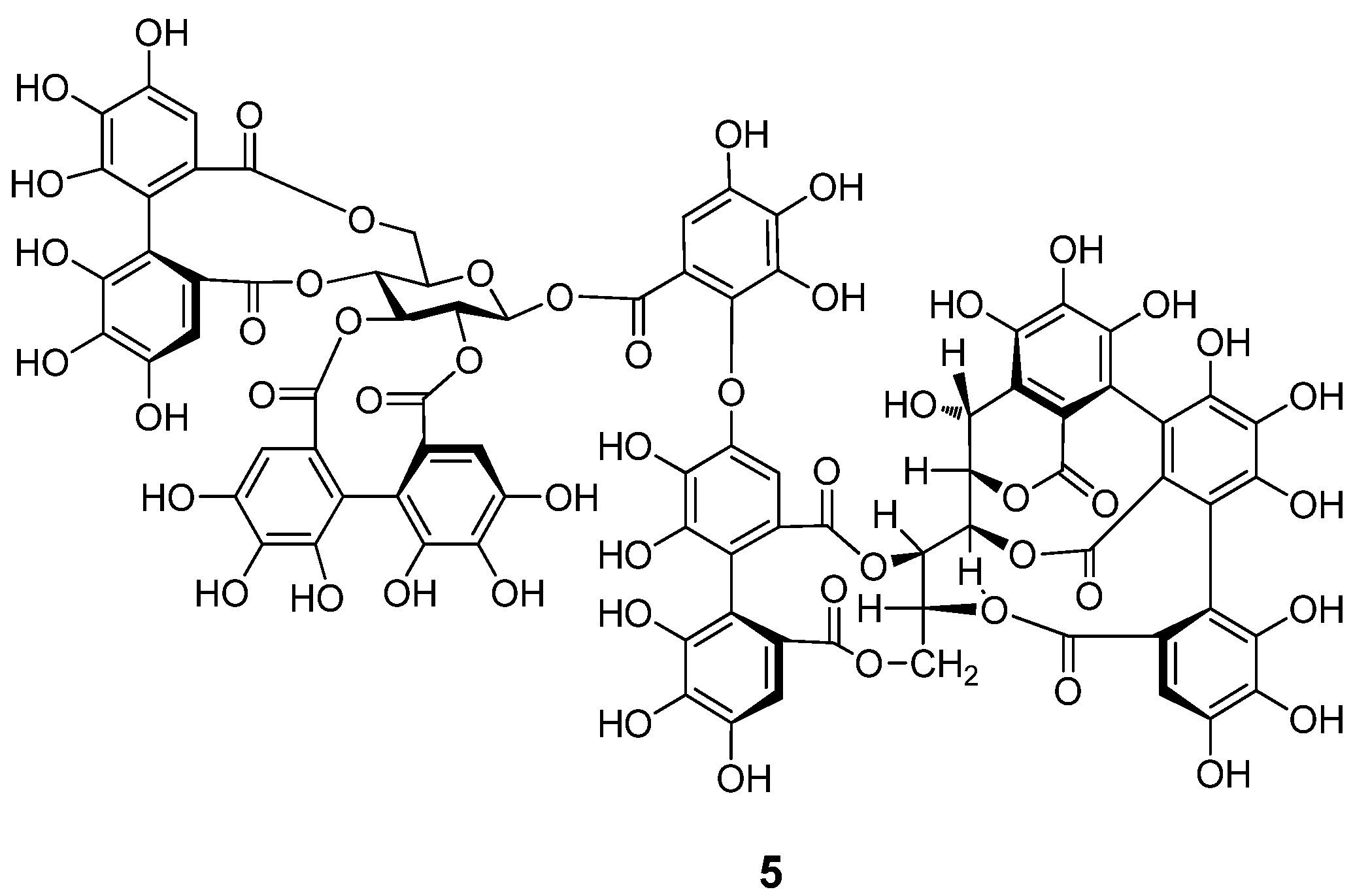
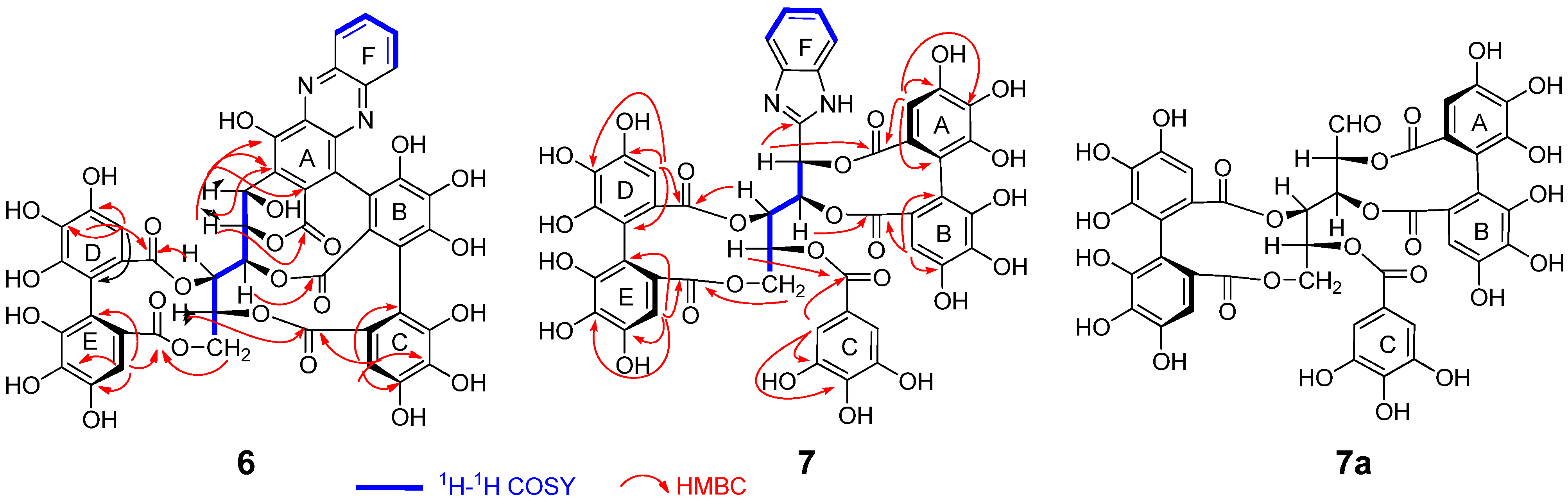
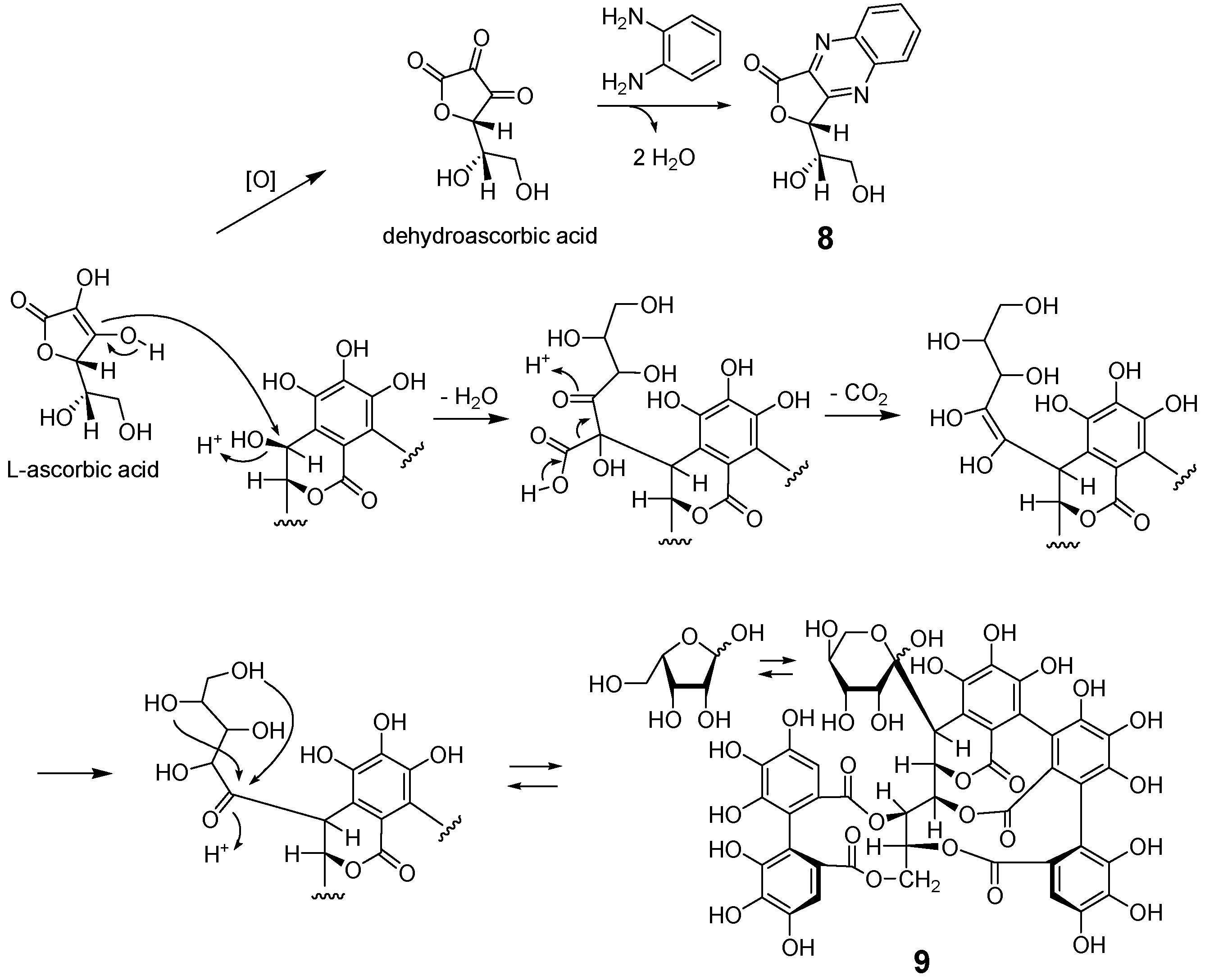
2.2. Separation and Structure Determination
3. Materials and Methods
3.1. General Information
3.2. Plant Material
3.3. HPLC Analysis
3.4. Extraction and Separation
3.4.1. Compound 6
3.4.2. Compound 7
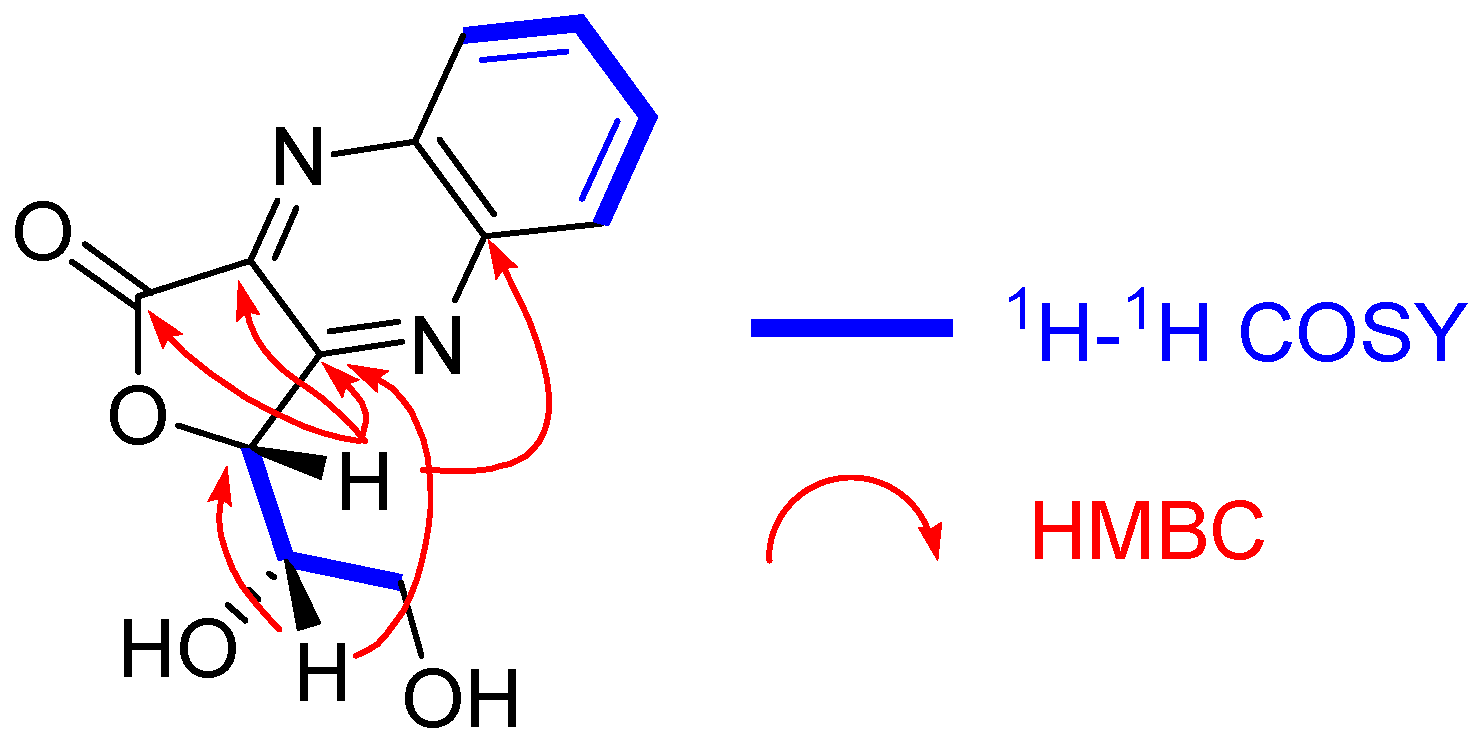
3.4.3. (R)-3-((S)-1,2-Dihydroxyethyl)furo [3,4-b]quinoxaline-1(3H)-one (8)
3.5. Preparation of 8 from L-ascorbic Acid
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Okuda, T.; Yoshida, T.; Hatano, T. Hydrolyzable tannins and related polyphenols. In Progress in the Chemistry of Organic Natural Products; Herz, W., Kirby, G.W., Moore, R.E., Steglich, W., Tamm, C., Eds.; Springer: New York, NY, USA, 1995; Volume 66, pp. 1–117. [Google Scholar]
- Haslam, E.; Cai, Y. Plant polyphenols (vegetable tannins): Gallic acid metabolism. Nat. Prod. Rep. 1994, 11, 41–66. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Hatano, T.; Ito, H.; Okuda, T. Structural diversity and antimicrobial activities of ellagitannins. In Chemistry and Biology of Ellagitannins, an Underestimated Class of Bioactive Plant Polyphenols; Quideau, S., Ed.; World Scientific Publishing: Singapore, 2009; pp. 55–93. [Google Scholar]
- Quideau, S.; Feldman, K.S. Ellagitannin chemistry. Chem. Rev. 1996, 96, 475–504. [Google Scholar] [CrossRef] [PubMed]
