Selection and Identification of an ssDNA Aptamer for Fibroblast Activation Protein
Abstract
:1. Introduction
2. Results
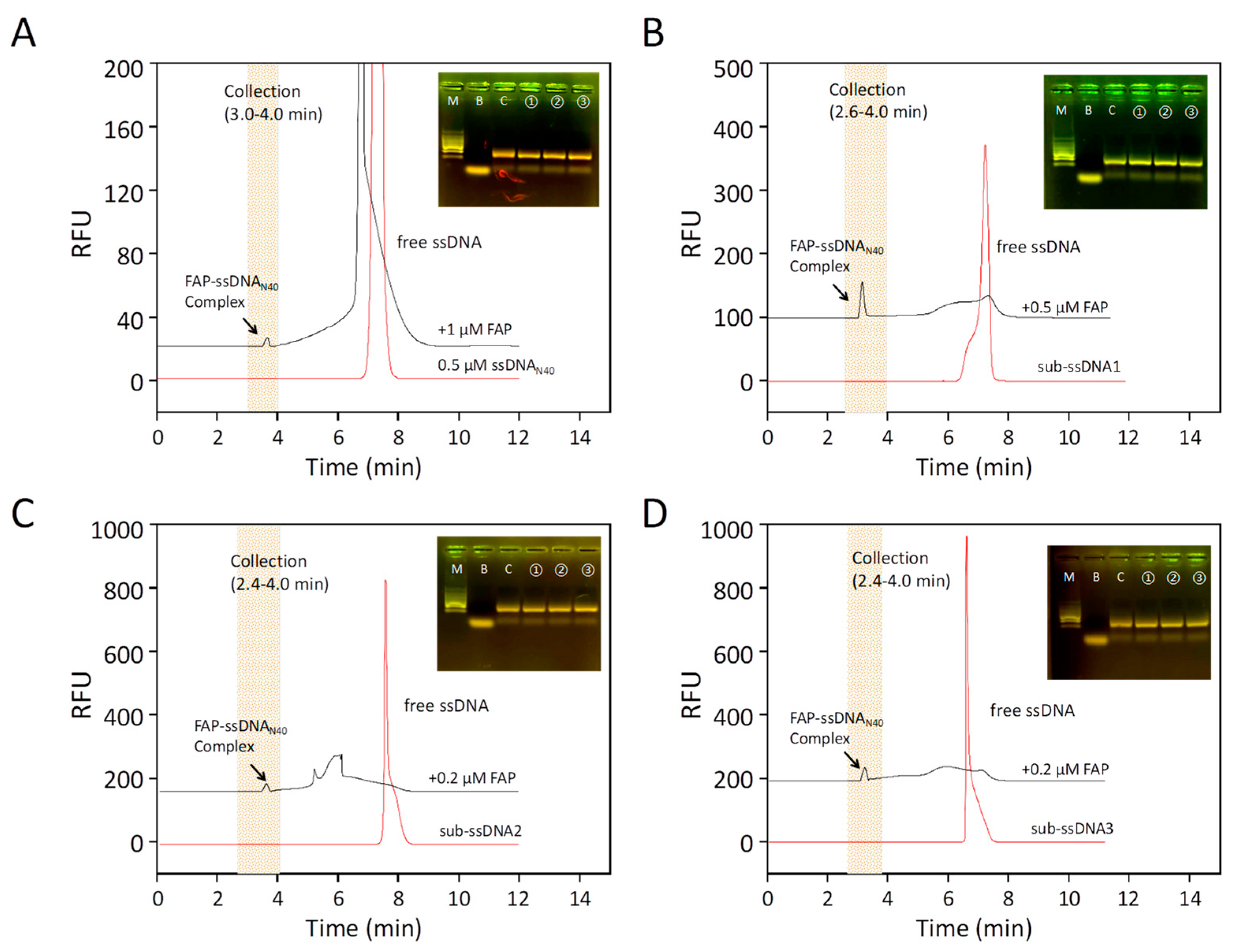
2.1. Aptamer Screening by CE-SELEX
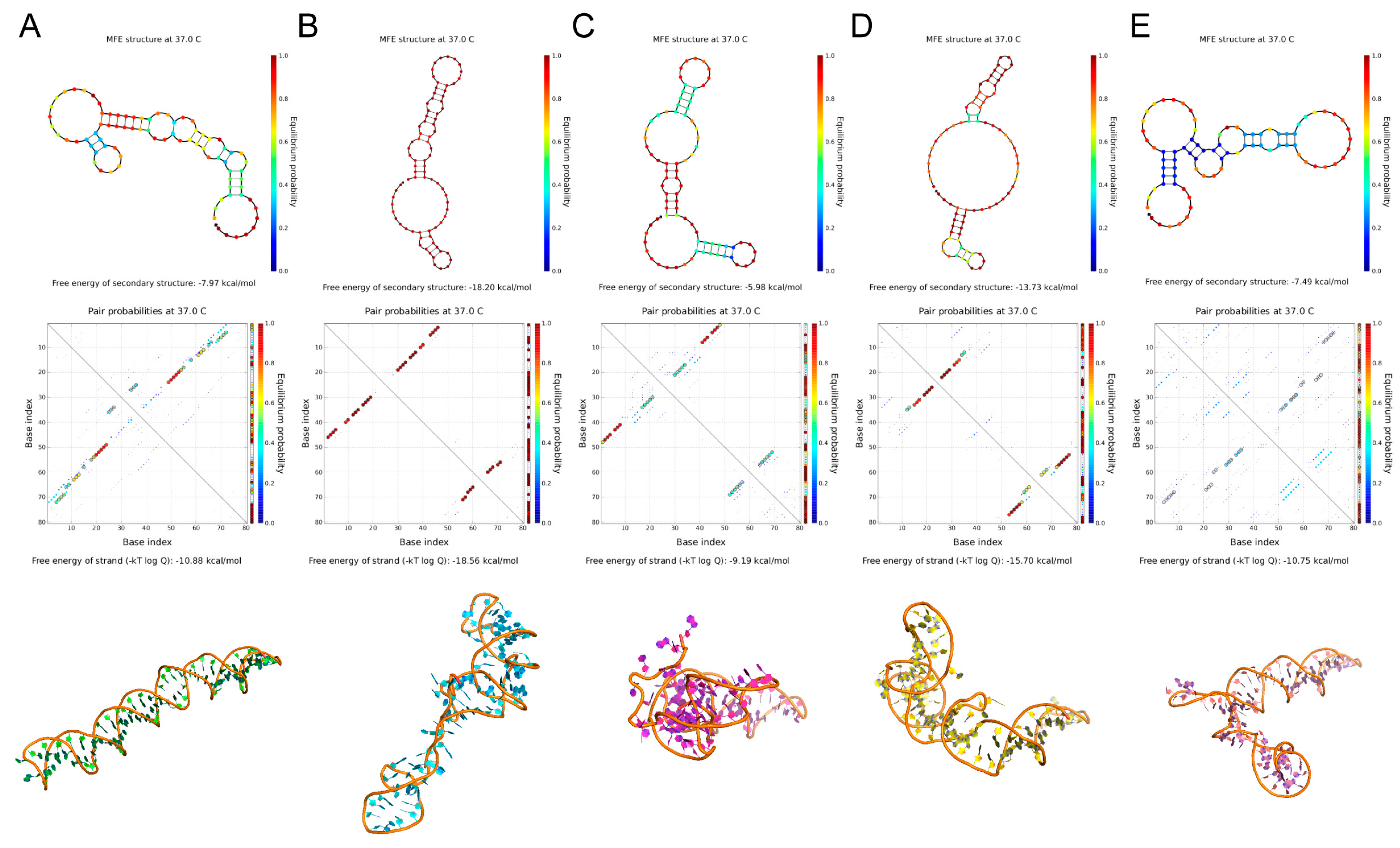
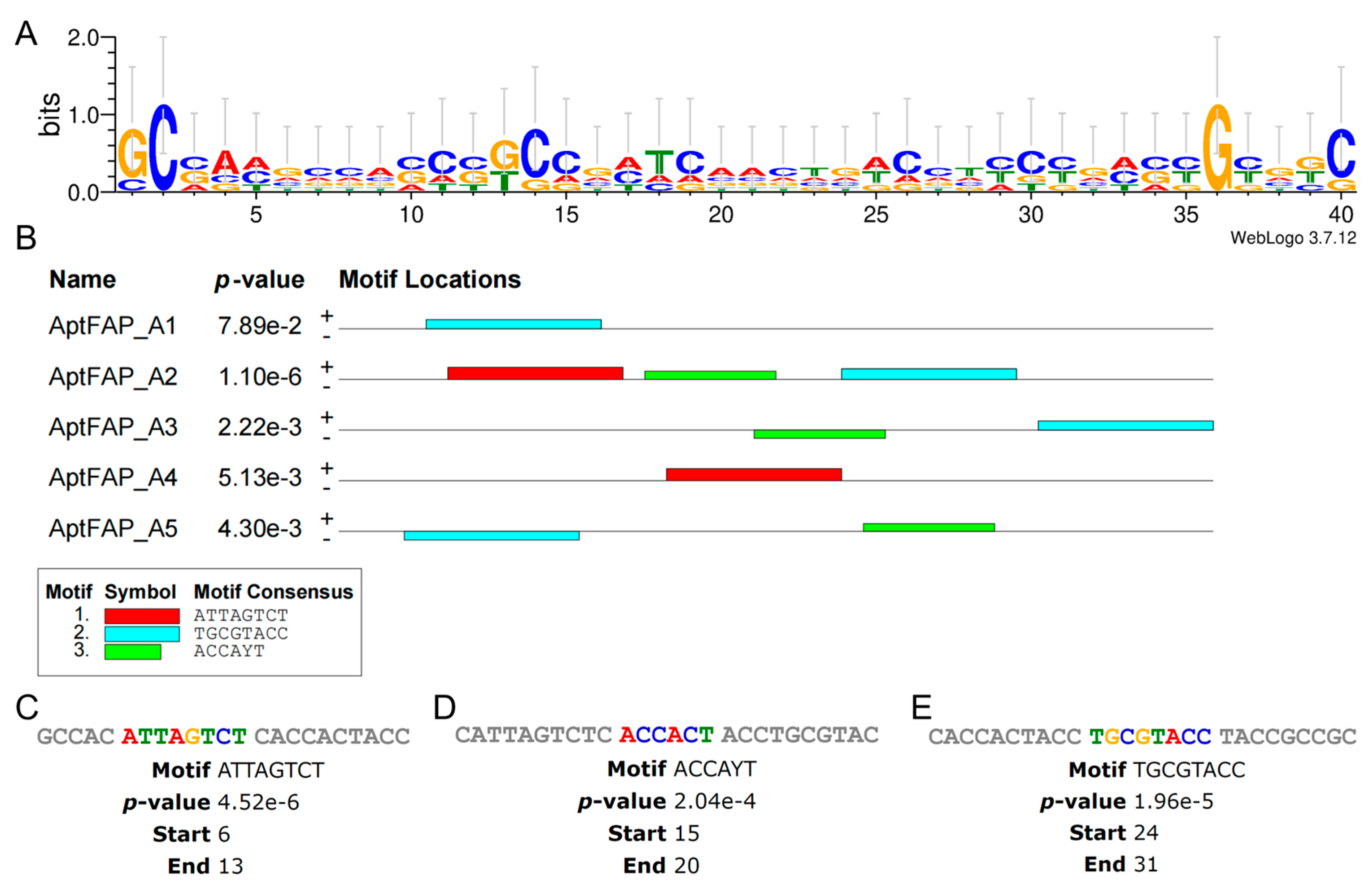
2.2. Predicting Structure and Motif of Candidate Aptamers
2.3. Molecular Docking and Kinetic Analysis
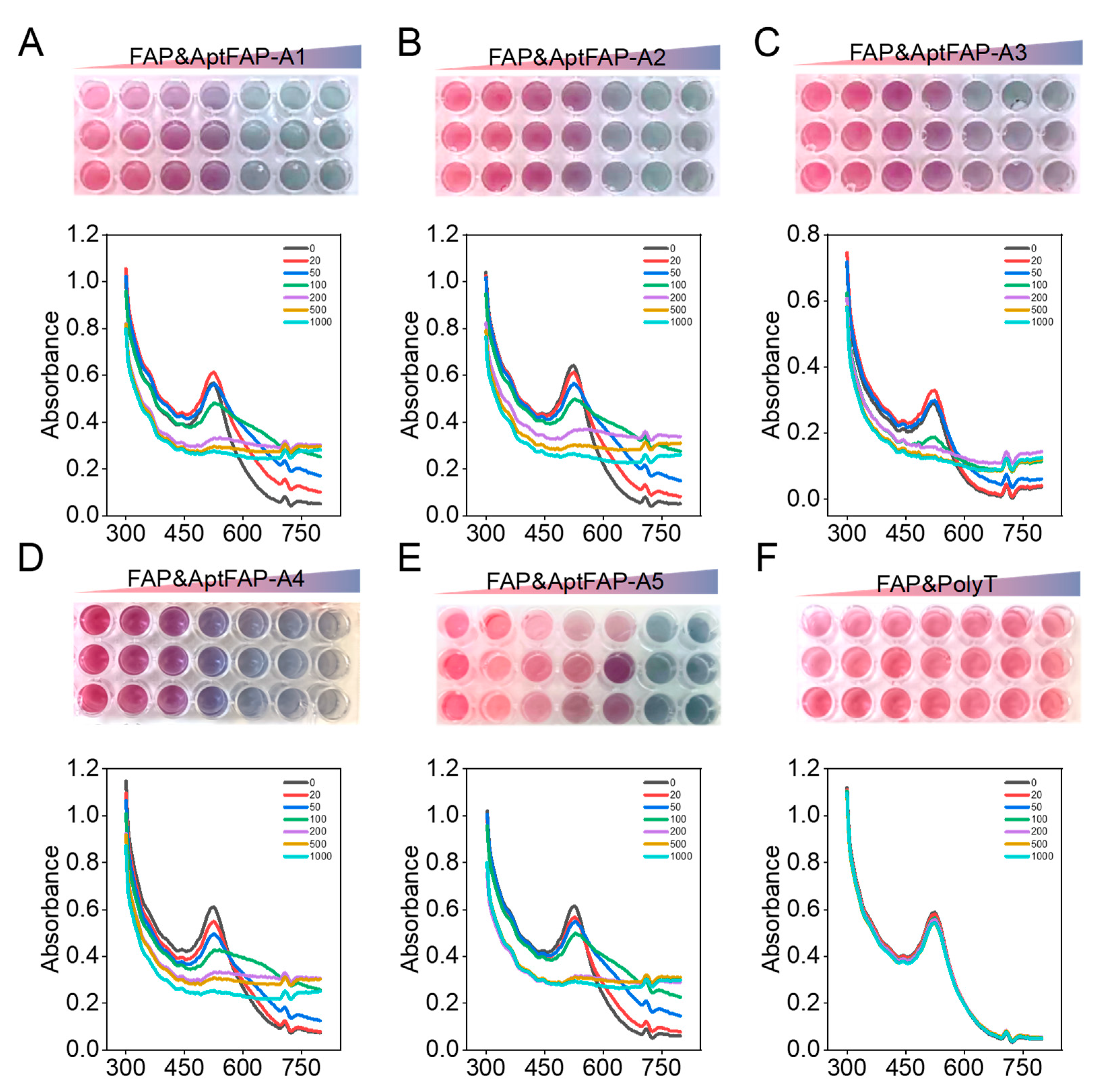
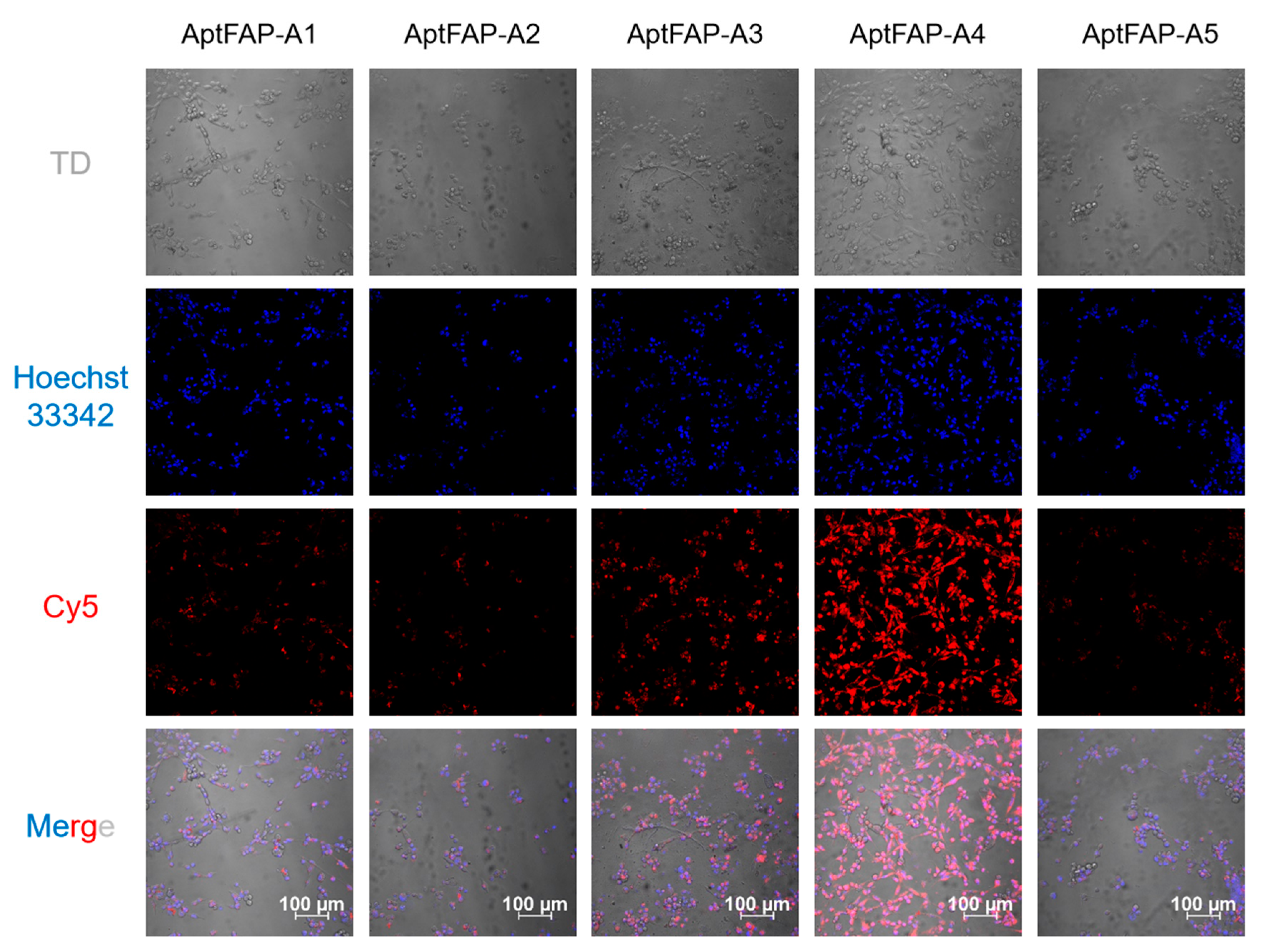
2.4. Affinity Characterization of Candidate Aptamers
2.5. Candidate Aptamers’ Affinity for Target Cells
3. Materials and Methods
3.1. Reagents and Cell Lines
3.2. CE-SELEX Assays
3.3. Cloning and Sequencing
3.4. Structure and Motif Predictions
3.5. Binding Mode Predictions
3.6. Affinity Characterization
3.7. Targeting of Aptamers to Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Rettig, W.J.; Chesa, P.G.; Beresford, H.R.; Feickert, H.J.; Jennings, M.T.; Cohen, J.; Oettgen, H.F.; Old, L.J. Differential expression of cell surface antigens and glial fibrillary acidic protein in human astrocytoma subsets. Cancer Res. 1986, 46 Pt 1, 6406–6412. [Google Scholar]
- Mei, S.; Zhang, Y.; Yu, L.; Chen, G.; Zi, F. Expression and role of fibroblast activation protein α in acute myeloid leukemia. Oncol. Rep. 2020, 45, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.J.; Alpaugh, R.K.; Palazzo, I.; Meropol, N.J.; Rogatko, A.; Xu, Z.; Hoffman, J.P.; Weiner, L.M.; Cheng, J.D. Fibroblast activation protein and its relationship to clinical outcome in pancreatic adenocarcinoma. Pancreas 2008, 37, 154–158. [Google Scholar] [CrossRef]
- Garin-Chesa, P.W.J.; Old, L.J.; Rettig, W.J. Cell surface glycoprotein of reactive stromal fibroblasts as a potential antibody target in human epithelial cancers. Proc. Natl. Acad. Sci. USA 1990, 87, 7235–7239. [Google Scholar] [CrossRef] [PubMed]
- LeBeau, A.M.; Nathaniel Brennen, W.; Aggarwal, S.; Denmeade, S.R. Targeting the cancer stroma with a fibroblast activation protein-activated promelittin protoxin. Mol. Cancer Ther. 2009, 8, 1378–1386. [Google Scholar] [CrossRef]
