Quantum Computational, Spectroscopic (FT-IR, FT-Raman, NMR, and UV–Vis) Hirshfeld Surface and Molecular Docking-Dynamics Studies on 5-Hydroxymethyluracil (Monomer and Trimer)
Abstract
1. Introduction
2. Results and Discussion
2.1. Optimized Geometry
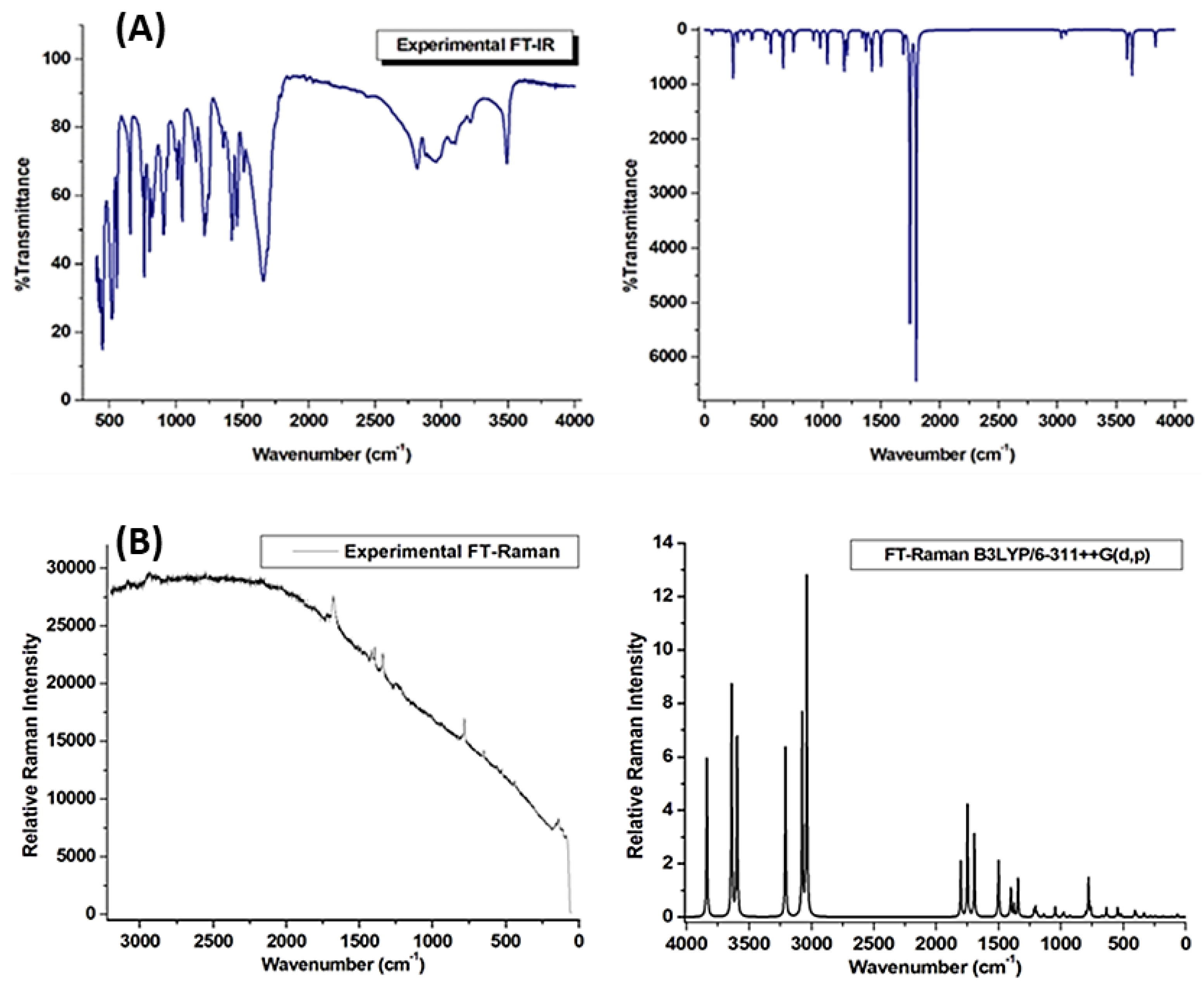
2.2. Vibrational Spectral Analysis
2.2.1. N-H and C-H Vibrations
2.2.2. C-N, H-C-C-C, and C-C Vibrations
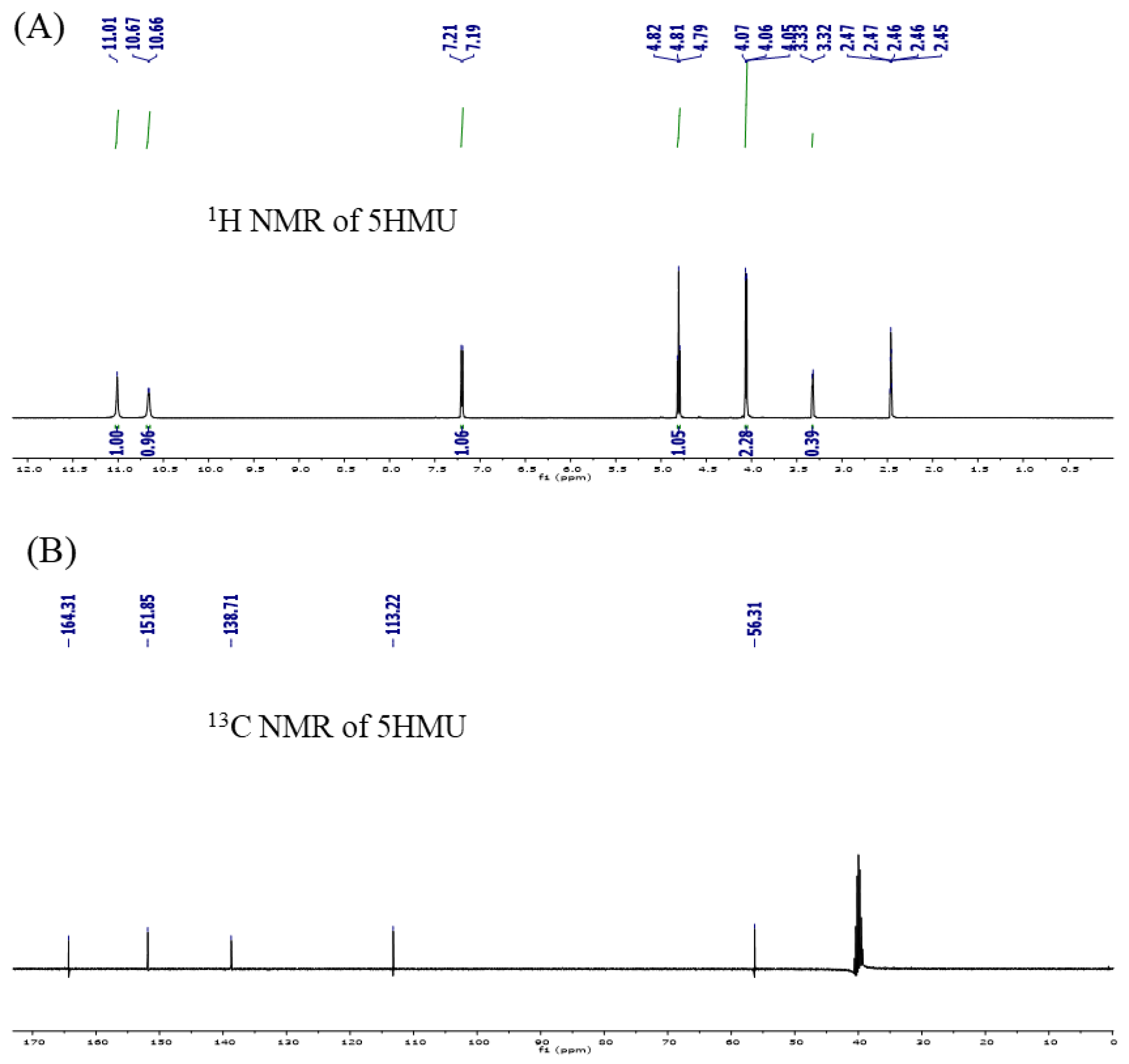
2.3. NMR Analysis
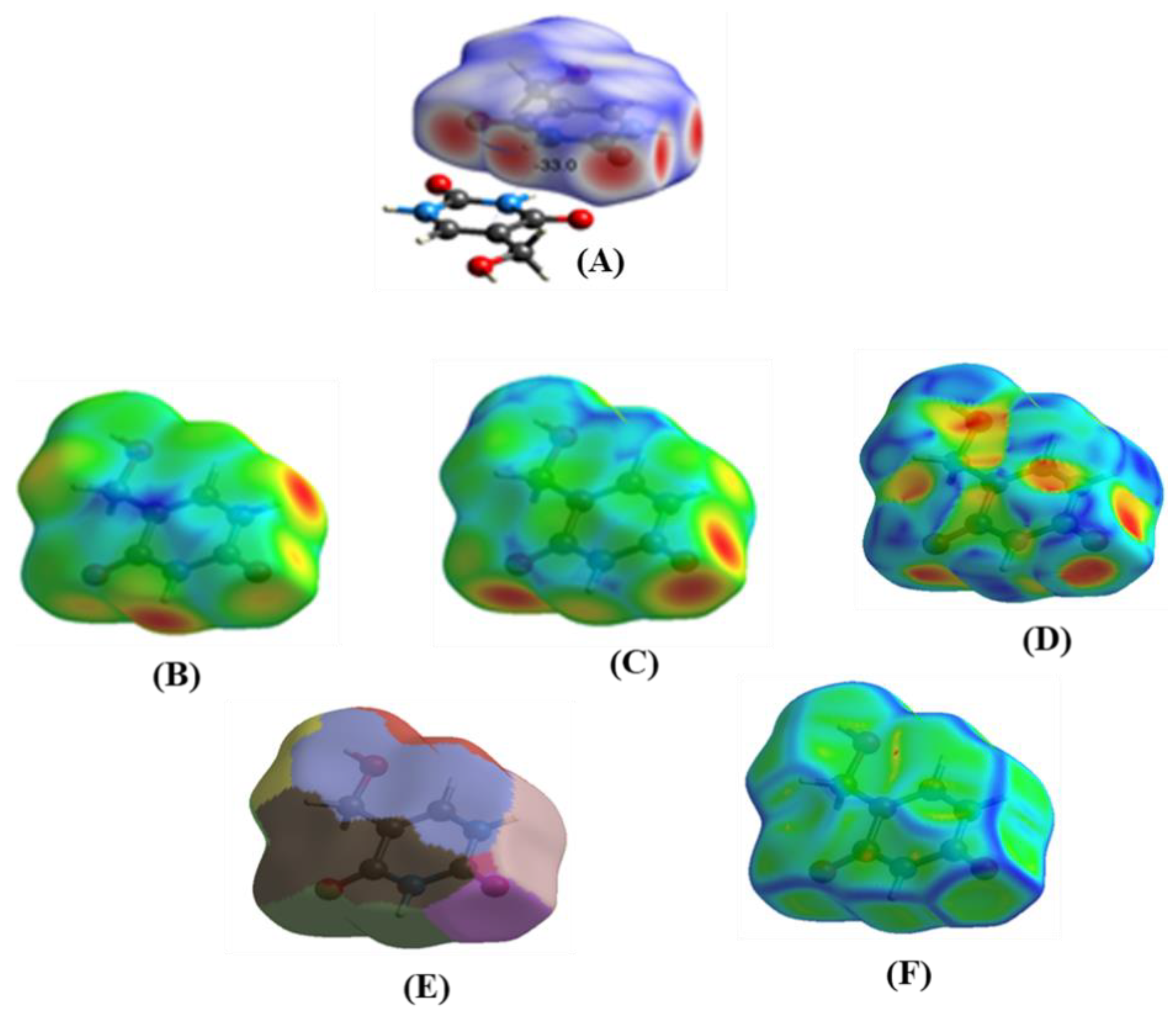
2.4. Hirshfeld Surface (HS) Analysis
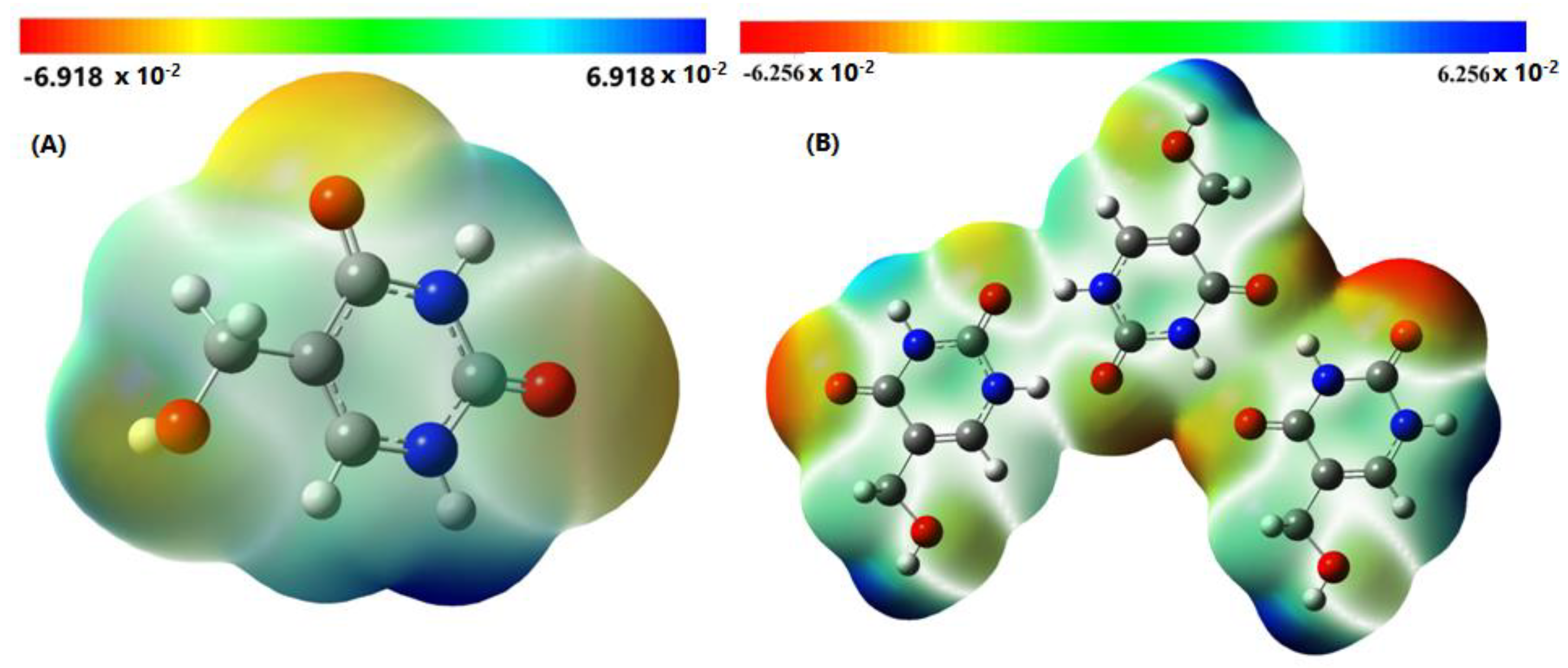
2.5. Molecular Electrostatic Potential Surface (MEPs)
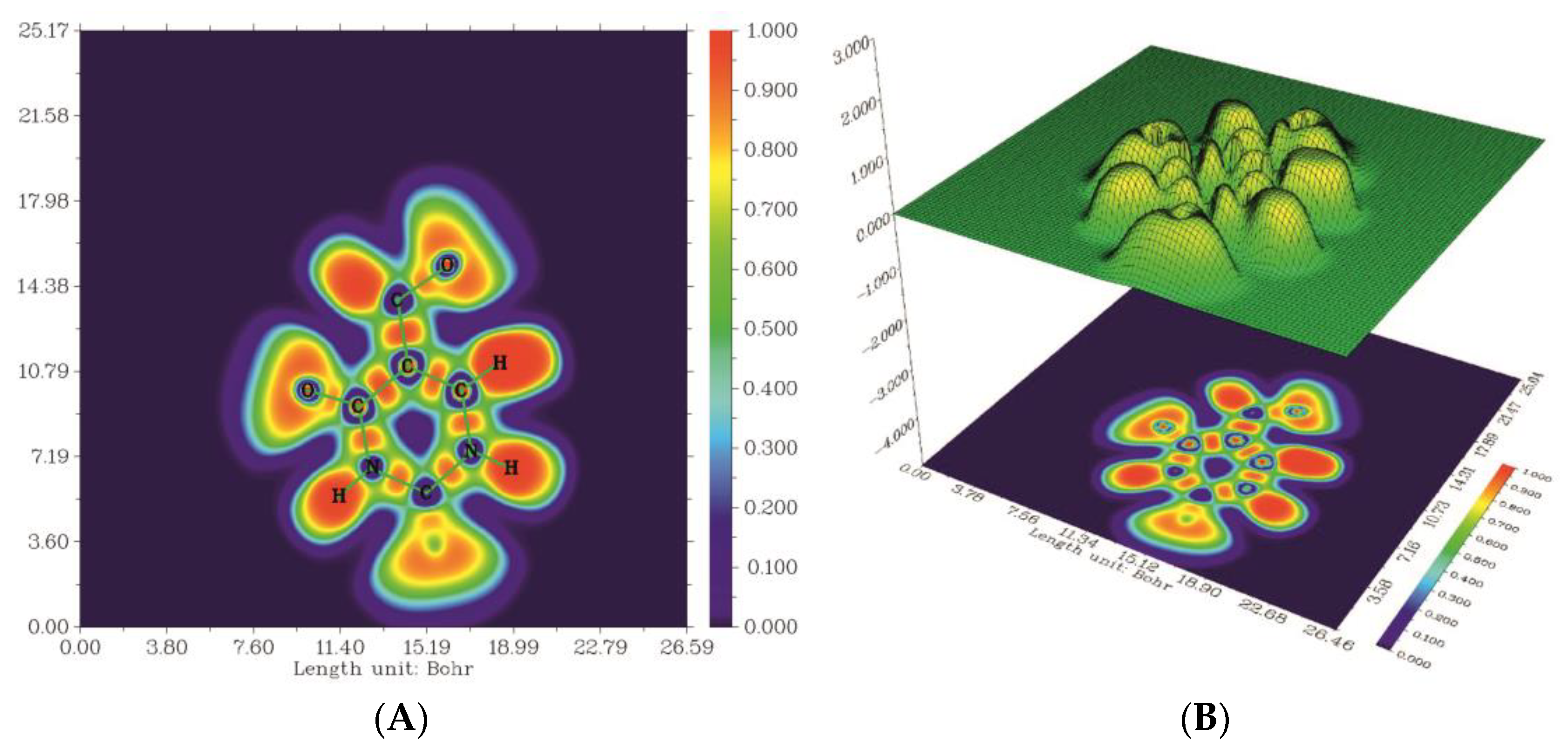
2.6. Electron Localization Function
2.7. Non-Linear Optical (NLO) Analysis
2.8. Natural Bond Orbital (NBO) and Natural Hybrid Orbital (NHO) Analysis
2.9. Population Analysis
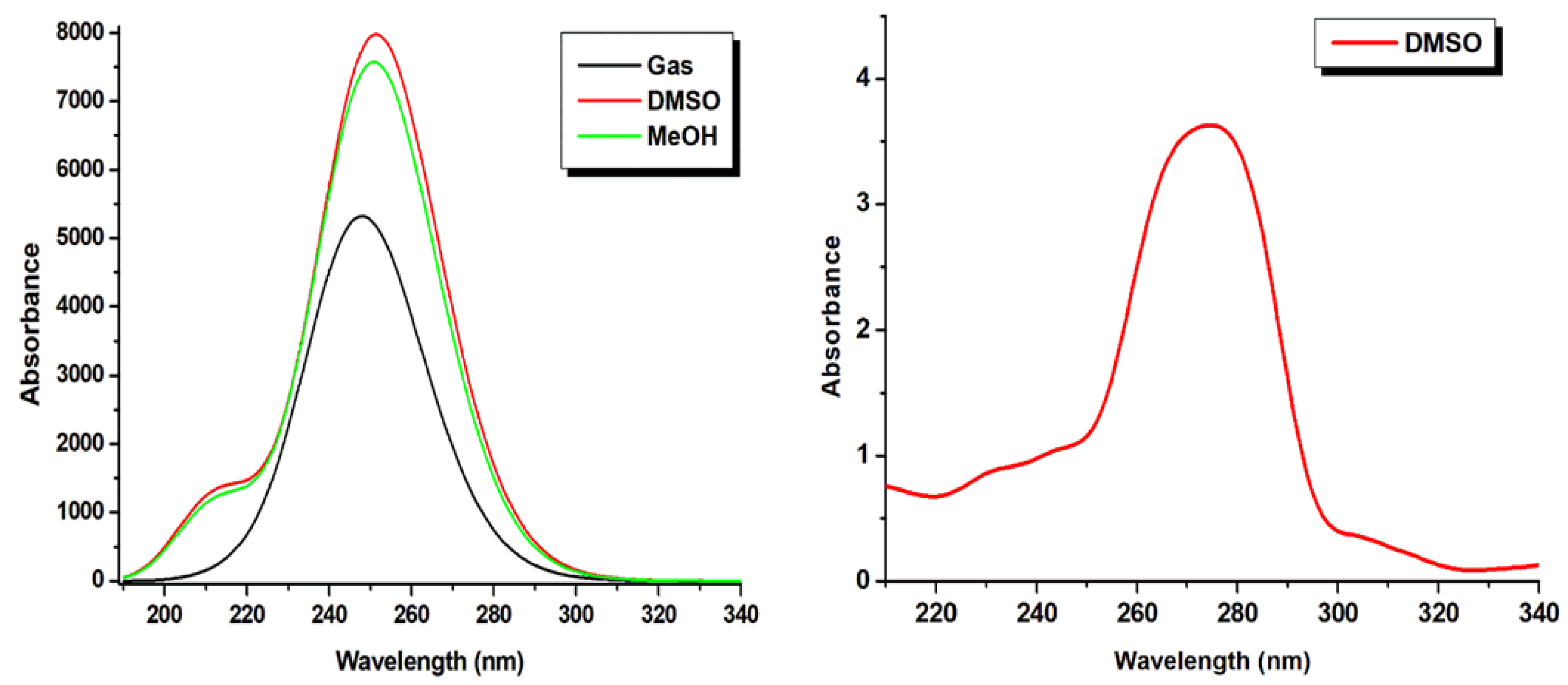
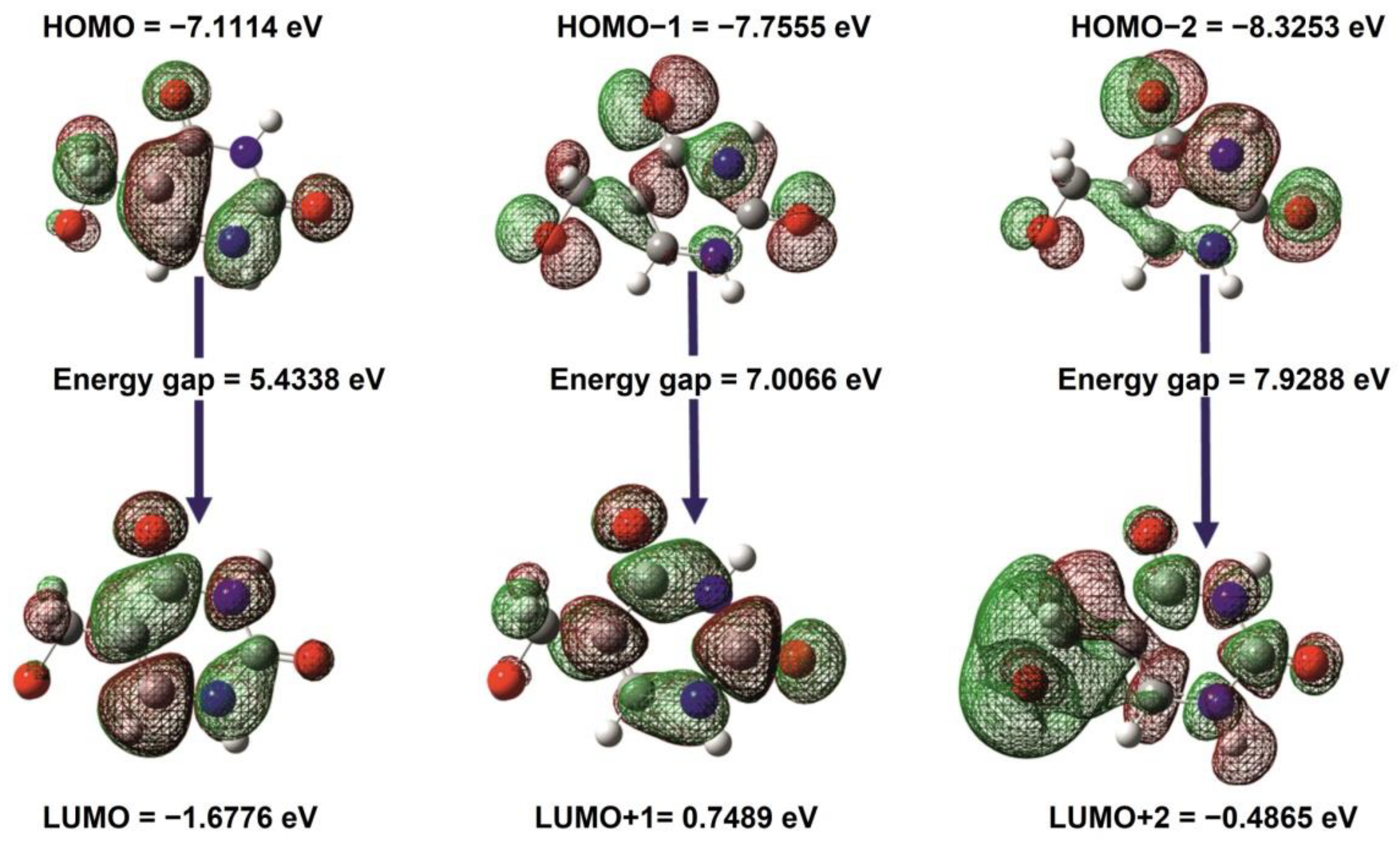
2.10. Frontier Molecular Orbital (FMO) and UV–Vis Spectra
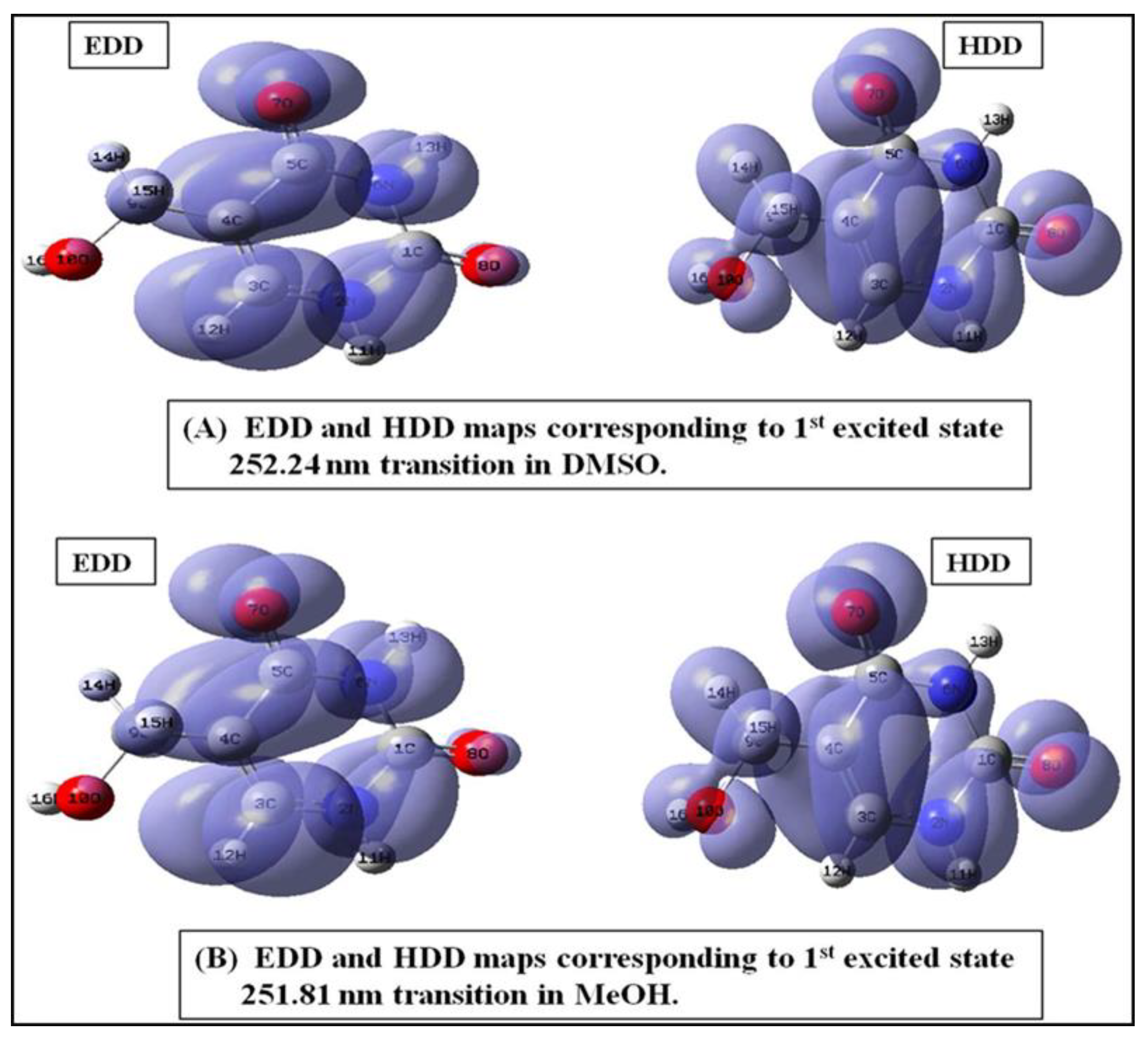
2.11. Electron Excitation (Electron and Hole Density Distribution) Analysis
2.12. Thermodynamic Properties
2.13. Molecular Docking
2.14. Drug-Likeness
2.15. Molecular Dynamic (MD) Simulation
3. Experimental Section
3.1. Materials and Instrumentations
3.2. Computational Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Çırak, Ç.; Sert, Y.; Ucun, F. Effect of intermolecular hydrogen bonding, vibrational analysis and molecular structure of a biomolecule: 5-Hydroxymethyluracil. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 127, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Bandekar, J.; Zundel, G. Normal coordinate analysis treatment on uracil in solid state. Spectrochim. Acta Part A Mol. Spectrosc. 1983, 39, 343–355. [Google Scholar] [CrossRef]
- Aruna, S.; Shanmugam, G. Vibrational assignments of six-membered heterocyclic compounds: Normal vibrations of 6-amino uracil and 6-amino 2-thio uracil. Spectrochim. Acta Part A Mol. Spectrosc. 1985, 41, 531–536. [Google Scholar] [CrossRef]
- Portalone, G.; Ballirano, P.; Maras, A. The crystal structure of 3-methyluracil from X-ray powder diffraction data. J. Mol. Struct. 2002, 608, 35–39. [Google Scholar] [CrossRef]
- Grosmaire, L.; Delarbre, J.-L. Vibrational spectra of 6-methyluracil, 6-methyl-2-thiouracil and their deuterated analogues. J. Mol. Struct. 2012, 1011, 42–49. [Google Scholar] [CrossRef]
- Yaniv, M.; Folk, W. The nucleotide sequences of the two glutamine transfer ribonucleic acids from Escherichia coli. J. Biol. Chem. 1975, 250, 3243–3253. [Google Scholar] [CrossRef]
- Amir, M.; Javed, S.; Kumar, H. Pyrimidine as antiinflammatory agent: A review. Indian J. Pharm. Sci. 2007, 69, 337. [Google Scholar] [CrossRef]
- Shigenaga, M.K.; Ames, B.N. Assays for 8-hydroxy-2′-deoxyguanosine: A biomarker of in vivo oxidative DNA damage. Free Radic. Biol. Med. 1991, 10, 211–216. [Google Scholar] [CrossRef]
- Rusmintratip, V.; Sowers, L.C. An unexpectedly high excision capacity for mispaired 5-hydroxymethyluracil in human cell extracts. Proc. Natl. Acad. Sci. USA 2000, 97, 14183–14187. [Google Scholar] [CrossRef]
- Djuric, Z.; Heilbrun, L.K.; Reading, B.A.; Boomer, A.; Valeriote, F.A.; Martino, S. Effects of a Low-Fat Diet on Levels of Oxidative Damage to DNA in Human Peripheral Nucleated Blood Cells. JNCI J. Natl. Cancer Inst. 1991, 83, 766–769. [Google Scholar] [CrossRef]
