Permethrin as a Potential Furin Inhibitor through a Novel Non-Competitive Allosteric Inhibition
Abstract
:1. Introduction
2. Results
2.1. Expression, Purification, and Activity Assay of Furin
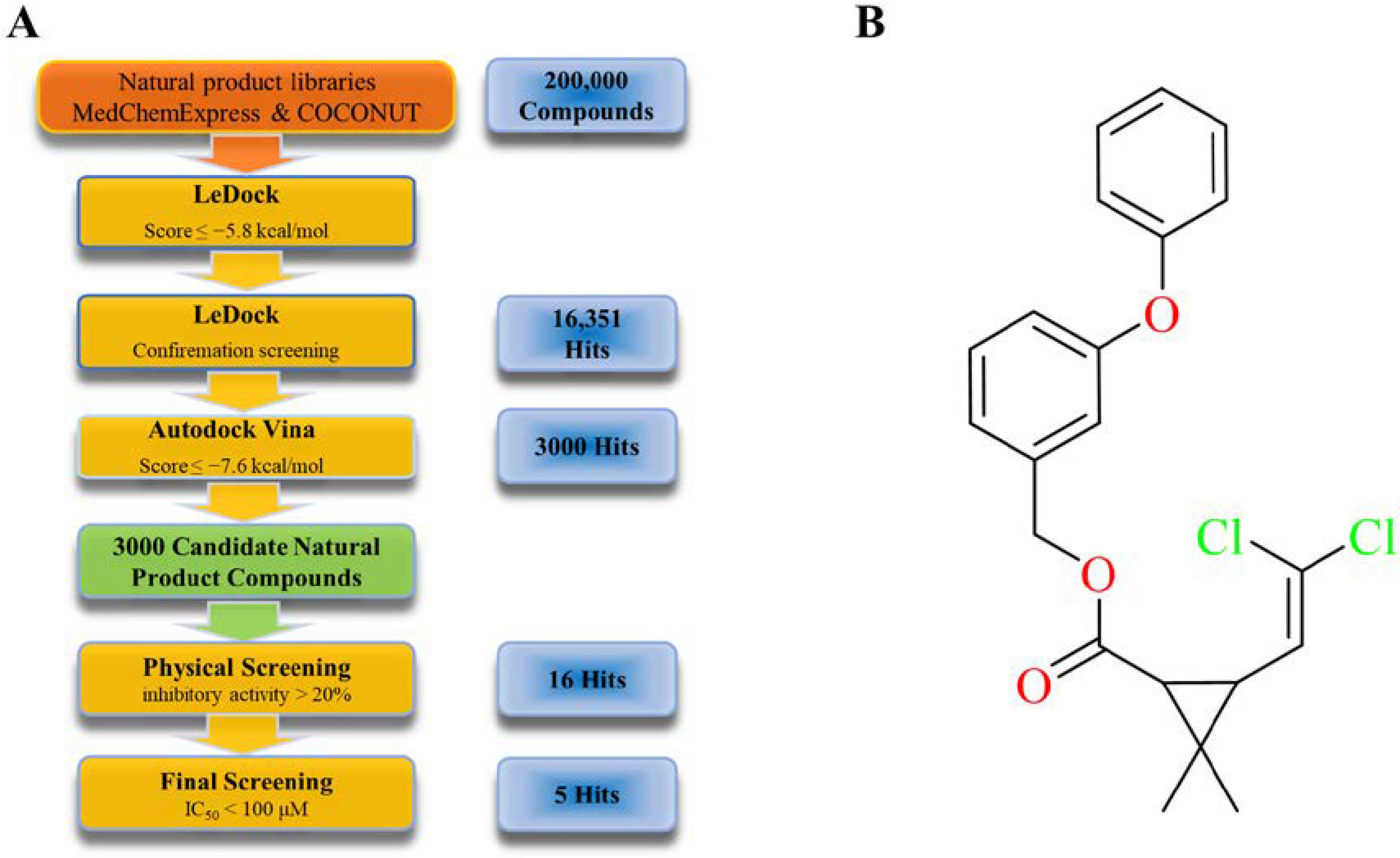
2.2. Virtual and Physical Screening of Furin Inhibitors
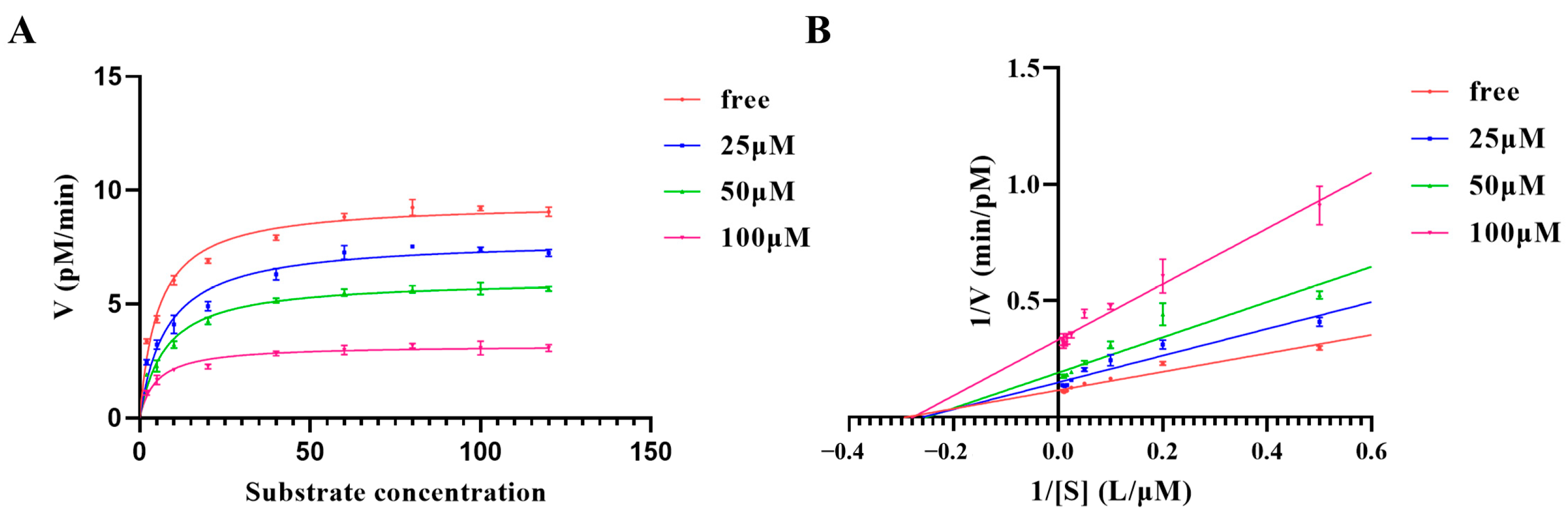
2.3. Kinetic Characterization of Furin
2.4. Interaction Mode of Permethrin with Furin
2.5. Molecular Dynamics Simulation
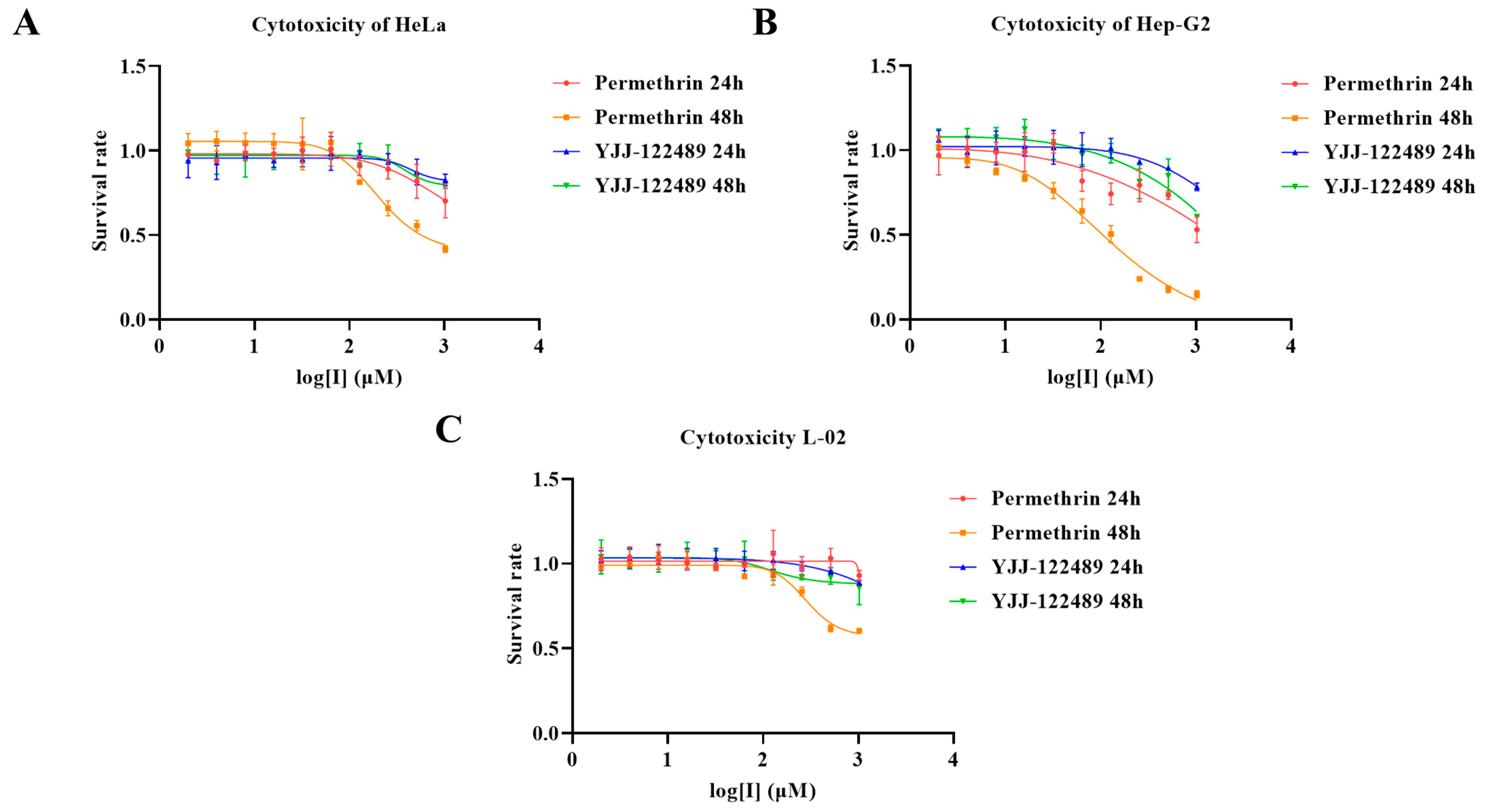
2.6. Cellular Experiment Results
3. Discussion
4. Materials and Methods
4.1. Expression and Purification of Furin
4.2. Virtual Inhibitor Screening and Molecular Dynamics Simulation
4.3. Furin Activity Assay and Enzymatic Kinetics
4.4. Allosteric Site Prediction
4.5. Cellular Experiments
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Roebroek, A.J.; Schalken, J.A.; Leunissen, J.A.; Onnekink, C.; Bloemers, H.P.; Van de Ven, W.J. Evolutionary conserved close linkage of the c-fes/fps proto-oncogene and genetic sequences encoding a receptor-like protein. EMBO J. 1986, 5, 2197–2202. [Google Scholar] [CrossRef] [PubMed]
- Roebroek, A.J.M.; Schalken, J.A.; Bussemakers, M.J.G.; van Heerikhuizen, H.; Onnekink, C.; Debruyne, F.M.J.; Bloemers, H.P.J.; Van de Ven, W.J.M. Characterization of human c-fes/fps reveals a new transcription unit (fur) in the immediately upstream region of the proto-oncogene. Mol. Biol. Rep. 1986, 11, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Dahms, S.O.; Hardes, K.; Becker, G.L.; Steinmetzer, T.; Brandstetter, H.; Than, M.E. X-ray structures of human furin in complex with competitive inhibitors. ACS Chem. Biol. 2014, 9, 1113–1118. [Google Scholar] [CrossRef] [PubMed]
- Artenstein, A.W.; Opal, S.M. Proprotein Convertases in Health and Disease. N. Engl. J. Med. 2011, 365, 2507–2518. [Google Scholar] [CrossRef]
