Abstract
Two new 4-hydroxy-2-pyridone alkaloids furanpydone A and B (1 and 2), along with two known compounds N-hydroxyapiosporamide (3) and apiosporamide (4) were isolated from the endophytic fungus Arthrinium sp. GZWMJZ-606 in Houttuynia cordata Thunb. Furanpydone A and B had unusual 5-(7-oxabicyclo[2.2.1]heptane)-4-hydroxy-2-pyridone skeleton. Their structures including absolute configurations were determined on the basis of spectroscopic analysis, as well as the X-ray diffraction experiment. Compound 1 showed inhibitory activity against ten cancer cell lines (MKN-45, HCT116, K562, A549, DU145, SF126, A-375, 786O, 5637, and PATU8988T) with IC50 values from 4.35 to 9.72 µM. Compounds 1, 3 and 4 showed moderate inhibitory effects against four Gram-positive strains (Staphylococcus aureus, methicillin-resistant S. aureus, Bacillus Subtilis, Clostridium perfringens) and one Gram-negative strain (Ralstonia solanacarum) with MIC values from 1.56 to 25 µM. However, compounds 1–4 showed no obvious inhibitory activity against two Gram-negative bacteria (Escherichia coli and Pseudomonas aeruginosa) and two pathogenic fungi (Candida albicans and Candida glabrata) at 50 µM. These results show that compounds 1–4 are expected to be developed as lead compounds for antibacterial or anti-tumor drugs.
1. Introduction
Since ricinine [1] and ilicicolin H [2] were found in the early 1970s, a series of 4-hydroxy-2-pyridinone alkaloids with diverse structures were reported [3,4,5,6]. These alkaloids were mainly isolated from plants and fungi and had good biological activity [7]. According to the position and type of substituents, C-3 was often replaced by alkanes (e.g., septoriamycin from Septoria pistaciarum [8]) or terpenes (e.g., tenelin from Beauveria tenella and Beauveria bassiana [9]), and C-5 was often replaced by phenyl (e.g., sambutoxin from Fusarium sambucinum [10]) or cyclohexyl (e.g., torrubiellone A-B from Torrubiella sp. [11]). There were also a small number of derivatives whose C-6 was replaced by alkanes (e.g., pyridomacrolidin from Beauveria basiana [12]). These kind of compounds usually have anti-inflammatory, antibacterial, cytotoxicity, antimalarial, antiviral, insecticidal, antioxidant, anti-fibrosis, neuroprotection, inhibition of protein tyrosine kinase, and so on [13,14,15,16], which have attracted widespread attention.
In the past few years, there were some new 4-hydroxy-2-pyridones discovered from fungi, such as (+)-didymellamide B, (±)-didymellamide E, (+)-N-hydroxyapiosporamide, and didymellamides F–H which were isolated from Coniochaeta cephalothecoides [17], and arthpyrones A and B with novel oxabicyclo[3.3.1]-nonane ring which were isolated from Arthrinium arundinis ZSDS1-F3 [18]. Three new 4-hydroxy-2-pyridone alkaloids citridones E–G with antibacterial activity were isolated from the endophytic fungus Penicillium sumatrense GZWMJZ-313 in our previous studies [19]. In order to obtain more compounds of this type from endophytic fungus, Arthrinium sp. GZWMJZ-606 was isolated from Houttuynia cordata Thunb. Further chemical investigation of this fungal strain led to the isolation of two new 4-hydroxy-2-pyridone derivatives (1 and 2) which we named furaprazone A and B (Figure 1), along with the known N-hydroxyapiosporamide (3) [18,20] and apiosporamide (4) [21]. Compounds 1 and 2 were the first reported compounds with 5-(7-oxabicyclo[2.2.1]heptane)-4-hydroxy-2-pyridone skeleton. Compared with the previously reported 1,2-epoxyhexane [17,21], 2-oxobicyclo [3.3.1] nonane [18] or benzene [22], 7-oxadicyclo [2.2.1] heptane can improve some biological activities [23,24,25]. Herein, the isolation, structure elucidation, the antimicrobial and cytotoxic activity of these compounds are described.
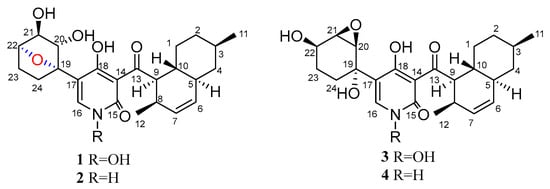
Figure 1.
Structures of compounds 1–4.