- Frenkel, K.; Karkoszka, J.; Kim, E.; Taioli, E. Recognition of oxidized DNA bases by sera of patients with inflammatory diseases. Free Radic. Biol. Med. 1993, 14, 483–494. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.-J.; Han, K.-L. Hydrogen Bonding in the Electronic Excited State. Acc. Chem. Res. 2012, 45, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.-J.; Han, K.-L. Early Time Hydrogen-Bonding Dynamics of Photoexcited Coumarin 102 in Hydrogen-Donating Solvents: Theoretical Study. J. Phys. Chem. A 2007, 111, 2469–2474. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.-J.; Han, K.-L. Ultrafast Hydrogen Bond Strengthening of the Photoexcited Fluorenone in Alcohols for Facilitating the Fluorescence Quenching. J. Phys. Chem. A 2007, 111, 9218–9223. [Google Scholar] [CrossRef]
- Fatima, A.; Ali, A.; Shabbir, S.; Khan, M.; Mehkoom, M.; Afzal, S.M.; Ahmad, M.; Althubeiti, K.; Siddiqui, N.; Singh, M.; et al. Synthesis, crystal structure, characterization, Hirshfeld analysis, molecular docking and DFT calculations of 5-Phenylamino-isophthalic acid: A good NLO material. J. Mol. Struct. 2022, 1261, 132791. [Google Scholar] [CrossRef]
- Manhas, F.M.; Fatima, A.; Verma, I.; Siddiqui, N.; Muthu, S.; AlSalem, H.S.; Savita, S.; Singh, M.; Javed, S. Quantum computational, spectroscopic (FT-IR, NMR and UV–Vis) profiling, Hirshfeld surface, molecular docking and dynamics simulation studies on pyridine-2,6-dicarbonyl dichloride. J. Mol. Struct. 2022, 1265, 133374. [Google Scholar] [CrossRef]
- Fatima, A.; Singh, M.; Abualnaja, K.M.; Althubeiti, K.; Muthu, S.; Siddiqui, N.; Javed, S. Experimental Spectroscopic, Structural (Monomer and Dimer), Molecular Docking, Molecular Dynamics Simulation and Hirshfeld Surface Analysis of 2-Amino-6-Methylpyridine. Polycycl. Aromat. Compd. 2022, 1–31. [Google Scholar] [CrossRef]
- Kumar, M.; Fatima, A.; Singh, M.; Verma, I.; Khanum, G.; Muthu, S.; Althubeiti, K.; Abualnaja, K.M.; Ahmad, M.; Siddiqui, N.; et al. Experimental Spectroscopic, Quantum Computational, Hirshfeld Surface, Molecular Docking, and Electronic Excitation Studies on an Antibiotic Agent: SDZ. Polycycl. Aromat. Compd. 2022, 1–25. [Google Scholar] [CrossRef]
- Sharma, A.; Fatima, A.; Malla, M.A.; Khanum, G.; Kumar, A.; Singh, M.; Abualnaja, K.M.; Althubeiti, K.; Muthu, S.; Siddiqui, N.; et al. Molecular Docking, Dynamic Simulation and DFT Approach to Noble “2-Hydrazinobenzothiazole” Compound. Polycycl. Aromat. Compd. 2022, 1–28. [Google Scholar] [CrossRef]
- Law, G.-L.; Szeto, L.; Wong, W.-T. 2,4-Dioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid monohydrate. Acta Cryst. 2004, E60, o1072–o1074. [Google Scholar] [CrossRef]
- Seiler, V.K.; Hützler, W.M.; Bolte, M. Eight new crystal structures of 5-(hy¬droxy¬meth¬yl)uracil, 5-carb¬oxy¬uracil and 5-carb¬oxy-2-thio-uracil: Insights into the hydrogen-bonded networks and the predominant conformations of the C5-bound residues. Acta Crystallogr. C Struct. Chem. 2016, 72, 379. [Google Scholar] [CrossRef]
- Stewart, R.F.; Jensen, L.H. Redetermination of the crystal structure of uracil. Acta Crystallogr. 1967, 23, 1102–1105. [Google Scholar] [CrossRef]
- Long, D.A. Infrared and Raman Characteristic Group Frequencies: Tables and Charts, 3rd ed.; Socrates, S., Ed.; John Wiley and Sons, Ltd.: Chichester, UK, 2001. [Google Scholar]