- Liu, F.; Qi, L.; Liu, B.; Liu, J.; Zhang, H.; Che, D.; Cao, J.; Shen, J.; Geng, J.; Bi, Y.; et al. Fibroblast activation protein overexpression and clinical implications in solid tumors: A meta-analysis. PLoS ONE 2015, 10, e0116683. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Tuveson, D. Diversity and Biology of Cancer-Associated Fibroblasts. Physiol. Rev. 2021, 101, 147–176. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Song, E. Turning foes to friends: Targeting cancer-associated fibroblasts. Nat. Rev. Drug Discov. 2019, 18, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Xu, J.; Wang, W.; Liang, C.; Hua, J.; Liu, J.; Zhang, B.; Meng, Q.; Yu, X.; Shi, S. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: New findings and future perspectives. Molecular cancer. Mol. Cancer 2021, 20, 131. [Google Scholar] [PubMed]
- Zheng, Y.; Han, Y.; Sun, Q.; Li, Z. Harnessing anti-tumor and tumor-tropism functions of macrophages via nanotechnology for tumor immunotherapy. Exploration 2022, 2, 20210166. [Google Scholar] [CrossRef]
- Fabre, M.; Ferrer, C.; Domínguez-Hormaetxe, S.; Bockorny, B.; Murias, L.; Seifert, O.; Eisler, S.A.; Kontermann, R.E.; Pfizenmaier, K.; Lee, S.Y.; et al. OMTX705, a Novel FAP-Targeting ADC Demonstrates Activity in Chemotherapy and Pembrolizumab-Resistant Solid Tumor Models. Clin. Cancer Res. 2020, 26, 3420–3430. [Google Scholar] [CrossRef]
- Sum, E.; Rapp, M.; Fröbel, P.; Clech, M.L.; Dürr, H.; Giusti, A.M.; Perro, M.; Speziale, D.; Kunz, L.; Menietti, E.; et al. Fibroblast activation protein α-targeted CD40 agonism abrogates systemic toxicity and enables administration of high doses to induce effective anti-tumor immunity. Clin. Cancer Res. 2021, 27, 4036–4053. [Google Scholar]
- Adams, S.; Miller, G.T.; Jesson, M.I.; Watanabe, T.; Jones, B.; Wallner, B.P. PT-100, a Small Molecule Dipeptidyl Peptidase Inhibitor, Has Potent Antitumor Effects and Augments Antibody-Mediated Cytotoxicity via a Novel Immune Mechanism. Cancer Res. 2004, 64, 5471–5480. [Google Scholar] [CrossRef]
