Virtual Screening of a Library of Naturally Occurring Anthraquinones for Potential Anti-Fouling Agents
Abstract
1. Introduction
2. Results and Discussion
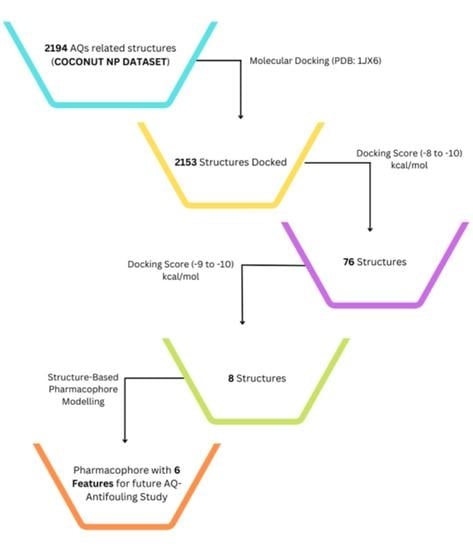
2.1. Virtual Screening by Molecular Docking Analysis Using a Natural Products Dataset
2.2. Pharmacophore Evaluation (Structure-Based Pharmacophore)
3. Materials and Methods
3.1. Origin of Compounds (for Future Anthraquinone Anti-Fouling Study)
3.2. 3D Pharmacophore Model Generation (Structure-Based)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Locatelli, M. Anthraquinones: Analytical techniques as a novel tool to investigate on the triggering of biological targets. Curr. Drug Targets 2011, 12, 366–380. [Google Scholar] [CrossRef] [PubMed]
- Stompor-Gorący, M. The health benefits of emodin, a natural anthraquinone derived from rhubarb—A summary update. Int. J. Mol. Sci. 2021, 22, 9522. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Munoz, G.; Miranda, I.L.; Sartori, S.K.; de Rezende, D.C.; Diaz, M.A. Chapter 11: Anthraquinones—An overview. Stud. Nat. Prod. Chem. 2018, 58, 313–338. [Google Scholar] [CrossRef]
- Malik, E.M.; Müller, C.E. Anthraquinones as pharmacological tools and drugs. MedResRev 2016, 36, 705–748. [Google Scholar] [CrossRef]
- Wuthi-udomlert, M.; Kupittayanant, P.; Gritsanapan, W. In vitro evaluation of antifungal activity of anthraquinone derivatives of Senna alata. J. Health Res. 2010, 24, 117–122. Available online: https://he01.tci-thaijo.org/index.php/jhealthres/article/view/156816 (accessed on 9 September 2022).
- Malmir, M.; Serrano, R.; Silva, O. Anthraquinones as potential antimicrobial agents—A review. In Antimicrobial Research: Novel Bio knowledge and Educational Programs; Mendez-Vilas, A., Ed.; Formatex Research Center S.L.: Badajoz, Spain, 2017; pp. 55–61. [Google Scholar]
- Osman, C.P.; Ismail, N.H. Antiplasmodial anthraquinones from medicinal plants: The chemistry and possible mode of actions. Nat. Prod. Commun. 2018, 13, 1934578X1801301207. [Google Scholar] [CrossRef]
- Chien, S.C.; Wu, Y.C.; Chen, Z.W.; Yang, W.C. Naturally occurring anthraquinones: Chemistry and therapeutic potential in autoimmune diabetes. Evid.-Based Complement Altern. Med. 2015, 2015, 357357. [Google Scholar] [CrossRef]
- Kshirsagar, A.D.; Panchal, P.V.; Harle, U.N.; Nanda, R.K.; Shaikh, H.M. Anti-inflammatory and antiarthritic activity of anthraquinone derivatives in rodents. Int. J. Inflam. 2014, 2014, 690596. [Google Scholar] [CrossRef]
- Wu, C.-M.; Wu, S.-C.; Chung, W.-J.; Lin, H.-C.; Chen, K.-T.; Chen, Y.-C.; Hsu, M.-F.; Yang, J.-M.; Wang, J.-P.; Lin, C.-N. Antiplatelet effect and selective binding to cyclooxygenase (COX) by molecular docking analysis of flavonoids and lignans. Int. J. Mol. Sci. 2007, 8, 830–841. [Google Scholar] [CrossRef]
- Seo, E.J.; Ngoc, T.M.; Lee, S.M.; Kim, Y.S.; Jung, Y.S. Chrysophanol-8O-glucoside, an anthraquinone derivative in rhubarb, has antiplatelet and anticoagulant activities. J. Pharmacol. Sci. 2012, 118, 245–254. [Google Scholar] [CrossRef]