2. Results and Discussion
Structure Elucidation
Compound 1 was obtained as a yellow crystal. The molecular formula was deduced as C24H31NO7 based on the HRESIMS ion peak at m/z 468.19861 [M + Na]+ (calcd. for C24H31NO7Na = 468.19927). Its IR (KBr) spectrum exhibited absorptions at 3434 cm−1 (hydroxy), 1649 cm−1 (carbonyl), and 1605/1552/1446 cm−1 (aromatic heterocycle). Compound 1 had the same molecular formula with N-hydroxyapiosporamide (3) and showed a high degree of similarity in UV absorption. The NMR spectra displayed two methyls, five sp3-methylenes, eight sp3-methines, three sp2-methines, one sp3-quaternary carbon, five sp2-quaternary carbons (including two carbonyls) (Table 1), which was also similar to those of compound 3, especially for the important 1H NMR signals, such as two methyl groups at H3-11(δH 0.94) and H3-12 (δH 0.82), a single special hydrogen signal at H-16 (δH 7.93), two olefinic protons at H-6 (δH 5.41) and H-7 (δH 5.60). The above evidence suggested that compound 1 has a similar skeleton with compound 3. The 1H-1H COSY correlations (Figure 2) from H2-1 (δH 0.88 and 1.95) to H-10 (δH 1.58), H-3 (δH 1.50) to H3-11, and H-8 (δH 2.85) to H-12 proved the existence of a decalin moiety. The relative configurations of this part were confirmed by the NOESY correlations (Figure 2) from H-10 to H3-11/H3-12 and H-5 (δH 1.83) to H-3/H-9 (δH 4.45), and indicated that compound 1 has the same decalin moiety as compound 3. The 1H-1H COSY correlations from H-20 (δH 3.87) to H2-24 (δH 1.63 and 2.25) and the HMBC correlations (Figure 2) from H-21 (δH 4.00) and H2-23 (δH 1.70 and 2.25) to C-19 (δC 89.4) confirmed the presence of an oxygenated cyclohexane moiety in compound 1. However, there was a large chemical shift difference between these two compounds at C-19/20/21/22 (δC 89.4, 82.1, 82.9, 78.6 for 1; 70.4, 60.5, 57.7, 67.2 for 3). Nevertheless, there is still one degree of unsaturation in the structure 1, implying an oxygen bridge in this cyclohexane moiety, but the HMBC correlations cannot be used to confirm it. The key HMBC correlations from H-9 to C-13 (δC 211.4), H-16 to C-15 (δC 159.9)/C-18 (δC 173.2)/C-19 indicated that the decalin and hexane moieties substituted at C-13 and C-17 of the 4-hydroxy-2-pyridinone part. The crystal of compound 1 was fortunately acquired in methanol/water (v/v, 1:1) solution. The results of the X-ray (Figure 3) analysis (Flack parameter = −0.15 (11), CCDC: 2218951) confirmed an oxygen bridge between C-19 and C-22 forming the furan ring and led to the final determination of its absolute configuration as 3R, 5S, 8R, 9R, 10R, 19S, 20S, 21S, 22S. This novel 4-hydroxy-2-pyridone was named furanpydone A.

Table 1.
1H (600 MHz) and 13C (150 MHz) NMR data of 1–4.
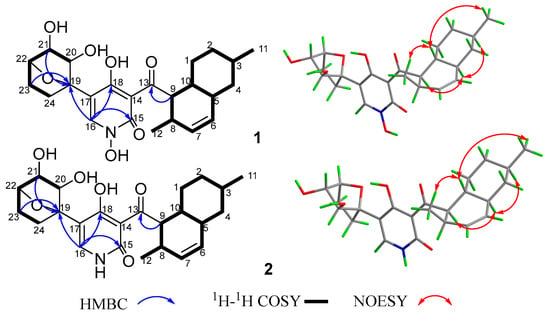
Figure 2.
The key 2D NMR correlations of compounds 1 and 2.
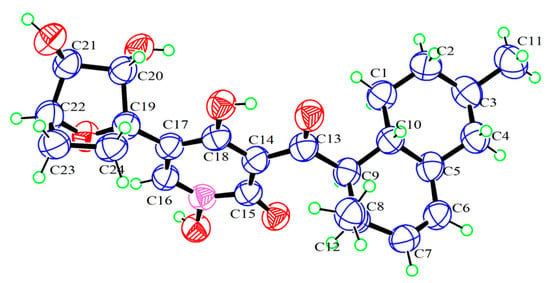
Figure 3.
ORTEP drawing of 1.