- Quideau, S.; Deffieux, D.; Douat-Casassus, C.; Pouységu, L. Plant polyphenols: Chemical properties, biological activities, and synthesis. Angew. Chem. Int. Ed. 2011, 50, 586–621. [Google Scholar] [CrossRef]
- Matsuo, Y.; Wakamatsu, H.; Omar, M.; Tanaka, T. Reinvestigation of the stereochemistry of the C-glycosidic ellagitannins, vescalagin and castalagin. Org. Lett. 2015, 17, 46–49. [Google Scholar] [CrossRef]
- Fernández de Simón, B.; Sanz, M.; Cadahía, E.; Poveda, P.; Broto, M. Chemical characterization of oak heartwood from Spanish forests of Quercus pyrenaica (Wild.). Ellagitannins, low molecular weight phenolic, and volatile compounds. J. Agric. Food Chem. 2006, 54, 8314–8321. [Google Scholar] [CrossRef]
- Fernández de Simón, B.; Cadahía, E.; Conde, E.; García-Vallejo, M.C. Evolution of phenolic compounds of Spanish oak wood during natural seasoning. first results. J. Agric. Food Chem. 1999, 47, 1687–1694. [Google Scholar] [CrossRef]
- Saucier, C.; Jourdes, M.; Glories, Y.; Quideau, S. Extraction, detection, and quantification of flavano-ellagitannins and ethylvescalagin in a Bordeaux red wine aged in oak barrels. J. Agric. Food Chem. 2006, 54, 7349–7354. [Google Scholar] [CrossRef]
- Winstel, D.; Capello, Y.; Quideau, S.; Marchal, A. Isolation of a new taste-active brandy tannin A: Structural elucidation, quantitation and sensory assessment. Food Chem. 2022, 377, 131963. [Google Scholar] [CrossRef]