- Watabe, T.; Liu, Y.; Kaneda-Nakashima, K.; Shirakami, Y.; Lindner, T.; Ooe, K.; Toyoshima, A.; Nagata, K.; Shimosegawa, E.; Haberkorn, U.; et al. Theranostics Targeting Fibroblast Activation Protein in the Tumor Stroma: 64Cu- and 225Ac-Labeled FAPI-04 in Pancreatic Cancer Xenograft Mouse Models. J. Nucl. Med. 2020, 61, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Tsai, T.Y.; Yeh, T.K.; Chen, X.; Hsu, T.; Jao, Y.C.; Huang, C.H.; Song, J.S.; Huang, Y.C.; Chien, C.H.; Chiu, J.H.; et al. Substituted 4-carboxymethylpyroglutamic acid diamides as potent and selective inhibitors of fibroblast activation protein. J. Med. Chem. 2010, 53, 6572–6583. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.; Guo, J.; Zhang, Z.; Zhou, Y.; Hua, Y. Polyphyllin I inhibits gastric cancer cell proliferation by downregulating the expression of fibroblast activation protein alpha (FAP) and hepatocyte growth factor (HGF) in cancer-associated fibroblasts. Biochem. Biophys. Res. Commun. 2018, 497, 1129–1134. [Google Scholar] [CrossRef] [PubMed]
- Altmann, A.; Haberkorn, U.; Siveke, J. The Latest Developments in Imaging of Fibroblast Activation Protein. J. Nucl. Med. 2021, 62, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Muhammad, I.; Zhang, X.; Yang, G.; Ma, Y.; Wei, B.; Qu, F. Magnetic hydrophilic polymer-based apta-sensing probe for sensitive detection of fetuin-A in serum. Sens. Actuators B Chem. 2022, 368, 132152. [Google Scholar] [CrossRef]
- Zhu, G.; Chen, X. Aptamer-based targeted therapy. Adv. Drug Deliv. Rev. 2018, 134, 65–78. [Google Scholar] [CrossRef]
- Qu, H.; Csordas, A.T.; Wang, J.; Oh, S.S.; Eisenstein, M.S.; Soh, H.T. Rapid and Label-Free Strategy to Isolate Aptamers for Metal Ions. ACS Nano. 2016, 10, 7558–7565. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Hu, X.; Zhu, C.; Ma, Q.; Liang, L.; Li, Z.; Yuan, Q. Dual-Aptamer-Conjugated Molecular Modulator for Detecting Bioactive Metal Ions and Inhibiting Metal-Mediated Protein Aggregation. Anal. Chem. 2019, 91, 823–829. [Google Scholar] [CrossRef]
- Razmi, N.; Baradaran, B.; Hejazi, M.; Hasanzadeh, M.; Mosafer, J.; Mokhtarzadeh, A.; de la Guardia, M. Recent advances on aptamer-based biosensors to detection of platelet-derived growth factor. Biosens. Bioelectron. 2018, 113, 58–71. [Google Scholar] [CrossRef]