- Marra, M.A.; Jones, S.J.M.; Astell, C.R.; Holt, R.A.; Brooks-Wilson, A.; Butterfield, Y.S.N.; Khattra, J.; Asano, J.K.; Barber, S.A.; Chan, S.Y.; et al. The Genome Sequence of the SARS-Associated Coronavirus. Science 2003, 300, 1399–1404. [Google Scholar] [CrossRef] [Green Version]
- Rota, P.A.; Oberste, M.S.; Monroe, S.S.; Nix, W.A.; Campagnoli, R.; Icenogle, J.P.; Peñaranda, S.; Bankamp, B.; Maher, K.; Chen, M.-h.; et al. Characterization of a Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. Science 2003, 300, 1394–1399. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Gao, X.; Bai, X.; Yao, S.; Chang, Y.Z.; Gao, G. The emerging role of furin in neurodegenerative and neuropsychiatric diseases. Transl. Neurodegener. 2022, 11, 39. [Google Scholar] [CrossRef]
- Osman, E.E.A.; Rehemtulla, A.; Neamati, N. Why All the Fury over Furin? J. Med. Chem. 2022, 65, 2747–2784. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Belouzard, S.; Millet, J.K.; Licitra, B.N.; Whittaker, G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 2012, 4, 1011–1033. [Google Scholar] [CrossRef] [Green Version]
- Puente, X.S.; Sánchez, L.M.; Overall, C.M.; López-Otín, C. Human and mouse proteases: A comparative genomic approach. Nat. Rev. Genet. 2003, 4, 544–558. [Google Scholar] [CrossRef]
- Tortorici, M.A.; Veesler, D. Structural insights into coronavirus entry. Adv. Virus Res. 2019, 105, 93–116. [Google Scholar] [CrossRef]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef] [Green Version]
- Meng, T.; Cao, H.; Zhang, H.; Kang, Z.; Xu, D.; Gong, H.; Wang, J.; Li, Z.; Cui, X.; Xu, H.; et al. The Insert Sequence in SARS-CoV-2 Enhances Spike Protein Cleavage by TMPRSS; Cold Spying Harbor Laboratory: Cold Spring Harbor, NY, USA, 2020. [Google Scholar] [CrossRef]
- Zhang, H.; Kang, Z.; Gong, H.; Xu, D.; Wang, J.; Li, Z.; Li, Z.; Cui, X.; Xiao, J.; Zhan, J.; et al. Digestive system is a potential route of COVID-19: An analysis of single-cell coexpression pattern of key proteins in viral entry process. Gut 2020, 69, 1010–1018. [Google Scholar] [CrossRef] [Green Version]