- Glabasnia, A.; Hofmann, T. Sensory-directed identification of taste-active ellagitannins in American (Quercus alba L.) and European oak wood (Quercus robur L.) and quantitative analysis in bourbon whiskey and oak-matured red wines. J. Agric. Food Chem. 2006, 54, 3380–3390. [Google Scholar] [CrossRef]
- Fujieda, M.; Tanaka, T.; Suwa, Y.; Koshimizu, S.; Kouno, I. Isolation and structure of whiskey polyphenols produced by oxidation of oak wood ellagitannins. J. Agric. Food Chem. 2008, 56, 7305–7310. [Google Scholar] [CrossRef]
- Wakamatsu, H.; Matsuo, Y.; Omar, M.; Saito, Y.; Nishida, K.; Tanaka, T. Oxidation of the oak ellagitannin, vescalagin. J. Nat. Prod. 2020, 83, 413–421. [Google Scholar] [CrossRef]
- Viriot, C.; Scalbert, A.; Hervé du Penhoat, C.L.M.; Moutounet, M. Ellagitannins in woods of sessile oak and sweet chestnut dimerization and hydrolysis during wood ageing. Phytochemistry 1994, 36, 1253–1260. [Google Scholar] [CrossRef]
- Nonaka, G.; Ishimaru, K.; Azuma, R.; Ishimatsu, M.; Nishioka, I. Tannins and related compounds. LXXXV.: Structures of novel C-glycosidic ellagitannins, grandinin and pterocarinins A and B. Chem. Pharm. Bull. 1989, 37, 2071–2077. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, T.; Ueda, N.; Shinohara, H.; Nonaka, G.; Fujioka, T.; Mihashi, K.; Kouno, I. C-Glycosidic ellagitannin metabolites in the heartwood of japanese chestnut tree (Castanea crenata Sieb. et Zucc.). Chem. Pharm. Bull. 1996, 44, 2236–2242. [Google Scholar] [CrossRef] [Green Version]