- Parashar, A.; Bhushan, V.; Mahanandia, N.C.; Kumar, S.; Mohanty, A.K. Non-SELEX method for aptamer selection against β-casomorphin-7 peptide. J. Dairy Sci. 2022, 105, 5545–5560. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Li, Z.; Mohammed, I.; Zhao, L.; Wei, W.; Xiao, H.; Guo, W.; Zhao, Y.; Qu, F.; Huang, Y. Identification of SARS-CoV-2-against aptamer with high neutralization activity by blocking the RBD domain of spike protein 1. Signal Transduct. Target. Ther. 2021, 6, 227. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef] [PubMed]
- Meyer, S.; Maufort, J.P.; Nie, J.; Stewart, R.; McIntosh, B.E.; Conti, L.R.; Ahmad, K.M.; Soh, H.T.; Thomson, J.A. Development of an efficient targeted cell-SELEX procedure for DNA aptamer reagents. PLoS ONE 2013, 8, e71798. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Hu, J.; Lu, F. Aptamers used for biosensors and targeted therapy. Biomed Pharm. 2020, 132, 110902. [Google Scholar] [CrossRef]
- Wen, J.; Tao, W.; Hao, S.; Iyer, S.P.; Zu, Y. A unique aptamer-drug conjugate for targeted therapy of multiple myeloma. Leukemia 2016, 30, 987–991. [Google Scholar] [CrossRef]
- Yang, S.; Wen, J.; Li, H.; Xu, L.; Liu, Y.; Zhao, N.; Zeng, Z.; Qi, J.; Jiang, W.; Han, W.; et al. Aptamer-Engineered Natural Killer Cells for Cell-Specific Adaptive Immunotherapy. Small 2019, 15, e1900903. [Google Scholar] [CrossRef]
- Yu, T.; Zhang, H.; Huang, Z.; Luo, Z.; Huang, N.; Ding, S.; Feng, W. A Simple Electrochemical Aptamer Cytosensor for Direct Detection of Chronic Myelogenous Leukemia K562 Cells. Electroanalysis 2017, 29, 828–834. [Google Scholar] [CrossRef]
- Song, K.M.; Lee, S.; Ban, C. Aptamers and their biological applications. Sensors 2012, 12, 612–631. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C. Methods developed for SELEX. Anal. Bioanal. Chem. 2007, 387, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Dembowski, S.K.; Bowser, M.T. Microfluidic methods for aptamer selection and characterization. Analyst 2017, 143, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Hybarger, G.; Bynum, J.; Williams, R.F.; Valdes, J.J.; Chambers, J.P. A microfluidic SELEX prototype. Anal. Bioanal. Chem. 2006, 384, 191–198. [Google Scholar] [CrossRef]