- Jennings, K.R. Spectrometric Identification of Organic Compounds, 5th ed.; Silverstein, R.M., Bassler, G.C., Morrill, T.C., Eds.; Wiley: New York, NY, USA, 1991; 430p. [Google Scholar]
- Sameut, A.; Zanndouche, S.Y.; Boumaza, C.; Dikes, C.; Ziani, B.E.C. Chemical Synthesis and Hemi-Synthesis of Novel Benzimidazole Derivatives Using Microwave-Assisted Process: Chemical Characterization, Bioactivities and Molecular Docking. Chem. Proc. 2021, 3, 71. [Google Scholar]
- Fuson, N.; Josien, M.-L. Infrared Spectra of Polynuclear Aromatic Compounds. I. 1,2-Benzanthracene, the Monomethyl-1,2-benzanthracenes and Some Dimethyl-1,2-benzanthracenes 1. J. Am. Chem. Soc. 1956, 78, 3049–3060. [Google Scholar] [CrossRef]
- Barnes, A.J.; Majid, M.A.; Stuckey, M.A.; Gregory, P.; Stead, C.V. The resonance Raman spectra of Orange II and Para Red: Molecular structure and vibrational assignment. Spectrochim. Acta Part A Mol. Spectrosc. 1985, 41, 629–635. [Google Scholar] [CrossRef]
- Nerli, S.; McShan, A.C.; Sgourakis, N.G. Chemical shift-based methods in NMR structure determination. Prog. Nucl. Magn. Reson. Spectrosc. 2018, 106–107, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Armaković, S.; Armaković, S.J. Atomistica.online—Web application for generating input files for ORCA molecular modelling package made with the Anvil platform. Mol. Simul. 2022, 49, 117–123. [Google Scholar] [CrossRef]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld surface analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Ashfaq, M.; Tahir, M.N.; Kuznetsov, A.; Mirza, S.H.; Khalid, M.; Ali, A. DFT and single crystal analysis of the pyrimethamine-based novel co-crystal salt: 2,4-diamino-5-(4-chloro-phenyl)-6-ethylpyrimidin-1-ium:4-hydroxybenzoate:methanol:hydrate (1:1:1:1) (DEHMH). J. Mol. Struct. 2020, 1199, 127041. [Google Scholar] [CrossRef]
- Tahir, M.N.; Ashfaq, M.; de la Torre, A.F.; Caballero, J.; Hernández-Rodríguez, E.W.; Ali, A. Rationalizing the stability and interactions of 2,4-diamino-5-(4-chlorophenyl)-6-ethylpyrimidin-1-ium 2-hydroxy-3,5-dinitrobenzoate salt. J. Mol. Struct. 2019, 1193, 185–194. [Google Scholar] [CrossRef]
- Ali, A.; Zukerman-Schpector, J.; Weber Paixão, M.; Jotani, M.M.; Tiekink, E.R.T. 7-Methyl-5-[(4-methylbenzene)sulfonyl]-2H,5H-[1,3]dioxolo[4,5-f]indole: Crystal structure and Hirshfeld analysis. Acta Crystallogr. Sect. E Crystallogr. Commun. 2018, 74, 184–188. [Google Scholar] [CrossRef]
- Ali, A.; Khalid, M.; ur Rehman, M.F.; Haq, S.; Ali, A.; Tahir, M.N.; Ashfaq, M.; Rasool, F.; Braga, A.A.C. Efficient Synthesis, SC-XRD, and Theoretical Studies of O-Benzenesulfonylated Pyrimidines: Role of Noncovalent Interaction Influence in Their Supramolecular Network. ACS Omega 2020, 5, 15115–15128. [Google Scholar] [CrossRef]
- Ashfaq, M.; Bogdanov, G.; Ali, A.; Tahir, M.N.; Abdullah, S. Pyrimethamine-Based Novel Co-Crystal Salt: Synthesis, Single-Crystal Investigation, Hirshfeld surface analysis and DFT inspection of the 2,4-diamino-5-(4-chlorophenyl)-6-ethylpyrimidin-1-ium 2,4-dichlorobenzoate (1:1) (DECB). J. Mol. Struct. 2021, 1235, 130215. [Google Scholar] [CrossRef]