- Jackson, T.C.; Verrier, J.D.; Kochanek, P.M. Anthraquinone-2-sulfonic acid (Aq2s) is a novel neurotherapeutic agent. Cell Death Dis. 2013, 4, e451. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Wang, C.; Li, D.; Hou, H. Novel anthraquinone compounds as anticancer agents and their potential mechanism. Future Med. Chem. 2020, 12, 627–644. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, G.; Babykutty, S.; Sathiadevan, P.P.; Srinivas, P. Molecular mechanism of emodin action: Transition from laxative ingredient to an antitumor agent. Med. Res. Rev. 2007, 27, 591–608. [Google Scholar] [CrossRef]
- Huang, Q.; Lu, G.; Shen, H.M.; Chung, M.C.; Ong, C.N. Anti-cancer properties of anthraquinones from rhubarb. Med. Res. Rev. 2007, 27, 609–630. [Google Scholar] [CrossRef] [PubMed]
- Malik, E.M.; Baqi, Y.; Müller, C.E. Syntheses of 2-substituted 1-amino-4-bromoanthraquinones (bromaminic acid analogues)—precursors for dyes and drugs. Beilstein J. Org. Chem. 2015, 11, 2326–2333. [Google Scholar] [CrossRef]
- Caro, Y.; Anamale, L.; Fouillaud, M.; Laurent, P.; Petit, T.; Dufosse, L. Natural hydroxyanthraquinoid pigments as potent food grade colorants: An overview. Nat. Prod. Bioprospect. 2012, 2, 174–193. [Google Scholar] [CrossRef]
- Dufossé, L. Anthraquinones, the Dr Jekyll and Mr Hyde of the food pigment family. Food Res. Int. 2014, 65, 132–136. [Google Scholar] [CrossRef]
- Furuta, T.; Hirayama, Y.; Iwamura, M. Anthraquinon-2-ylmethoxycarbonyl (Aqmoc): A new photochemically removable protecting group for alcohols. Org. Lett. 2001, 3, 1809–1812. [Google Scholar] [CrossRef]
- Patnaik, S.; Swami, A.; Sethi, D.; Pathak, A.; Garg, B.S.; Gupta, K.C.; Kumar, P. N-(Iodoacetyl)-N′-(anthraquinon-2-oyl)-ethylenediamine (IAED): A new heterobifunctional reagent for the preparation of biochips. Bioconjug. Chem. 2007, 18, 8–12. [Google Scholar] [CrossRef]
- Ballinger, K. Use of Quinone Compound in Building Materials. WO2015116668A1, 6 August 2015. [Google Scholar]
- Najmi, A.; Javed, S.A.; Al Bratty, M.; Alhazmi, H.A. Modern Approaches in the Discovery and Development of Plant-Based Natural Products and Their Analogues as Potential Therapeutic Agents. Molecules 2022, 27, 349. [Google Scholar] [CrossRef]
- Shanghai Min Xuan Steel Structure Work Co., Ltd. Anti-Fouling Anticorrosion Coating for Surfaces of Steel Structures. CN104592861A, 28 September 2016. [Google Scholar]
- No 750 Test Field of China Shipbuilding Industry Corp. Equipment and Method for Long-Acting Environment-Friendly Inhibition of Marine Organism Adhesion of Underwater Structure. CN111074871A, 28 April 2020. [Google Scholar]
- Li, Y.; Ning, C. Latest research progress of marine microbiological corrosion and bio-fouling, and new approaches of marine anti-corrosion and anti-fouling. Bioact. Mater. 2019, 4, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Banderas, J. Marine natural products: A promising source of environmentally friendly anti-fouling agents for the maritime industries. Front. Mar. Sci. 2022, 9, 858757. [Google Scholar] [CrossRef]
- Schultz, M.P.; Bendick, J.A.; Holm, E.R.; Hertel, W.M. Economic impact of biofouling on a naval surface ship. Biofouling 2011, 27, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Molnar, J.L.; Gamboa, R.L.; Revenga, C.; Spalding, M.D. Assessing the global threat of invasive species to marine biodiversity. Fron. Ecol. Environ. 2008, 6, 485–492. [Google Scholar] [CrossRef]
- Dobretsov, S.; Rittschof, D. Love at first taste: Induction of larval settlement by marine microbes. Int. J. Mol. Sci. 2020, 21, 731. [Google Scholar] [CrossRef]