- Huang, C.J.; Lin, H.I.; Shiesh, S.C.; Lee, G.B. Integrated microfluidic system for rapid screening of CRP aptamers utilizing systematic evolution of ligands by exponential enrichment (SELEX). Biosens. Bioelectron. 2010, 25, 1761–1766. [Google Scholar] [CrossRef]
- Ngubane, N.A.; Gresh, L.; Pym, A.; Rubin, E.J.; Khati, M. Selection of RNA aptamers against the M. tuberculosis EsxG protein using surface plasmon resonance-based SELEX. Biochem. Biophys. Res. Commun. 2014, 449, 114–119. [Google Scholar] [CrossRef]
- Han, X.; Zhang, Y.; Nie, J.; Zhao, S.; Tian, Y.; Zhou, N. Gold nanoparticle based photometric determination of tobramycin by using new specific DNA aptamers. Mikrochim. Acta 2017, 185, 4. [Google Scholar] [CrossRef]
- Gu, H.; Duan, N.; Xia, Y.; Hun, X.; Wang, H.; Wang, Z. Magnetic Separation-Based Multiple SELEX for Effectively Selecting Aptamers against Saxitoxin, Domoic Acid, and Tetrodotoxin. J. Agric. Food Chem. 2018, 66, 9801–9809. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Yang, G.; Ghulam, M.; Li, L.; Qu, F. Evolution of multi-functional capillary electrophoresis for high-efficiency selection of aptamers. Biotechnol. Adv. 2019, 37, 107432. [Google Scholar] [CrossRef]
- Mosing, R.K.; Mendonsa, S.D.; Bowser, M.T. Capillary electrophoresis-SELEX selection of aptamers with affinity for HIV-1 reverse transcriptase. Anal. Chem. 2005, 77, 6107–6112. [Google Scholar] [CrossRef]
- Ruff, P.; Pai, R.B.; Storici, F. Real-Time PCR-Coupled CE-SELEX for DNA Aptamer Selection. ISRN Mol. Biol. 2012, 2012, 939083. [Google Scholar] [CrossRef]
- Rozing, G. Microfluidic selection and applications of aptamers. J. Sep. Sci. 2007, 30, 1420–1426. [Google Scholar]
- Mosing, R.K.; Bowser, M.T. Isolating aptamers using capillary electrophoresis-SELEX (CE-SELEX). Methods Mol. Biol. 2009, 535, 33–43. [Google Scholar] [PubMed]
- Dong, L.; Tan, Q.; Ye, W.; Liu, D.; Chen, H.; Hu, H.; Wen, D.; Liu, Y.; Cao, Y.; Kang, J. Screening and Identifying a Novel ssDNA Aptamer against Alpha-fetoprotein Using CE-SELEX. Sci. Rep. 2015, 5, 15552. [Google Scholar] [CrossRef]
- Kanoatov, M.; Galievsky, V.A.; Krylova, S.M.; Cherney, L.T.; Jankowski, H.K.; Krylov, S.N. Using nonequilibrium capillary electrophoresis of equilibrium mixtures (NECEEM) for simultaneous determination of concentration and equilibrium constant. Anal. Chem. 2015, 87, 3099–3106. [Google Scholar] [CrossRef] [PubMed]