- Taia, A.; Ibrahimi, B.E.; Benhiba, F.; Ashfaq, M.; Tahir, M.N.; Essaber, M.; Aatif, A.; Hökelek, T.; Mague, J.T.; Sebbar, N.K.; et al. Syntheses, single crystal X-ray structure, Hirshfeld surface analyses, DFT computations and Monte Carlo simulations of New Eugenol derivatives bearing 1,2,3-triazole moiety. J. Mol. Struct. 2021, 1234, 130189. [Google Scholar] [CrossRef]
- Ali, A.; Khalid, M.; Tahir, M.N.; Imran, M.; Ashfaq, M.; Hussain, R.; Assiri, M.A.; Khan, I. Synthesis of Diaminopyrimidine Sulfonate Derivatives and Exploration of Their Structural and Quantum Chemical Insights via SC-XRD and the DFT Approach. ACS Omega 2021, 6, 7047–7057. [Google Scholar] [CrossRef] [PubMed]
- Muthu, S.; Renuga, S. Vibrational spectra and normal coordinate analysis of 2-hydroxy-3-(2-methoxyphenoxy) propyl carbamate. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 132, 313–325. [Google Scholar] [CrossRef]
- Spackman, M.A.; Byrom, P.G. A novel definition of a molecule in a crystal. Chem. Phys. Lett. 1997, 267, 215–220. [Google Scholar] [CrossRef]
- Gadre, S.R.; Babu, K.; Rendell, A.P. Electrostatics for Exploring Hydration Patterns of Molecules. 3. Uracil. J. Phys. Chem. A 2000, 104, 8976–8982. [Google Scholar] [CrossRef]
- Prabavathi, N.; Nilufer, A.; Krishnakumar, V. Vibrational spectroscopic (FT-IR and FT-Raman) studies, natural bond orbital analysis and molecular electrostatic potential surface of Isoxanthopterin. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 114, 101–113. [Google Scholar] [CrossRef]
- Poater, J.; Duran, M.; Solà, M.; Silvi, B. Theoretical Evaluation of Electron Delocalization in Aromatic Molecules by Means of Atoms in Molecules (AIM) and Electron Localization Function (ELF) Topological Approaches. Chem. Rev. 2005, 105, 3911–3947. [Google Scholar] [CrossRef]
- Hermann, J.P.; Ricard, D.; Ducuing, J. Optical nonlinearities in conjugated systems: β-carotene. Appl. Phys. Lett. 1973, 23, 178–180. [Google Scholar] [CrossRef]
- Ishow, E.; Bellaïche, C.; Bouteiller, L.; Nakatani, K.; Delaire, J.A. Versatile Synthesis of Small NLO-Active Molecules Forming Amorphous Materials with Spontaneous Second-Order NLO Response. J. Am. Chem. Soc. 2003, 125, 15744–15745. [Google Scholar] [CrossRef] [PubMed]
- Reed, A.E.; Curtiss, L.A.; Weinhold, F. Intermolecular interactions from a natural bond orbital, donor-acceptor viewpoint. Chem. Rev. 1988, 88, 899–926. [Google Scholar] [CrossRef]
- Tobey, S.W.; West, R. Pentachlorocyclopropane 1. J. Am. Chem. Soc. 1966, 88, 2478–2481. [Google Scholar] [CrossRef]
- Sekino, H.; Bartlett, R.J. Frequency dependent nonlinear optical properties of molecules. J. Chem. Phys. 1986, 85, 976–989. [Google Scholar] [CrossRef]
- Kosar, B.; Albayrak, C. Spectroscopic investigations and quantum chemical computational study of (E)-4-methoxy-2-[(p-tolylimino)methyl]phenol. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2011, 78, 160–167. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, D.; Jiang, W.; Wang, Z. The electron density delocalization of hydrogen bond systems. Adv. Phys. X 2018, 3, 1428915. [Google Scholar] [CrossRef]
- Al-Ahmary, K.M.; Habeeb, M.M.; Aljahdali, S.H. Synthesis, spectroscopic studies and DFT/TD-DFT/PCM calculations of molecular structure, spectroscopic characterization and NBO of charge transfer complex between 5-amino-1,3-dimethylpyrazole (5-ADMP) with chloranilic acid (CLA) in different solvents. J. Mol. Liq. 2019, 277, 453–470. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09, Revision A.02; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Parr, R.G.; Yang, W. Density functional approach to the frontier-electron theory of chemical reactivity. J. Am. Chem. Soc. 1984, 106, 4049–4050. [Google Scholar] [CrossRef]