- Liu, L.L.; Wu, C.H.; Qian, P.Y. Marine natural products as anti-fouling molecules—A mini-review (2014–2020). Biofouling 2020, 36, 1210–1226. [Google Scholar] [CrossRef]
- Wang, K.L.; Dou, Z.R.; Gong, G.F.; Li, H.F.; Jiang, B.; Xu, Y. Anti-Larval and Anti-Algal Natural Products from Marine Microorganisms as Sources of Anti-Biofilm Agents. Mar. Drugs 2022, 20, 90. [Google Scholar] [CrossRef]
- Xu, Y.; He, H.; Schulz, S.; Liu, X.; Fusetani, N.; Xiong, H.; Xiao, X.; Qian, P.Y. Potent anti-fouling compounds produced by marine Streptomyces. Bioresour. Technol. 2010, 101, 1331–1336. [Google Scholar] [CrossRef]
- Piazza, V.; Roussis, V.; Garaventa, F.; Greco, G.; Smyrniotopoulos, V.; Vagias, C.; Faimali, M. Terpenes from the red alga Sphaerococcus coronopifolius inhibit the settlement of barnacles. Mar. Biotechnol. 2011, 13, 764–772. [Google Scholar] [CrossRef]
- Umezawa, T.; Oguri, Y.; Matsuura, H.; Yamazaki, S.; Suzuki, M.; Yoshimura, E.; Furuta, T.; Nogata, Y.; Serisawa, Y.; Matsuyama-Serisawa, K.; et al. Omaezallene from red alga Laurencia sp.: Structure elucidation, total synthesis, and anti-fouling activity. Angew. Chem. 2014, 53, 3909–3912. [Google Scholar] [CrossRef]
- Dahms, H.U.; Dobretsov, S. Anti-fouling compounds from marine macroalgae. Mar. Drugs 2017, 15, 265. [Google Scholar] [CrossRef]
- Hanssen, K.Ø.; Cervin, G.; Trepos, R.; Petitbois, J.; Haug, T.; Hansen, E.; Andersen, J.H.; Pavia, H.; Hellio, C.; Svenson, J. The bromotyrosine derivative ianthelline isolated from the arctic marine sponge Stryphnus fortis inhibits marine micro-and macrobiofouling. Mar. Biotechnol. 2014, 16, 684–694. [Google Scholar] [CrossRef] [PubMed]
- Ortlepp, S.; Sjögren, M.; Dahlström, M.; Weber, H.; Ebel, R.; Edrada, R.; Thoms, C.; Schupp, P.; Bohlin, L.; Proksch, P. Anti-fouling activity of bromotyrosine-derived sponge metabolites and synthetic analogues. Mar. Biotechnol. 2007, 9, 776–785. [Google Scholar] [CrossRef]
- Tintillier, F.; Moriou, C.; Petek, S.; Fauchon, M.; Hellio, C.; Saulnier, D.; Ekins, M.; Hooper, J.N.; Al-Mourabit, A.; Debitus, C. Quorum sensing inhibitory and anti-fouling activities of new bromotyrosine metabolites from the Polynesian sponge Pseudoceratina n. sp. Mar. Drugs 2020, 18, 272. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Gao, Y.; Huang, W.; Shao, Z.; Shi, J.; Liu, Z. A novel bioassay for high-throughput screening microorganisms with N-acyl homoserine lactone degradint activity. Appl. Biochem. Biotechnol. 2012, 167, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Vyshnava, S.S.; Kander, D.K.; Panjala, S.P.; Pandian, K.; Bontha, R.R.; Goukanapalle, P.K.; Banganapalli, B. Effect of silver nanoparticles against the formation of biofilm by Pseudomonas aeruginosa an in-silico approach. Appl. Biochem. Biotechnol. 2016, 180, 426–437. [Google Scholar] [CrossRef]
- Federle, M.J. Autoinducer-2-based chemical communication in bacteria: Complexities of interspecies signalling. Contrib. Microbiol. 2009, 16, 18–32. [Google Scholar]
- Atkinson, S.; Williams, P. Quorum sensing and social networking in the microbial world. J. R. Soc. Interface 2009, 6, 959–978. [Google Scholar] [CrossRef]