- Krylov, S.N.; Berezovski, M. Non-equilibrium capillary electrophoresis of equilibrium mixtures-appreciation of kinetics in capillary electrophoresis. Analyst 2003, 128, 571–575. [Google Scholar] [CrossRef]
- Berezovski, M.; Krylov, S.N. Nonequilibrium Capillary Electrophoresis of Equilibrium Mixtures—A Single Experiment Reveals Equilibrium and Kinetic Parameters of Protein-DNA Interactions. J. Am. Chem. Soc. 2002, 124, 13674–13675. [Google Scholar] [CrossRef]
- Wu, F.; Yang, J.; Liu, J.; Wang, Y.; Mu, J.; Zeng, Q.; Deng, S.; Zhou, H. Signaling pathways in cancer-associated fibroblasts and targeted therapy for cancer. Signal Transduct. Target. 2021, 6, 218. [Google Scholar]
- Nurmik, M.; Ullmann, P.; Rodriguez, F.; Haan, S.; Letellier, E. In search of definitions: Cancer-associated fibroblasts and their markers. Int. J. Cancer 2020, 146, 895–905. [Google Scholar] [CrossRef]
- Kraman, M.; Bambrough, P.J.; Arnold, J.N.; Roberts, E.W.; Magiera, L.; Jones, J.O.; Gopinathan, A.; Tuveson, D.A.; Fearon, D.T. Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science 2010, 330, 827–830. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.N.; Jackson, K.W.; Christiansen, V.J.; Lee, C.S.; Chun, J.G.; McKee, P.A. Antiplasmin-cleaving enzyme is a soluble form of fibroblast activation protein. Blood 2006, 107, 1397–1404. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, S.; Brennen, W.N.; Kole, T.P.; Schneider, E.; Topaloglu, O.; Yates, M.; Cotter, R.J.; Denmeade, S.R. Fibroblast activation protein peptide substrates identified from human collagen I derived gelatin cleavage sites. Biochemistry 2008, 47, 1076–1086. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Gong, Q.; Zou, R.; Li, Z.; Xia, Y.; Yu, Z.; Ye, Y.; Xiang, L.; Wu, A. A novel fibroblast activation protein-targeted near-infrared fluorescent off-on probe for cancer cell detection, in vitro and in vivo imaging. J. Mater. Chem. B 2018, 6, 1449–1451. [Google Scholar] [CrossRef] [PubMed]