- Karabacak, M.; Asiri, A.M.; Al-Youbi, A.O.; Qusti, A.H.; Cinar, M. Identification of structural and spectral features of synthesized cyano-stilbene dye derivatives: A comparative experimental and DFT study. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 120, 144–150. [Google Scholar] [CrossRef]
- Hughbanks, T.; Hoffmann, R. Chains of trans-edge-sharing molybdenum octahedra: Metal-metal bonding in extended systems. J. Am. Chem. Soc. 1983, 105, 3528–3537. [Google Scholar] [CrossRef]
- Miller, G.J. Solids and Surfaces: A Chemist’s View of Bonding in Extended Structures; VonR. Hoffmann. VCH Publishers: New York, NY, USA; VCH Verlagsgesellschaft: Weinheim, Germany, 1988. [Google Scholar]
- Balachandran, V.; Karpagam, V.; Revathi, B.; Kavimani, M.; Santhi, G. Conformational stability, vibrational and NMR analysis, chemical potential and thermodynamical parameter of 3-tert-butyl-4-hydroxyanisole. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 135, 1039–1051. [Google Scholar] [CrossRef] [PubMed]
- Saral, A.; Sudha, P.; Muthu, S.; Sevvanthi, S.; Sangeetha, P.; Selvakumari, S. Vibrational spectroscopy, quantum computational and molecular docking studies on 2-chloroquinoline-3-carboxaldehyde. Heliyon 2021, 7, e07529. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Chen, F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef]
- Roy, D.R.; Parthasarathi, R.; Maiti, B.; Subramanian, V.; Chattaraj, P.K. Electrophilicity as a possible descriptor for toxicity prediction. Bioorg. Med. Chem. 2005, 13, 3405–3412. [Google Scholar] [CrossRef]
- Shahraki, S.; Saeidifar, M.; Shiri, F.; Heidari, A. Assessment of the interaction procedure between Pt(IV) prodrug [Pt(5,5′-dmbpy)Cl 4 and human serum albumin: Combination of spectroscopic and molecular modeling technique. J. Biomol. Struct. Dyn. 2017, 35, 3098–3106. [Google Scholar] [CrossRef]
- Ben Issa, T.; Sagaama, A.; Issaoui, N. Computational study of 3-thiophene acetic acid: Molecular docking, electronic and intermolecular interactions investigations. Comput. Biol. Chem. 2020, 86, 107268. [Google Scholar] [CrossRef]
- Zhang, R.; Du, B.; Sun, G.; Sun, Y. Experimental and theoretical studies on o-, m- and p-chlorobenzylideneaminoantipyrines. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2010, 75, 1115–1124. [Google Scholar] [CrossRef]
- Ruszkowski, M.; Sliwiak, J.; Ciesielska, A.; Barciszewski, J.; Sikorski, M.; Jaskolski, M. Specific binding of gibberellic acid by Cytokinin-Specific Binding Proteins: A new aspect of plant hormone-binding proteins with the PR-10 fold. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 2032–2041. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A Knowledge-Based Approach in Designing Combinatorial or Medicinal Chemistry Libraries for Drug Discovery. 1. A Qualitative and Quantitative Characterization of Known Drug Databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Bekker, H.; Berendsen, H.J.C.; Dijkstra, E.J.; Achterop, S.; van Drunen, R.; van der Spoel, D.; Sijbers, A.; Keegstra, H.; Renardus, M.K.R. Gromacs: A parallel computer for molecular dynamics simulations. In Physics Computing 92, Proceedings of the 4th International Conference, Prague, Czech Republic, 24–28 August 1992; de Groot, R.A., Nadrchal, J., Eds.; World Scientific: Singapore, 1993; pp. 252–256. [Google Scholar]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Petersson, G.A.; Al-Laham, M.A. A complete basis set model chemistry. II. Open-shell systems and the total energies of the first-row atoms. J. Chem. Phys. 1991, 94, 6081–6090. [Google Scholar] [CrossRef]