- Tome, M.; Pappalardo, A.; Soulet, F.; Lopez, J.J.; Olaizola, J.; Leger, Y.; Dubreuil, M.; Mouchard, A.; Fessart, D.; Delom, F.; et al. Inactivation of Proprotein Convertases in T Cells Inhibits PD-1 Expression and Creates a Favorable Immune Microenvironment in Colorectal Cancer. Cancer Res. 2019, 79, 5008–5021. [Google Scholar] [CrossRef] [Green Version]
- Khatib, A.M.; Siegfried, G.; Prat, A.; Luis, J.; Chretien, M.; Metrakos, P.; Seidah, N.G. Inhibition of proprotein convertases is associated with loss of growth and tumorigenicity of HT-29 human colon carcinoma cells: Importance of insulin-like growth factor-1 (IGF-1) receptor processing in IGF-1-mediated functions. J. Biol. Chem. 2001, 276, 30686–30693. [Google Scholar] [CrossRef] [Green Version]
- Declercq, J.; Brouwers, B.; Pruniau, V.P.; Stijnen, P.; Tuand, K.; Meulemans, S.; Prat, A.; Seidah, N.G.; Khatib, A.M.; Creemers, J.W. Liver-Specific Inactivation of the Proprotein Convertase FURIN Leads to Increased Hepatocellular Carcinoma Growth. Biomed Res. Int. 2015, 2015, 148651. [Google Scholar] [CrossRef] [Green Version]
- Scamuffa, N.; Siegfried, G.; Bontemps, Y.; Ma, L.; Basak, A.; Cherel, G.; Calvo, F.; Seidah, N.G.; Khatib, A.M. Selective inhibition of proprotein convertases represses the metastatic potential of human colorectal tumor cells. J. Clin. Investig. 2008, 118, 352–363. [Google Scholar] [CrossRef] [Green Version]
- Couture, F.; Kwiatkowska, A.; Dory, Y.L.; Day, R. Therapeutic uses of furin and its inhibitors: A patent review. Expert Opin. Ther. Pat. 2015, 25, 379–396. [Google Scholar] [CrossRef]
- Dahms, S.O.; Haider, T.; Klebe, G.; Steinmetzer, T.; Brandstetter, H. OFF-State-Specific Inhibition of the Proprotein Convertase Furin. ACS Chem. Biol. 2021, 16, 1692–1700. [Google Scholar] [CrossRef]