- Moran-Zorzano, M.T.; Montero, M.; Munoz, F.J.; Alonso-Casajus, N.; Viale, A.M.; Eydallin, G.; Sesma, M.T.; Baroja-Fernandez, E.; Pozueta-Romero, J. Cytoplasmic Escherichia coli ADP sugar pyrophosphatase binds to cell membranes in response to extracellular signals as the cell population density increases. FEMS Microbiol. Lett. 2008, 288, 25–32. [Google Scholar] [CrossRef]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef]
- Sifri, C.D. Quorum sensing: Bacteria talk sense. Healthc. Epidemiol. 2008, 47, 1070–1076. [Google Scholar]
- Roux, A.; Payne, S.M.; Gilmore, M.S. Microbial telesensing: Probing the environment for friends, foes, and food. Cell Host Microbe 2009, 6, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Bassler, B.L. Small talk: Cell to cell communication in bacteria. Cell 2002, 109, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Schauder, S.; Shokat, K.; Surette, M.G.; Bassler, B.L. The LuxS family of bacterial autoinducers: Biosynthesis of a novel quorum sensing signal molecule. Mol. Microbiol. 2001, 41, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Neiditch, M.B.; Federie, M.J.; Miller, S.T.; Bassler, B.L.; Hughson, F.M. Regulation of LuxPQ receptor activity by the quorum sensing signal autoinducer-2. Mol. Cell 2005, 18, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.Y.; Nega, M.; Wolfle, M.; Plener, L.; Grond, S.; Jung, K.; Gotz, F. A new class of quorum quenching molecules from Staphylococcus species affects communication and growth of gram-negative bacteria. PLoS Pathog. 2013, 9, e1003654. [Google Scholar] [CrossRef] [PubMed]
- Waters, C.; Bassler, B.L. Quorum sensing: Cell to cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef]
- Neiditch, M.B.; Federle, M.J.; Pompeani, A.J.; Kelly, R.C.; Swem, D.L.; Jeffrey, P.D.; Bassler, B.L.; Hughson, F.M. Ligand induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell 2006, 126, 1095–1108. [Google Scholar] [CrossRef]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef]
- Preet, G.; Gomez-Banderas, J.; Ebel, R.; Jaspars, M. A structure-activity relationship analysis of anthraquinones with anti-fouling activity against marine biofilm-forming bacteria. Front. Nat. Prod. 2022, 1, 990822. [Google Scholar] [CrossRef]
- Sorokina, M.; Merseburger, P.; Rajan, K.; Yirik, M.A.; Steinbeck, C. COCONUT online: Collection of Open Natural Products database. J. Cheminform. 2021, 13, 2. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Modeling 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimisation and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Chemical Computing Group ULC. Molecular Operating Environment (MOE) (2022.02); Chemical Computing Group ULC: Montreal, QC, Canada, 2022. [Google Scholar]
- Edelsbrunner, H.; Liang, J.; Fu, P.; Facello, M. Measuring proteins and voids in proteins. In Proceedings of the 2014 47th Hawaii International Conference on System Sciences, Waikoloa, HI, USA, 6–9 January 2014; p. 256. [Google Scholar] [CrossRef]
- Chen, X.; Schauder, S.; Potier, N.; Van Dorsselaer, A.; Pelczer, I.; Bassler, B.; Hughson, F. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 2002, 415, 545–549. [Google Scholar] [CrossRef]
- SAMSON: Software for Adaptive Modeling and Simulation Of Nanosystems. Available online: https://www.samson-connect.net (accessed on 26 August 2022).