- Ito, H.; Yamaguchi, K.; Kim, T.-H.; Khennouf, S.; Gharzouli, K.; Yoshida, T. Dimeric and trimeric hydrolyzable tannins from Quercus coccifera and Quercus suber. J. Nat. Prod. 2002, 65, 339–345. [Google Scholar] [CrossRef]
- Yoshizaki, M.; Shingu, T.; Okuda, T.; Hatano, T.; Kaneda, T. Liquidambin, an ellagitannin from Liquidambar formosana. Phytochemistry 1987, 26, 2053–2055. [Google Scholar] [CrossRef]
- Gupta, R.K.; Al-Shafi, S.M.K.; Layden, K.; Haslam, E. The metabolism of gallic acid and hexahydroxydiphenic acid in plants. Part 2. Esters of (S)-hexahydroxydiphenic acid with D-glucopyranose (4C1). J. Chem. Soc., Perkin Trans. 1 1982, 2525–2534. [Google Scholar] [CrossRef]
- Era, M.; Matsuo, Y.; Shii, T.; Saito, Y.; Tanaka, T.; Jiang, Z.-H. Diastereomeric ellagitannin isomers from Penthorum chinense. J. Nat. Prod. 2015, 78, 2104–2109. [Google Scholar] [CrossRef]
- Okuda, T.; Yoshida, T.; Hatano, T.; Koga, T.; Toh, N.; Kuriyama, K. Circular dichroism of hydrolysable tannins-I ellagitannins and gallotannins. Tetrahedron Lett. 1982, 23, 3937–3940. [Google Scholar] [CrossRef]
- Okuda, T.; Yoshida, T.; Hatano, T. Classification of oligomeric hydrolysable tannins and specificity of their occurrence in plants. Phytochemistry 1993, 32, 507–521. [Google Scholar] [CrossRef]
- Esteban, M.R.; Ho, C.-N. Enzymatic spectrophotometric determination of ascorbic acid in commercial vitamin C tablets. Microchem. J. 1997, 56, 122–129. [Google Scholar] [CrossRef]
| Position | δH | (J in Hz) | δc | Position | δH | (J in Hz) | δc | ||