Compound 2 was obtained as a yellow powder. The molecular formula was deduced as C24H31O6N based on the HRESIMS peak at m/z 452.20319 ([M + Na]+, calcd. for 452.20436), which has ten degrees of unsaturation as furanpydone A (1), but one less oxygen atom than it. According to IR (KBr) spectrum data, they seem to have similar functional groups at 3445 cm−1 (hydroxy), 1652 cm−1 (carbonyl), and 1604/1557/1456 cm−1 (aromatic heterocycle). According to 1D NMR and HSQC data, compound 2 displayed two methyl (δH/C 0.74/17.9, 0.87/22.5), five sp3-methylene (δH/C 1.50 and 2.06/23.0; 0.81 and 1.83/29.6; 1.47 and 2.04/31.8; 0.96 and 1.67/35.1; 0.75 and 1.70/41.4), eight sp3-methines (δH/C 2.74/30.6, 1.46/32.6, 1.45/35.8, 1.75/41.4, 4.33/51.8, 4.30/76.6, 3.58/80.5, 3.78/81.2), three sp2-methines (δH/C 5.37/130.4, 5.57/131.7, 7.33/139.1), one sp3-quaternary carbon (δC 88.0), and five sp2-quaternary carbons (δC 106.7, 110.8, 161.8, 175.7, 209.6), which were extremely similar to compound 1 (Table 1 and Table S1) suggested the similar structure of these two compounds. The 1H-1H COSY correlations (Figure 2) from H2-1 (δH 0.81 and 1.83) to H-10 (δH 1.45), H-3 (δH 1.46) to H3-11 (δH 0.87), H-8 (δH 2.74) to H3-12 (δH 0.74), H-20 (δH 3.58) to H2-24 (δH 1.47 and 2.04), the key HMBC correlations (Figure 2) from H-9 (δH 4.33) to C-13 (δC 209.6), H-16 (δH 7.33) to C-15 (δC 161.8)/18 (δC 175.7)/19 (δC 88.0), H-21 (δH 3.78) to C-19 further confirmed that compound 2 and 1 have the same skeleton structure. Analysis of the NMR spectral data revealed that the chemical shift of C-15 (δC 161.8) moved to a lower field, which was similar to compound 4, the key 1H-1H COSY correlation between H-16 and H-NH (δH 11.38) confirmed the absence of an N-hydroxy group in 2. The key NOESY correlations from H-10 to H3-11/H3-12, H-5 (δH 1.75) to H-3 (δH 1.47)/H-9 (δH 4.33), as well as the same chemical shift for C-19/20/21/22/23/24 with furanpydone A (1) suggested that these two compounds had the same relative configuration. The similarity of electronic circular dichroism (ECD) curve of compound 2 (213 (−5.83), 228 (−11.74), 265 (+5.40), 310 (+7.09), 341 (−1.63) to 1 (217 (−2.73), 242 (−3.51), 270 (+2.00), 316 (+2.75), 343 (−0.27)) (Figure 4) along with the similar optical rotation values (1: −89.7, 2: −80.0) indicated the same absolute configuration for 2 and 1. This novel 4-hydroxy-2-pyridone was named furanpydone B.
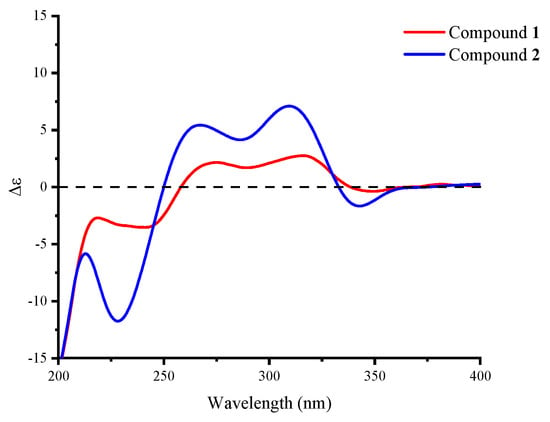
Figure 4.
Experimental CD spectra of compounds 1 and 2.
We propose a possible biosynthetic pathway for compounds 1–4. Didymellamide B was the key intermediate in the biosynthesis of these compounds [18]. The intermediates a and b were obtained by reduction from didymellamide B. Compound 2 was obtained by oxidation, hydration and cyclization reaction from a, and compound 1 was syntheszed by further oxidation. Compound 4 was obtained by two oxidation reactions from b, and compound 3 was syntheszed by further oxidation. (Figure 5).
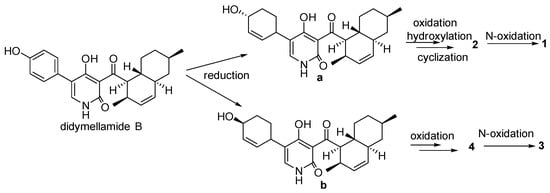
Figure 5.