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera- a visualisation system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Wolber, G.; Thierry, L. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J. Chem. Inf. Model 2005, 45, 160–169. [Google Scholar] [CrossRef]















| Residues | 465D | 302D | 52D | 310D | 100D | 298D | 950D | 132D |
|---|---|---|---|---|---|---|---|---|
| Ala239A | X | X | X | XX | ||||
| Arg215A | X | X | X | |||||
| Arg310A | X | X | X | X | ||||
| Asn159A | X | X | ||||||
| Asp136A | X | X | ||||||
| Asp267A | X | X | ||||||
| Gln77A | X | |||||||
| Gly288A | X | |||||||
| His180A | X | |||||||
| Ile211A | X | X | X | X | X | X | ||
| Phe178A | X | X | ||||||
| Phe206A | X | X | X | X | X | X | X | |
| Pro74A | X | |||||||
| Pro109A | XX | |||||||
| Ser79A | X | X | ||||||
| Ser265A | X | |||||||
| Thr134A | X | X | XX | X | X | |||
| Thr266A | X | X | X | X | X | X | ||
| Trp82A | XX | XX | X | X | ||||
| Trp289A | X | X | X | |||||
| Tyr81A | X | X | X | X | ||||
| Tyr210A | X | |||||||
| Val78A | X | |||||||
| Val268A | XX |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Preet, G.; Astakala, R.V.; Gomez-Banderas, J.; Rajakulendran, J.E.; Hasan, A.H.; Ebel, R.; Jaspars, M. Virtual Screening of a Library of Naturally Occurring Anthraquinones for Potential Anti-Fouling Agents. Molecules 2023, 28, 995. https://doi.org/10.3390/molecules28030995
Preet G, Astakala RV, Gomez-Banderas J, Rajakulendran JE, Hasan AH, Ebel R, Jaspars M. Virtual Screening of a Library of Naturally Occurring Anthraquinones for Potential Anti-Fouling Agents. Molecules. 2023; 28(3):995. https://doi.org/10.3390/molecules28030995
Chicago/Turabian StylePreet, Gagan, Rishi Vachaspathy Astakala, Jessica Gomez-Banderas, Joy Ebenezer Rajakulendran, Ahlam Haj Hasan, Rainer Ebel, and Marcel Jaspars. 2023. "Virtual Screening of a Library of Naturally Occurring Anthraquinones for Potential Anti-Fouling Agents" Molecules 28, no. 3: 995. https://doi.org/10.3390/molecules28030995
APA StylePreet, G., Astakala, R. V., Gomez-Banderas, J., Rajakulendran, J. E., Hasan, A. H., Ebel, R., & Jaspars, M. (2023). Virtual Screening of a Library of Naturally Occurring Anthraquinones for Potential Anti-Fouling Agents. Molecules, 28(3), 995. https://doi.org/10.3390/molecules28030995