|---|---|---|---|---|---|---|---|---|---|
| Glc | 1 | 5.35 | d (2.1) | 64.6 | D/E | 1/1′ | 124.4 a/124.2 a | ||
| 2 | 5.47 | br. s | 77.2 | 2/2′ | 115.2/114.2 | ||||
| 3 | 4.63 | br. d (7.2) | 67.4 | 3/3′ | 144.1 c/143.9 c | ||||
| 4 | 5.23 | t (7.2) | 68.4 | 4/4′ | 135.9/135.0 | ||||
| 5 | 5.72 | br. d (7.4) | 70.2 | 5/5′ | 144.6/144.5 | ||||
| 6 | 5.12 | dd (13.0, 2.4) | 64.8 | 6/6′ | 6.88/6.61 | s/s | 107.7/106.6 | ||
| 4.04 | d (13.0) | COO | 165.5/168.2 | ||||||
| A/B | 1/1′ | 135.1/127.0 a | F | 1 | 143.7 | ||||
| 2/2′ | 116.1/115.6 b | 2 | 141.6 | ||||||
| 3/3′ | 151.7/144.3 c | 3 | 8.22 | d (7.2) | 129.2 | ||||
| 4/4′ | 135.2 e/134.1 e | 4 | 7.93 | m | 131.8 | ||||
| 5/5′ | 143.7/144.6 c | 5 | 7.89 | m | 131.6 | ||||
| 6/6′ | 126.5 a/111.9 b | 6 | 8.11 | dd (8.5) | 129.6 | ||||
| COO | 163.4/164.8 | ||||||||
| C | 1 | 126.1 a | |||||||
| 2 | 113.5 | ||||||||
| 3 | 143.8 c | ||||||||
| 4 | 135.6 | ||||||||
| 5 | 144.4 | ||||||||
| 6 | 6.76 | s | 107.8 | ||||||
| COO | 166 | ||||||||
| Position | δH | (J in Hz) | δc | Position | δH | (J in Hz) | δc | ||
|---|---|---|---|---|---|---|---|---|---|
| Glc | 1 | 146.7 | D/E | 1/1′ | 125.2 a/124.1 a | ||||
| 2 | 6.35 | br. d (9.7) | 68.7 | 2/2′ | 115.3/114.1 | ||||
| 3 | 6.29 | dd (9.7, 1.3) | 74.1 | 3/3′ | 143.6 b/143.5 b | ||||
| 4 | 5.62 | dd (9.0, 1.3) | 68.8 | 4/4′ | 136.0/134.9 | ||||
| 5 | 5.69 | dd (9.0, 3.8) | 69.7 | 5/5′ | 144.2/144.2 | ||||