- Dahms, S.O.; Schnapp, G.; Winter, M.; Buttner, F.H.; Schleputz, M.; Gnamm, C.; Pautsch, A.; Brandstetter, H. Dichlorophenylpyridine-Based Molecules Inhibit Furin through an Induced-Fit Mechanism. ACS Chem. Biol. 2022, 17, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Dahms, S.O.; Creemers, J.W.; Schaub, Y.; Bourenkov, G.P.; Zogg, T.; Brandstetter, H.; Than, M.E. The structure of a furin-antibody complex explains non-competitive inhibition by steric exclusion of substrate conformers. Sci. Rep. 2016, 6, 34303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albakri, L.; Goldman, R.D. Permethrin for scabies in children. Can. Fam. Physician 2010, 56, 1005. [Google Scholar]
- Tarbox, M.; Walker, K.; Tan, M. Scabies. JAMA 2018, 320, 612. [Google Scholar] [CrossRef] [PubMed]
- Ellis, E.; Scheinfeld, N. Eosinophilic Pustular Folliculitis. Am. J. Clin. Dermatol. 2004, 5, 189–197. [Google Scholar] [CrossRef]
- Kim, I.Y.; Han, S.Y.; Kang, T.S.; Lee, B.M.; Choi, K.S.; Moon, H.J.; Kim, T.S.; Kang, I.H.; Kwack, S.J.; Moon, A.; et al. Pyrethroid insecticides, fenvalerate and permethrin, inhibit progesterone-induced alkaline phosphatase activity in T47D human breast cancer cells. J. Toxicol. Environ. Health A 2005, 68, 2175–2186. [Google Scholar] [CrossRef]
- Angliker, H.; Wikstrom, P.; Shaw, E.; Brenner, C.; Fuller, R.S. The synthesis of inhibitors for processing proteinases and their action on the Kex2 proteinase of yeast. Biochem. J. 1993, 293, 75–81. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Zhao, H. Enriching screening libraries with bioactive fragment space. Bioorg. Med. Chem. Lett. 2016, 26, 3594–3597. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Belouzard, S.; Chu, V.C.; Whittaker, G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. USA 2009, 106, 5871–5876. [Google Scholar] [CrossRef] [Green Version]
- Belouzard, S.; Madu, I.; Whittaker, G.R. Elastase-mediated activation of the SARS coronavirus spike protein at discrete sites within the S2 domain. J. Biol. Chem. 2010, 285, 22758–22763. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, R.; Matsuyama, S.; Shirato, K.; Maejima, M.; Fukushi, S.; Morikawa, S.; Taguchi, F. Entry from the Cell Surface of Severe Acute Respiratory Syndrome Coronavirus with Cleaved S Protein as Revealed by Pseudotype Virus Bearing Cleaved S Protein. J. Virol. 2008, 82, 11985–11991. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Martínez, M.-A.; Dai, M.; Chen, D.; Ares, I.; Romero, A.; Castellano, V.; Martínez, M.; Rodríguez, J.L.; Martínez-Larrañaga, M.-R.; et al. Permethrin-induced oxidative stress and toxicity and metabolism. A review. Environ. Res. 2016, 149, 86–104. [Google Scholar] [CrossRef]
- Choi, J.; Rose, R.L.; Hodgson, E. In vitro human metabolism of permethrin: The role of human alcohol and aldehyde dehydrogenases. Pestic. Biochem. Physiol. 2002, 74, 117–128. [Google Scholar] [CrossRef]
- Thomas, G.; Couture, F.; Kwiatkowska, A. The Path to Therapeutic Furin Inhibitors: From Yeast Pheromones to SARS-CoV-2. Int. J. Mol. Sci. 2022, 23, 3435. [Google Scholar] [CrossRef]
- He, Z.; Khatib, A.-M.; Creemers, J.W.M. The proprotein convertase furin in cancer: More than an oncogene. Oncogene 2022, 41, 1252–1262. [Google Scholar] [CrossRef]
- Garten, W. Characterization of Proprotein Convertases and Their Involvement in Virus Propagation. In Activation of Viruses by Host Proteases; Böttcher-Friebertshäuser, E., Garten, W., Klenk, H.D., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 205–248. [Google Scholar] [CrossRef]
- Sorokina, M.; Merseburger, P.; Rajan, K.; Yirik, M.A.; Steinbeck, C. COCONUT online: Collection of Open Natural Products database. J. Cheminform. 2021, 13, 2. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]

| Compound | IC50 (μM) | Inhibition Rate (50 μM) |
|---|---|---|
| YJJ-N2376 | 83.36 | 29.41% |
| YJJ-N3443 | 75.82 | 33.49% |
| YJJ-N0522 | 73.78 | 43.31% |
| YJJ-B0887 | 38.72 | 55.89% |
| YJJ-18963 | 46.97 | 48.64% |
| YJJ-N0120 | 101.83 | 25.61% |
| YJJ-N0290 | 112.54 | 22.13% |
| YJJ-14855B | 128.33 | 25.04% |
| YJJ-N4184 | 134.62 | 27.28% |
| YJJ-N2350 | 141.99 | 26.45% |
| YJJ-N0594 | 102.81 | 31.00% |