- De Sostoa, J.; Fajardo, C.A.; Moreno, R.; Ramos, M.D.; Farrera-Sal, M.; Alemany, R. Targeting the tumor stroma with an oncolytic adenovirus secreting a fibroblast activation protein-targeted bispecific T-cell engager. J. Immunother. Cancer 2019, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.C.; Lo, A.; Scholler, J.; Sun, J.; Majumdar, R.S.; Kapoor, V.; Antzis, M.; Cotner, C.E.; Johnson, L.A.; Durham, A.C.; et al. Targeting fibroblast activation protein in tumor stroma with chimeric antigen receptor T cells can inhibit tumor growth and augment host immunity without severe toxicity. Cancer Immunol. Res. 2014, 2, 154–166. [Google Scholar] [CrossRef]
- Sterner, R.C.; Sterner, R.M. CAR-T cell therapy: Current limitations and potential strategies. Blood Cancer J. 2021, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wu, Z.; Liu, Y.; Han, W. New development in CAR-T cell therapy. J. Hematol. Oncol. 2017, 10, 53. [Google Scholar] [CrossRef] [PubMed]
- Malecka, K.E.; Mikuła, E.; Ferapontova, E.E. Design Strategies for Electrochemical Aptasensors for Cancer Diagnostic Devices. Sensors 2021, 21, 736. [Google Scholar] [CrossRef]
- Omage, J.I.; Easterday, E.; Rumph, J.T.; Brula, I.; Hill, B.; Kristensen, J.; Ha, D.T.; Galindo, C.L.; Danquah, M.K.; Sims, N. Cancer Diagnostics and Early Detection Using Electrochemical Aptasensors. Micromachines 2022, 13, 522. [Google Scholar] [CrossRef]
- Hosseinzadeh, L.; Mazloum-Ardakani, M. Advances in aptasensor technology. Adv. Clin. Chem. 2020, 99, 237–279. [Google Scholar]
- Liu, L.S.; Wang, F.; Ge, Y.; Lo, P.K. Recent Developments in Aptasensors for Diagnostic Applications. ACS Appl. Mater. Interfaces 2021, 13, 9329–9358. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zheng, Y.; Lin, Z.; Liu, X.; Li, J.; Yang, H.; Tan, W. Equipping Natural Killer Cells with Specific Targeting and Checkpoint Blocking Aptamers for Enhanced Adoptive Immunotherapy in Solid Tumors. Angew. Chem. Int. Ed. Engl. 2020, 59, 12022–12028. [Google Scholar] [CrossRef]