Proposed biosynthetic pathway for compounds 1–4.
Compounds 1–4 were tested for their antimicrobial activities against nine pathogenic microorganisms. As shown in Table 2, compound 4 exhibited broad inhibitory activities against Staphylococcus aureus, methicillin-resistant S. aureus (MRSA), Bacillus subtilis, Clostridium perfringens, and Ralstonia solanacarum with the MIC values ranging from 1.56 to 6.25 µM. Compounds 1 and 3 showed moderate selective activities against S. aureus and MRSA with the MIC values of 12.5–25.0 µM. Compounds 1–4 showed no obvious inhibitory activity against two Gram-negative bacteria (E. coli and P. aeruginosa) and two pathogenic fungi (C. albicans and C. glabrata) at 50 µM. According to the results, it seems that the compounds with ternary epoxide showed better antibacterial activity than those with furan ring, but the effect of N-OH needs more research to determine.

Table 2.
Antimicrobial activity of 1–4 (MIC, µM), n = 3.
The antiproliferative activities against 18 cancer cell lines and one normal cell line were assayed by the CCK-8 method. Compound 1 showed significant cytotoxicity against 10 cancer cell lines, compound 3 showed activities against HCT116 and 786-O cell lines (Table 3). The compounds with furan ring showed better antiproliferative activities than those with ternary epoxide. At the same time, nitrogen hydroxyl is the necessary group for maintaining the inhibitory activity.

Table 3.
Cytotoxic activity (µM, IC50 ± SD), n = 3.
3. Materials and Methods
3.1. General Experimental Procedures
The NMR spectra were recorded on Bruker Advance NEO 600 spectrometer (Bruker Corporation, Zurich, Switzerland) using TMS as an internal standard. MS analysis were carried out on Agilent 1100 instrument (Agilent Technologies, Santa Clara, CA, USA) and Thermo ultimate 3000/Q EXACTIVE FOCUS mass spectrometers (Thermo Scientific™, Waltham, MA, USA), respectively. Optical rotations were determined on Rudolph Autopol1 automatic polarimeter (Rudolph Research Analytical, Hackettstown, NJ, USA). UV spectra were detected on a Cary 60-UV-Vis spectrometer (Agilent Technologies, Santa Clara, CA, USA). IR spectra were determined on an iCAN 9 infrared spectrophotometer (Tianjin Nengpu Technology Co., Ltd, Tianjin, China) with KBr disks. X-ray data were generated using a Bruker Smart-1000 CCD (Bruker Corporation, Billerica, MA, USA) area detector diffractometer with graphite monochromatic Cu-Kα radiation. Column chromatography was performed on silica gel (200–300 mesh; Qingdao Puke Parting Materials Co., Ltd., Qingdao, China), Sephadex LH-20 gel (Amersham Biosciences, Uppsala, Sweden). HPLC separation was performed on HITACHI Primaide with an ODS-A column (YMC-pack ODS-A, 10 × 250 mm, 5 μm, 4 mL/min). Melting point instrument (SGW X-4).
3.2. Fungal Material
The endophytic fungus Arthrinium sp. GZWMJZ-606 was isolated from the leaves of Houttuynia cordata Thunb., which was collected from Longli, Guizhou, China. The leaves were treated with 75% alcohol for 30 s, and the residual alcohol was washed with sterile water. Then 1 g of fresh leaves was grinded into a pulp and 10 mL sterile water added. The suspension (100 μL) was deposited on a rice agar plate, which was prepared from rice powder (10 g), agar (18 g), and 1 L water containing chloramphenicol (0.3%) as a bacterial inhibitor, and incubated at 28 °C for 5 days. Monoclonal was selected and streaked to purity using the same agar medium. This strain was determined as Arthrinium sp. by the phylogenetic tree (Figure S1) of the ITS sequence (GenBank No. OP810989). The strain was deposited in our laboratory of Guizhou in 20% glycerol at −80 °C.
3.3. Fermentation and Extraction
The fungal strain GZWMJZ-606 was cultured on PDA at 28 °C for 3 days and then was cut into 100 × 1000 mL Erlenmeyer flasks, each containing a solid medium prepared from 100 g rice and 110 mL distilled water. These flasks were incubated at room temperature under static conditions for 40 days. The cultures were extracted three times by EtOAc (each 500 mL) and the combined EtOAc solutions were dried in vacuo to yield the extract (480.0 g).