- Petersson, G.A.; Bennett, A.; Tensfeldt, T.G.; Al-Laham, M.A.; Shirley, W.A.; Mantzaris, J. A complete basis set model chemistry. I. The total energies of closed-shell atoms and hydrides of the first-row elements. J. Chem. Phys. 1988, 89, 2193–2218. [Google Scholar] [CrossRef]
- Neese, F. The ORCA program system. WIREs Comput. Mol. Sci. 2012, 2, 73–78. [Google Scholar] [CrossRef]
- Jamróz, M.H. Vibrational Energy Distribution Analysis (VEDA): Scopes and limitations. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 114, 220–230. [Google Scholar] [CrossRef]
- Safin, D.A.; Robeyns, K.; Garcia, Y. 1,2,4-Triazole-based molecular switches: Crystal structures, Hirshfeld surface analysis and optical properties. CrystEngComm 2016, 18, 7284–7296. [Google Scholar] [CrossRef]
- Wang, W.; Ling, Y.; Yang, L.-J.; Liu, Q.-L.; Luo, Y.-H.; Sun, B.-W. Crystals of 4-(2-benzimidazole)-1,2,4-triazole and its hydrate: Preparations, crystal structure and Hirshfeld surfaces analysis. Res. Chem. Intermed. 2016, 42, 3157–3168. [Google Scholar] [CrossRef]
- Shyamapada, S.; Christoph, M.; Mitra, S. Synthesis, Crystal structure, and Hirshfeld Surface Analysis of a New Mixed Ligand Copper(II) Complex. Acta Chim. Slov. 2016, 63, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Chohan, Z.H.; Youssoufi, M.H.; Jarrahpour, A.; Ben Hadda, T. Identification of antibacterial and antifungal pharmacophore sites for potent bacteria and fungi inhibition: Indolenyl sulfonamide derivatives. Eur. J. Med. Chem. 2010, 45, 1189–1199. [Google Scholar] [CrossRef] [PubMed]
- Ben M’leh, C.; Brandán, S.A.; Issaoui, N.; Roisnel, T.; Marouani, H. Synthesis, molecular structure, vibrational and theoretical studies of a new non-centrosymmetric organic sulphate with promising NLO properties. J. Mol. Struct. 2018, 1171, 771–785. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera, A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Yang, W.; Mortier, W.J. The use of global and local molecular parameters for the analysis of the gas-phase basicity of amines. J. Am. Chem. Soc. 1986, 108, 5708–5711. [Google Scholar] [CrossRef]



| (A) | |||||
| Bond length (Å) | Bond angle (˚) | ||||
| Parameter | B3LYP/6-311++G(d,p) | Experimental | Parameter | B3LYP/6-311++G(d,p) | Experimental |
| C1-N2 | 1.39 | 1.35 | C1-N2-H11 | 120.93 | 117.15 |
| C1-N6 | 1.38 | 1.36 | C3-N2-H11 | 122.36 | 120.83 |
| C1-O8 | 1.21 | 1.23 | N2-C3-C4 | 116.13 | 122.70 |
| N2-C3 | 1.37 | 1.37 | N2-C3-H12 | 121.50 | 118.62 |
| N2-H11 | 1.00 | 0.87 | C4-C3-H12 | 118.77 | 118.66 |
| C3-C4 | 1.34 | 1.35 | C3-C4-C5 | 122.99 | 118.01 |
| C3-H12 | 1.08 | 0.95 | C3-C4-C9 | 118.21 | 122.87 |
| C4-C5 | 1.46 | 1.45 | C5-C4-C9 | 114.34 | 119.09 |
| C4-C9 | 1.51 | 1.50 | C4-C5-6 | 125.20 | 115.13 |
| C5-N6 | 1.40 | 1.38 | C4-C5-O7 | 120.45 | 124.63 |
| C5-O7 | 1.21 | 1.23 | N6-C5-O7 | 128.00 | 120.23 |