- Qian, H.; Fu, Y.; Guo, M.; Chen, Y.; Zhang, D.; Wei, Y.; Jin, F.; Zeng, Q.; Wang, Y.; Chai, C. Dual-aptamer-engineered M1 macrophage with enhanced specific targeting and checkpoint blocking for solid-tumor immunotherapy. Mol. Ther. 2022, 30, 2817–2827. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.G.; Wang, Y.; Liu, P.; Yao, Q.L.; Zhou, Y.Y.; Li, C.F.; Zhao, Q.; Liu, G.H.; Zhang, X.L. Aptamer-T Cell Targeted Therapy for Tumor Treatment Using Sugar Metabolism and Click Chemistry. ACS Chem. Biol. 2020, 15, 1554–1565. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zeng, Z.; Wan, Q.; Liu, X.; Qi, J.; Zu, Y. Targeted immunotherapy of triple-negative breast cancer by aptamer-engineered NK cells. Biomaterials 2022, 280, 121259. [Google Scholar] [CrossRef] [PubMed]


| Name | Sequence (5′-3′) | ∆G (kcal/mol) | ∆H (kcal/mol) | ∆S (cal/K·mol) | Tm (°C) | Seq-Frequency |
|---|---|---|---|---|---|---|
| AptFAP-A1 | P1-GCGAAGCGTACCGGCTACCCAGTGACAGTCGCCGTGGGTC-P2 | −2.45 | −59.20 | −182.9 | 50.3 | 347 |
| AptFAP-A2 | P1-GCCACATTAGTCTCACCACTACCTGCGTACCTACCGCCGC-P2 | −1.78 | −55.00 | −171.5 | 47.3 | 311 |
| AptFAP-A3 | P1-GCCATCCCCCCGTCCGATGAGTGGTGCTCCTATGCGTTCC-P2 | −1.42 | −53.30 | −167.2 | 45.4 | 59 |
| AptFAP-A4 | P1-CCGCAGGCAGCTGCCATTAGTCTCTATCCGTGACGGTATG-P2 | −7.29 | −95.90 | −285.7 | 62.5 | 18 |
| AptFAP-A5 | P1-GCAGCTAAGCAGGCGGCTCACAAAACCATTCGCATGCGGC-P2 | −3.07 | −83.00 | −257.7 | 48.9 | 17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Yang, G.; Zhao, Y.; Dai, X.; Liu, W.; Qu, F.; Huang, Y. Selection and Identification of an ssDNA Aptamer for Fibroblast Activation Protein. Molecules 2023, 28, 1682. https://doi.org/10.3390/molecules28041682
Zhang X, Yang G, Zhao Y, Dai X, Liu W, Qu F, Huang Y. Selection and Identification of an ssDNA Aptamer for Fibroblast Activation Protein. Molecules. 2023; 28(4):1682. https://doi.org/10.3390/molecules28041682
Chicago/Turabian StyleZhang, Xiaomin, Ge Yang, Yi Zhao, Xuyan Dai, Wenjing Liu, Feng Qu, and Yuanyu Huang. 2023. "Selection and Identification of an ssDNA Aptamer for Fibroblast Activation Protein" Molecules 28, no. 4: 1682. https://doi.org/10.3390/molecules28041682
APA StyleZhang, X., Yang, G., Zhao, Y., Dai, X., Liu, W., Qu, F., & Huang, Y. (2023). Selection and Identification of an ssDNA Aptamer for Fibroblast Activation Protein. Molecules, 28(4), 1682. https://doi.org/10.3390/molecules28041682