3.4. Isolation and Purification
The EtOAc extract (480.0 g) was fractionated into 19 fractions (Fr.1–Fr.19) by chromatography on a silica gel column using step gradient elution of petroleum ether (PE)-EtOAc (v/v, 100:1–1:1) and CH2Cl2-MeOH (v/v, 20:1–1:1). Fr.17 (9.4 g) was further separated into 15 subfractions (Fr.17.1–Fr.17.15) by Sephadex LH-20 (CH2Cl2-MeOH, v/v, 1:1). Fr.17.11 (207.6 mg) was purified by semipreparative HPLC on an ODS-A column eluting with 60% MeCN-H2O containing 0.05% trifluoroacetic acid (TFA) to yield compound 1 (35.6 mg, tR 11.1 min). Fr.17.14 (75.8 mg) was purified by semipreparative HPLC on an ODS-A column (60% MeCN-H2O containing 0.05% TFA) to yield compound 2 (6.8 mg, tR 9.3 min). Fr.17.2 (830.5 mg) was further separated into 7 subfractions (Fr.17.2.1–Fr.17.2.7). Compound 4 (12.2 mg, tR 10.1 min) was obtained from Fr.17.2.1 (57.1 mg) by semipreparative HPLC (55% MeCN-H2O containing 0.05% TFA). Fr.16 (1.6 g) was further separated into 6 subfractions (Fr.16.1–Fr.16.6), and Fr.16.4 (120.5 mg) was performed on a semipreparative ODS-A column (61% MeCN-H2O containing 0.05% TFA) to yield compound 3 (38.6 mg, tR 8.4 min).
3.5. Physical Properties and Spectral Data of 1–4
Compound 1: yellow crystal; m.p. 167.5–168.5 °C; ECD (1.12 mM, MeOH) λmax (Δε) 217 (−2.73), 242 (−3.51), 270 (+2.00), 316 (+2.75), 343 (−0.27) nm; [α−89.7 (c 0.58, MeOH); UV (MeOH) λmax (log ε) 281 (0.75), 341 (0.72) nm; IR (KBr) νmax 3434, 2913, 2953, 1649, 1605, 1446 cm−1; 1H NMR and 13C NMR data see Table 1 and Table S1 and Figures S3–S10; HRESIMS m/z 468.19861 [M + Na]+ (Figure S2), molecular formula: C24H31NO7.
X-ray crystallographic analyses of 1: C24H31NO7·CH3OH, orthorhombic, M = 477.54, a = 7.6539 (3) Å, b = 14.5093 (6) Å, c = 21.7608 (11) Å, α = 90°, β = 90°, γ = 90°, V = 2416.60 (18) Å3, T = 150 K, space group P21 21 21, Z = 4, μ (Cu Kα) = 0.807 mm−1, 8074 reflections measured, 4534 independent reflections (Rint = 0.019). The final R1 values were 0.0724 (I > 2σ (I)). The final wR (F2) values were 0.1921 (I > 2σ (I)). The final R1 values were 0.0767 (all data). The final wR(F2) values were 0.1991 (all data). The goodness of fit on F2 was 1.022. Flack parameter = −0.15 (11). CCDC: 2218951.
Compound 2: yellow powder; ECD (1.17 mM, MeOH) λmax (Δε) 213 (−5.83), 228 (−11.74), 265 (+5.40), 310 (+7.09), 341 (−1.63) [α−80.0 (c 0.20, MeOH); UV (MeOH) λmax (log ε) 235 (1.05), 270 (0.50), 338 (0.74) nm; IR (KBr) νmax 3445, 2909, 1652, 1604, 1456 cm−1; 1H NMR and 13C NMR data see Table 1 and Figures S12–S17; HRESIMS m/z 452.20319 [M + Na]+ (Figure S11), molecular formula: C24H31NO6.
Compound 3: yellow solid; the molecular formula is C24H31NO7 (m/z 444.1 [M − H]−) determined by ESIMS. [α−57.4 (c 2.3, MeOH); based on 1H NMR and 13C NMR data (Table 1, Figures S18 and S19) proved that compound 3 was N-hydroxyapiosporamide.
Compound 4: faint yellow solid powder; the molecular formula is C24H31NO6 (m/z 452.5 [M + Na]+) determined by ESIMS. [α−32.2 (c 0.87, MeOH); based on 1H NMR and 13C NMR data (Table 1, Figures S20 and S21) proved that compound 4 was apiosporamide.