| N6-H13 | 1.01 | 0.89 | C1-N6-C5 | 115.70 | 126.54 |
| C9-O10 | 1.42 | 1.40 | C1-N6-H13 | 116.28 | 117.05 |
| C9-H14 | 1.09 | 0.98 | C5-N6-H13 | 112.81 | 116.36 |
| C9-H15 | 1.09 | 0.99 | C4-C9-O10 | 109.30 | 108.30 |
| O10-H16 | 0.96 | 0.84 | C4-C9-H1C4 | 109.94 | 110.07 |
| Bond angle (˚) | C4-C9-H15 | 111.89 | 110.01 | ||
| N2-C1-N6 | 112.66 | 115.48 | O10-C9-H14 | 106.07 | 110.04 |
| N2-C1-O8 | 123.22 | 122.75 | O10-C9-H15 | 106.58 | 110.02 |
| 6-C1-O8 | 124.11 | 121.76 | H14-C9-H15 | 108.72 | 108.38 |
| C1-N2-C3 | 115.22 | 122.01 | C9-O10-H16 | 108.72 | 109.83 |
| R2 | 0.97 | R2 | 0.91 | ||
| (B) | |||||
| Bond length | Bond angle | ||||
| Parameter | B3LYP/6-311++G(d,p) | Parameter | B3LYP/6-311++G(d,p) | ||
| O17-H2 | 1.81 | N1-H2-O17 | 174.17 | ||
| O5-H18 | 1.78 | N22-H18-O5 | 174.54 | ||
| O12-H20 | 1.84 | N24-H20-O12 | 170.45 | ||
| O19-H9 | 1.84 | N6-H9-O19 | 170.46 | ||
| N22-C21 | 1.37 | C21-O17-H2 | 125.56 | ||
| C21-O17 | 1.23 | C3-O5-H18 | 126.78 | ||
| N22-H18 | 1.03 | C23-O19-H9 | 132.13 | ||
| Mode No | Experimental | Theoretical | IIR | IRAMAN | Assignments (PED) | ||
|---|---|---|---|---|---|---|---|
| FT-IR | FT-Raman | Unscaled | Scaled | ||||
| 42 | 3651 | 3836 | 3686 | 5 | 48 | γOH(100) | |
| 41 | 3491 | 3637 | 3495 | 13 | 69 | γNH(50) | |
| 40 | 3596 | 3455 | 9 | 54 | γNH(50) | ||
| 39 | 3078 | 3068 | 3208 | 3083 | 1 | 51 | γCH(99) |
| 38 | 2960 | 2980 | 3074 | 2954 | 2 | 61 | γCH(94) |
| 37 | 2917 | 3038 | 2919 | 3 | 100 | γCH(95) | |
| 36 | 1732 | 1801 | 1731 | 100 | 17 | γOC(48) + βHNC(10) | |
| 35 | 1658 | 1694 | 1748 | 1679 | 84 | 34 | γOC(44) |
| 34 | 1692 | 1626 | 7 | 26 | γCC(65) + βHCC(11) | ||
| 33 | 1458 | 1503 | 1444 | 2 | 6 | γNC(14) + βHNC(16) + βHCH(27) | |
| 32 | 1421 | 1432 | 1497 | 1438 | 4 | 16 | γNC(10) + βHNC(11) + βHCH(52) |
| 31 | 1365 | 1425 | 1369 | 1 | 1 | γNC(15) + βHNC(15) + βHCC(11) | |
| 30 | 1357 | 1357 | 1409 | 1354 | 4 | 1 | γNC(16) + βHNC(46) |
| 29 | 1399 | 1345 | 1 | 9 | γHC(10) + βHNC(11) + τHCCC(42) | ||
| 28 | 1374 | 1320 | 7 | 5 | βHOC(42) + βHCO(32) | ||
| 27 | 1215 | 1269 | 1342 | 1290 | 3 | 12 | γCC(15) + βHCC(43) |
| 26 | 1175 | 1213 | 1166 | 7 | 3 | γNC(25) + γCC(23) | |
| 25 | 1151 | 1201 | 1154 | 2 | 4 | βHOC(40) + βHCO(36) + τHCCC(11) | |
| 24 | 1148 | 1188 | 1142 | 12 | 2 | γNC(24) + βHNC(14) +βHCC(19) | |
| 23 | 1047 | 1090 | 1136 | 1091 | 1 | 1 | γNC(27) + γCC(11) + γOC(13) |
| 22 | 1013 | 1035 | 1044 | 1003 | 10 | 4 | γOC(72) |
| 21 | 945 | 984 | 945 | 5 | 1 | βHOC(10) + τHCCC(43) | |
| 20 | 906 | 975 | 937 | 1 | 2 | γNC(13) + βCNC(23) | |
| 19 | 800 | 898 | 926 | 890 | 3 | 1 | τHCCC(65) |
| 18 | 761 | 791 | 799 | 768 | 1 | 2 | γCC(28) + βCNC(12) + βCCN(10) |
| 17 | 776 | 746 | 1 | 12 | γNC(34) | ||
| 16 | 758 | 728 | 2 | 3 | τOCCC(30) | ||
| 15 | 755 | 726 | 5 | 1 | τOCCC(10) | ||
| 14 | 659 | 654 | 668 | 641 | 11 | 1 | τHNCC(66) |
| 13 | 636 | 611 | 2 | 3 | τHNCC(24) | ||
| 12 | 559 | 536 | 563 | 541 | 7 | 1 | τHNCC(69) |
| 11 | 520 | 514 | 546 | 525 | 2 | 3 | γNC(10) + βONC(25) + βCNC(29) + βCNC(10) |
| 10 | 451 | 520 | 499 | 3 | 1 | βCNC(12) + βNCN(32) + βOCC(22) | |
| 9 | 407 | 391 | 3 | 2 | βOCN(37) + βCNC(11) | ||
| 8 | 398 | 382 | 3 | 1 | τCCNC(42) | ||
| 7 | 334 | 321 | 2 | 2 | βCNC(12) + βOCN(16) + βCCN(14) + βNCN(10) + βOCC(15) | ||
| 6 | 282 | 271 | 4 | 1 | τHOCC(20) | ||