| YJJ-N0824 | 113.56 | 23.56% |
| YJJ-N1478 | 130.16 | 27.18% |
| YJJ-N1967 | 142.36 | 25.87% |
| YJJ-B2176A | 123.76 | 19.31% |
| YJJ-122489 | 145.27 | 21.24% |
| Residue | Interaction Energy (kcal/mol) | VDW Interaction Energy (kcal/mol) | Electrostatic Interaction Energy (kcal/mol) |
|---|---|---|---|
| Glu271 | −5.359410 | −3.310534 | −2.048877 |
| Phe275 | −0.127889 | −2.632041 | 2.504151 |
| Ile312 | −4.050157 | −1.933321 | −2.116836 |
| Tyr313 | 0.804807 | −4.178134 | 4.982942 |
| Lys449 | −25.798443 | −2.144799 | −23.653643 |
| Gln488 | −1.351767 | −2.007794 | 0.656026 |
| Arg490 | −14.920117 | −2.468614 | −12.451504 |
| Trp531 | 0.748106 | −1.649797 | 2.397903 |
| Ala532 | −1.503742 | −1.748421 | 0.244680 |
| Tyr571 | 1.961132 | −1.538561 | 3.499692 |
| Total | −49.59748 | −23.61202 | −25.98546 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, D.; Ren, L.; Wu, J.; Guo, L.; Han, Z.; Yang, J.; Xie, W.; Wang, Y.; Xu, F.; Su, X.; et al. Permethrin as a Potential Furin Inhibitor through a Novel Non-Competitive Allosteric Inhibition. Molecules 2023, 28, 1883. https://doi.org/10.3390/molecules28041883
Feng D, Ren L, Wu J, Guo L, Han Z, Yang J, Xie W, Wang Y, Xu F, Su X, et al. Permethrin as a Potential Furin Inhibitor through a Novel Non-Competitive Allosteric Inhibition. Molecules. 2023; 28(4):1883. https://doi.org/10.3390/molecules28041883
Chicago/Turabian StyleFeng, Dongyan, Le Ren, Jiaqi Wu, Lingling Guo, Zhitao Han, Jingjing Yang, Wei Xie, Yanbing Wang, Fanxing Xu, Xin Su, and et al. 2023. "Permethrin as a Potential Furin Inhibitor through a Novel Non-Competitive Allosteric Inhibition" Molecules 28, no. 4: 1883. https://doi.org/10.3390/molecules28041883
APA StyleFeng, D., Ren, L., Wu, J., Guo, L., Han, Z., Yang, J., Xie, W., Wang, Y., Xu, F., Su, X., Li, D., & Cao, H. (2023). Permethrin as a Potential Furin Inhibitor through a Novel Non-Competitive Allosteric Inhibition. Molecules, 28(4), 1883. https://doi.org/10.3390/molecules28041883