3.6. Antimicrobial Activities Assay
The isolated compounds were evaluated for antibacterial activity against pathogenic microorganisms including three Gram-negative strains (Escherichia. coli ATCC11775, Pseudomonas aeruginosa ATCC10145, Ralstonia solanacarum [26]), and four Gram-positive strains (Staphylococcus aureus ATCC6538, methicillin-resistant S. aureus ATCC43300 MRSA, Clostridium perfringens ATCC13124, and Bacillus subtilis ATCC6051), and two pathogenic fungi (Candida albicans ATCC10231 and Candida glabrata ATCC2001). The tested bacterial suspensions were incubated in Luria–Bertani (LB) medium and fungi in Mueller–Hinton agar (HMA) medium at 28 °C for 12 h and diluted to be 1 × 106 CFU/mL by the same medium. Then, the DMSO solution of each compound was diluted into the corresponding concentration using the LB or MHA medium; 100 µL solution of compound was added into the first well of a 96-well plate and resulted the initial tested concentration of each compound to be 50 µmol/L (DMSO < 0.5‰ in each well) and the concentration of each compound to be 25 µmol/L (DMSO < 0.5‰ in each well) in the second well of a 96-well plate after then following this method in sequence, adding 100 µL microbial suspension into a 96-well plate. The ciprofloxacin and DMSO were used as the positive and negative controls, respectively. All experiments were repeated three times. MIC values were assessed by whether compounds can inhibit the growth of microorganisms [19].
3.7. Cytotoxic Activity Assay
Cell proliferation was measured with the CCK-8 method. By the dye of WST-8 (2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4disulfophenyl)-2H-etrazolium, monosodium salt) was reduced by dehydrogenase in cells to form a water-soluble tetrazolium salt product (formazan dye) with orange color. In the measurement, the amount of the formazan dye is proportional to the number of living cells. Finally, the cell viability can be estimated by recording the optical density (OD) of formazan dye at 450 nm using a microplate reader [27].
A cell suspension of 100 μL was dispensed (adherent cell viewed 5 × 104/mL and suspension cell viewed 9 × 104/mL) in 96-well plates. With doxorubicin hydrochloride as positive drug and DMSO as control, plates were pre-cultured for 24 h, followed by treatments with various concentrations of compound (eight concentration gradients were set for each sample for IC50 determination and three multiple holes were set for each concentration, n = 3). Keep the 96-well plates at 37 °C in an incubator with 5% CO2 for 72 h. After the aspiration of the old medium, the 10-fold diluted CCK-8 (100 μL) solution was added to each well of the plate, which was then incubated for another 3 h. An absorbance microplate reader was used to measure the absorbance at 450 nm. The optical density values (OD) of each well represented the survival/proliferation of cells. The toxicity is expressed by cell inhibition. The half inhibitory concentration (IC50) was defined as the concentration causing 50% inhibition, each group of data has 8 concentration gradient responses. The IC50 value is calculated by curve fitting using the software GraphPad Prism 8 (version 8.0.2, from GraphPad Software Inc., Boston, MA, USA), the experimental results are expressed in IC50 ± SD [28,29].
Cell inhibition rate = (ODControl − ODDrug)/(ODControl − ODBlank) × 100%.
The tested cell lines: A549: human lung cancer cells; MKN-45: human gastric cancer cells; HCT116: human colon cancer cells; K562: human chronic myeloid leukemia cells; DU145: human prostate cancer cells; SF126: human brain tumor cells; A-375: human malignant melanoma cells; MCF-7: human breast cancer cells; 786-O: human renal clear cell adenocarcinoma cells; PATU8988T: human pancreatic cancer cells; 5637: human bladder cancer cells; HeLa: human cervical cancer cells; TE-1: human esophageal cancer cells; GBC-SD: human gallbladder cancer cells; HepG2: human hepatoma cells; CAL-62: human thyroid cancer cells; HOS: human osteosarcoma cells; A-673: human rhabdomyosarcoma cells; L-02: human normal liver cells.
4. Conclusions
Two new 4-hydroxy-2-pyridone alkaloids were isolated from an endophytic fungus Aspergillus sp. GZWMJZ-606, which was obtained from Houttuynia cordata Thunb. Compounds 1 and 2 are the first example of 4-hydroxy-2-pyridone alkaloids possessing novel 7-oxidicyclo[2.2.1]heptane part. Compound 1 exhibited broad-spectrum cytotoxicity against 10 cancer cell lines with the IC50 values of 4.35–9.72 µM, and showed selective activities against S. aureus and MRSA S. aureus with MIC values of 12.5 µM. The discovery of novel 4-hydroxy-2-pyridinone alkaloids can provide a material basis for the discovery of potential drug molecules.