| 5 | 245 | 235 | 14 | 1 | τHOCC(75) | ||
| 4 | 182 | 175 | 1 | 1 | βCCC(63) + βOCC(19) | ||
| 3 | 147 | 141 | 1 | 1 | τCNCN(77) | ||
| 2 | 110 | 106 | 1 | 1 | τCCNC(18) + τCNCN(58) | ||
| 1 | 66 | 64 | 2 | 1 | βCNCN(13) + βOCCC(67) | ||
| Atoms | Experimental | Calculated (B3LYP/6-311++G(d,p)) | Calculated (aug-cc-pV5Z def2/J) |
|---|---|---|---|
| C5 | 164.31 | 165.946 | 151.95 |
| C1 | 151.85 | 153.5959 | 128.18 |
| C3 | 138.71 | 141.7179 | 102.59 |
| C4 | 113.22 | 120.2056 | 70.18 |
| C9 | 56.31 | 62.2347 | 61.45 |
| H12 | 11.01 | 7.3857 | 19.54 |
| H13 | 10.66 | 7.061 | 8.46 |
| H11 | 7.20 | 6.7589 | 6.43 |
| H14 | 4.81 | 4.6287 | 6.43 |
| H15 | 4.06 | 4.2636 | 5.03 |
| H16 | 3.33 | 0.5378 | 3.19 |
| Parameters | B3LYP/6-311++G(d,p) (5-HMU) | B3LYP (Urcail) | Parameters | B3LYP/6-311++G(d,p) |
|---|---|---|---|---|
| μx | −1.4356 | βxxx | −41.6733 | |
| μy | −3.0482 | βyxx | 21.8308 | |
| μz | −0.989 | βxyy | 19.7825 | |
| μ(D) | 3.5115 | 1.7615 | βyyy | −41.3496 |
| αxx | −71.1752 | 93.36 | βzxx | −13.9748 |
| αxy | 2.0043 | βxyz | −7.1467 | |
| αyy | −58.1887 | 75.72 | βzyy | −2.1224 |
| αxz | 4.4834 | βxzz | −8.2926 | |
| αyz | 1.4686 | βyzz | −0.7765 | |
| αzz | −55.6042 | 70.11 | βzzz | −2.9145 |
| α0 (e.s.u) | −0.914 × 10−23 | βtot (e.s.u) | 0.3545 × 10−30 |
| Protein (PDB ID) | No of Residues | Bond Distance (Å) | Inhibition Constant (Micromolar) | Binding Energy (kcal/mol) | Reference RMSD (Å) |
|---|---|---|---|---|---|
| 4P6T | 3 | 2.092 | 200.89 | −5.0 | 3.620 |
| 4P6S | 3 | 2.484 | 101.67 | −5.4 | 3.690 |
| 6QXD | 3 | 2.270 | 43.40 | −5.9 | 0.000 |
| 6EI4 | 3 | 2.541 | 61.01 | −5.7 | 0.000 |
| 5OAE | 3 | 2.747 | 61.01 | −5.7 | 2.256 |
| 5M67 | 3 | 2.156 | 61.01 | −5.7 | 4.035 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, M.; Jaiswar, G.; Afzal, M.; Muddassir, M.; Alarifi, A.; Fatima, A.; Siddiqui, N.; Ayub, R.; Abduh, N.A.Y.; Saeed, W.S.; et al. Quantum Computational, Spectroscopic (FT-IR, FT-Raman, NMR, and UV–Vis) Hirshfeld Surface and Molecular Docking-Dynamics Studies on 5-Hydroxymethyluracil (Monomer and Trimer). Molecules 2023, 28, 2116. https://doi.org/10.3390/molecules28052116
Kumar M, Jaiswar G, Afzal M, Muddassir M, Alarifi A, Fatima A, Siddiqui N, Ayub R, Abduh NAY, Saeed WS, et al. Quantum Computational, Spectroscopic (FT-IR, FT-Raman, NMR, and UV–Vis) Hirshfeld Surface and Molecular Docking-Dynamics Studies on 5-Hydroxymethyluracil (Monomer and Trimer). Molecules. 2023; 28(5):2116. https://doi.org/10.3390/molecules28052116
Chicago/Turabian StyleKumar, Mohit, Gautam Jaiswar, Mohd. Afzal, Mohd. Muddassir, Abdullah Alarifi, Aysha Fatima, Nazia Siddiqui, Rashid Ayub, Naaser A. Y. Abduh, Waseem Sharaf Saeed, and et al. 2023. "Quantum Computational, Spectroscopic (FT-IR, FT-Raman, NMR, and UV–Vis) Hirshfeld Surface and Molecular Docking-Dynamics Studies on 5-Hydroxymethyluracil (Monomer and Trimer)" Molecules 28, no. 5: 2116. https://doi.org/10.3390/molecules28052116
APA StyleKumar, M., Jaiswar, G., Afzal, M., Muddassir, M., Alarifi, A., Fatima, A., Siddiqui, N., Ayub, R., Abduh, N. A. Y., Saeed, W. S., & Javed, S. (2023). Quantum Computational, Spectroscopic (FT-IR, FT-Raman, NMR, and UV–Vis) Hirshfeld Surface and Molecular Docking-Dynamics Studies on 5-Hydroxymethyluracil (Monomer and Trimer). Molecules, 28(5), 2116. https://doi.org/10.3390/molecules28052116