| 6 | 4.98 | dd (13.2,3.8) | 63.7 | 6/6′ | 6.73/6.44 | s/s | 107.3/106.5 | ||
| 4.07 | br. d (13.2) | COO | 167.0/166.9 | ||||||
| A/B | 1/1′ | 126.0 a/125.9 a | F | 1 | 137.4 | ||||
| 2/2′ | 113.2/112.8 | 2 | 137.4 | ||||||
| 3/3′ | 143.6 b/143.8 b | 3 | 7.29 | m | 123.4 | ||||
| 4/4′ | 135.3/135.4 | 4 | 7.69 | dd (6.0, 3.2) | 115.7 | ||||
| 5/5′ | 144.3/144.6 | 5 | 7.69 | dd (6.0, 3.2) | 115.7 | ||||
| 6/6′ | 6.40/6.84 | s/s | 106.8/106.7 | 6 | 7.29 | m | 123.4 | ||
| COO | 167.9/167.5 | ||||||||
| C | 1 | 119.6 | |||||||
| 2, 6 | 7.27 | s | 109.7 | ||||||
| 3, 5 | 145.1 | ||||||||
| 4 | 138.5 | ||||||||
| COO | 165.1 | ||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.-B.; Matsuo, Y.; Saito, Y.; Huang, Y.-L.; Li, D.-P.; Tanaka, T. Identification of Unstable Ellagitannin Metabolites in the Leaves of Quercus dentata by Chemical Derivatization. Molecules 2023, 28, 1246. https://doi.org/10.3390/molecules28031246
Liu Z-B, Matsuo Y, Saito Y, Huang Y-L, Li D-P, Tanaka T. Identification of Unstable Ellagitannin Metabolites in the Leaves of Quercus dentata by Chemical Derivatization. Molecules. 2023; 28(3):1246. https://doi.org/10.3390/molecules28031246
Chicago/Turabian StyleLiu, Zhang-Bin, Yosuke Matsuo, Yoshinori Saito, Yong-Lin Huang, Dian-Peng Li, and Takashi Tanaka. 2023. "Identification of Unstable Ellagitannin Metabolites in the Leaves of Quercus dentata by Chemical Derivatization" Molecules 28, no. 3: 1246. https://doi.org/10.3390/molecules28031246