Supplementary Materials
The following supporting information can be downloaded at: www.mdpi.com/article/10.3390/molecules28052192/s1, ITS1 gene sequences of Arthrinium sp. GZWMJZ-606; Figure S1: Species identification of endophytic fungi strain; Table S1: 1H (600 MHz) and 13C (150 MHz) NMR data of compound 1 DMSO-d6; Figure S2: HRESIMS spectrum of 1; Figure S3: 1H NMR spectrum (600 MHz, Methanol-d4) of 1; Figure S4: 13C NMR spectrum (150 MHz, Methanol-d4) s of 1; Figure S5: HSQC spectrum (Methanol-d4) of 1; Figure S6: HMBC spectrum (Methanol-d4) of 1; Figure S7: 1H-1H COSY spectrum (Methanol-d4) of 1; Figure S8: NOESY spectrum (Methanol-d4) of 1; Figure S9: 1H NMR spectrum (600 MHz, DMSO-d6) of 1; Figure S10: 13C NMR spectrum (150 MHz, DMSO-d6) of 1; Figure S11: HRESIMS spectrum of 2; Figure S12: 1H NMR spectrum (600 MHz, DMSO-d6) of 2; Figure S13: 13C NMR spectrum (150 MHz, DMSO-d6) of 2; Figure S14: HSQC spectrum (DMSO-d6) of 2; Figure S15: HMBC spectrum (DMSO-d6) of 2; Figure S16: 1H-1H COSY spectrum (DMSO-d6) of 2; Figure S17: NOESY spectrum (DMSO-d6) of 2; Figure S18: 1H NMR spectrum (600 MHz, Methanol-d4) of 3; Figure S19: 13C NMR spectrum (150 MHz, Methanol-d4) s of 3; Figure S20: 1H NMR spectrum (600 MHz, Methanol-d4) of 4; Figure S21: 13C NMR spectrum (150 MHz, Methanol-d4) of 4.
Author Contributions
Y.Y. performed the experiments and wrote the original draft; D.W. (Dongyang Wang) analyzed the data and confirmed the structure; D.W. (Dan Wu) and M.Z. performed the biological activity test; W.H. performed the fermentation and extraction; W.Z. helped to modify the manuscript; Y.X. isolated the strain and directed the implementation of the study; L.W. designed the study and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China, grant numbers 22207020 and U1812403; Guizhou Provincial Science and Technology Projects, grant numbers QKHJC-ZK[2021]ZD017, QKHZC[2022]YB191, QKHJC-ZK [2022]YB392, and QKHZYD[2022]4015; “Light of the West” Talent Cultivation Program of Chinese Academy of Sciences, grant numbers RZ [2022]4 for L. Wang, GMU (J [2020]006, 19NSP078, 20NSP065), and 100 Leading Talents of Guizhou Province for W. Zhu.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not available.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are available from the authors.
References
- Tuson, R.V. XII.-Note on an Alkaloïd contained in the seeds of the Ricinus communis, or Castor-oil Plant. J. Chem. Soc. 1864, 17, 195–197. [Google Scholar] [CrossRef]
- Hayakawa, S.; Minato, H.; Katagori, K. The ilicicolins antibiotics from Cylindrocladium ilicicola. J. Antibiot. 1971, 24, 653–654. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Zhang, X.; Feng, H.; Che, Q.; Zhu, T.; Gu, Q.; Li, D. Campyridones A-D, pyridone alkaloids from a mangrove endophytic fungus Campylocarpon sp. HDN13-307. Tetrahedron 2016, 72, 5679–5683. [Google Scholar] [CrossRef]
- Zhang, W.Y.; Zhong, Y.; Yu, Y.; Shi, D.F.; Huang, H.Y.; Tang, X.L.; Wang, Y.H.; Chen, G.D.; Zhang, H.P.; Liu, C.L.; et al. 4-Hydroxy Pyridones from Heterologous Expression and Cultivation of the Native Host. J. Nat. Prod. 2020, 83, 3338–3346. [Google Scholar] [CrossRef] [PubMed]
- Li, L.N.; Wang, L.; Cheng, Y.N.; Cao, Z.Q.; Zhang, X.K.; Guo, X.L. Discovery and Characterization of 4-Hydroxy-2-pyridone Derivative Sambutoxin as a Potent and Promising Anticancer Drug Candidate: Activity and Molecular Mechanism. Mol. Pharm. 2018, 15, 4898–4911. [Google Scholar] [CrossRef]
- Tang, Y.; Li, J.; Zhao, S. Progress in the Study of 4-Hydroxy-2-pyridone Natural Alkaloids. Chin. J. Org. Chem. 2011, 31, 9–21. [Google Scholar]
- Sarita, S.; Neelam, Y.; Ravi, K.; Sonu, C.; Vidhi, D.; Pooja, W.; Anil, D. A score years’ update in the synthesis and biological evaluation of medicinally important 2-pyridones. Eur. J. Med. Chem. 2022, 232, 114199. [Google Scholar]
- Ando, K.; Suzuki, S.; Saeki, T.; Tamura, G.; Arima, K. Funiculosin, A New Antibiotic. I Isolation, Biological and Chemical Properties. J. Antibiot. 1969, 22, 189–194. [Google Scholar] [CrossRef]
- Wat, C.K.; Mcinnes, A.G.; Smith, D.G.; Wright, J.L.C.; Vining, L.C. The yellow pigments of Beauveria species. Structures of tenellin and bassianin. Can. J. Chem. 1977, 55, 4090–4098. [Google Scholar] [CrossRef]
- Kim, J.C.; Lee, Y.W.; Tamura, H.; Yoshizawa, T. Sambutoxin: A new mycotoxin isolated from Fusarium sambucinum. Tetrahedron Lett. 1995, 36, 1047–1050. [Google Scholar] [CrossRef]
- Isaka, M.; Chinthanom, P.; Supothina, S.; Tobwor, P.; Hywel-Jones, N.L. Pyridone and Tetramic Acid Alkaloids from the Spider Pathogenic FungusTorrubiella sp. BCC 2165. J. Nat. Prod. 2010, 73, 2057–2060. [Google Scholar] [CrossRef]
- Takahashi, S.; Kakinuma, N.; Uchida, K.; Hashimoto, R.; Yanagisawa, T.; Nakagawa, A. Pyridovericin and pyridomacrolidin: Novel metabolites from entomopathogenic fungi, Beauveria bassiana. J. Antibiot. 1998, 51, 596–598. [Google Scholar] [CrossRef]
- Li, M.; Zhang, A.; Qi, X.; Yu, R.; Li, J. A novel inhibitor of PGK1 suppresses the aerobic glycolysis and proliferation of hepatocellular carcinoma. Biomed. Pharmacother. 2023, 158, 114115. [Google Scholar] [CrossRef]
- Zhu, Y.X.; Peng, C.; Ding, W.; Hu, J.F.; Li, J. Chromenopyridin A, a new N-methoxy-1-pyridone alkaloid from the endophytic fungus Penicillium nothofagi P-6 isolated from the critically endangered conifer Abies beshanzuensis. Nat. Prod. Res. 2022, 36, 2049–2055. [Google Scholar] [CrossRef]
- Chicca, A.; Berg, R.; Jessen, H.J.; Marck, N.; Schmind, F.; Burch, P.; Gertch, J.; Gademann, K. Biological evaluation of pyridone alkaloids on the endocannabinoid system. Bioorgan. Med. Chem. 2017, 25, 6102–6114. [Google Scholar] [CrossRef]
- Huang, B.; Lu, H.; Zhang, Y.; Gan, X.; Wang, X.; Liu, Y.; Luo, X. Bioactive Alkaloids from the Beibu Gulf Coral-associated Fungus Acremonium sclerotigenum GXIMD 02501. Rec. Nat. Prod. 2023, 17, 165–169. [Google Scholar]
- Han, J.; Liu, C.; Li, L.; Zhou, H.; Liu, L.; Bao, L.; Chen, Q.; Song, F.; Zhang, L.; Li, E.; et al. Decalin-Containing Tetramic Acids and 4-Hydroxy-2-pyridones with Antimicrobial and Cytotoxic Activity from the Fungus Coniochaeta cephalothecoides Collected in Tibetan Plateau (Medog). J. Org. Chem. 2017, 82, 11474–11486. [Google Scholar] [CrossRef]
- Wang, J.; Wei, X.; Qin, X.; Lin, X.; Zhou, X.; Liao, S.; Yang, B.; Liu, J.; Tu, Z.; Liu, Y. Arthpyrones A−C, Pyridone Alkaloids from a Sponge-Derived Fungus Arthrinium arundinis ZSDS1-F3. Org. Lett. 2015, 17, 656–659. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, L.; Zhu, G.; Zuo, M.; Gong, Q.; He, W.; Li, M.; Yuan, C.; Hao, X.; Zhu, W. New phenylpyridone derivatives from the Penicillium sumatrense GZWMJZ-313, a fungal endophyte of Garcinia multiflora. Chin. Chem. Lett. 2019, 30, 431–434. [Google Scholar] [CrossRef]
- Williams, D.R.; Kammler, D.C.; Donnell, A.F.; Goundry, W.R.F. Total Synthesis of (+)-Apiosporamide: Assignment of Relative and Absolute Configuration. Angew. Chem. Int. Ed. 2005, 44, 6715–6718. [Google Scholar] [CrossRef]
- Bao, J.; Zhai, H.; Zhu, K.; Yu, J.H.; Zhang, Y.; Wang, Y.; Jiang, C.S.; Zhang, X.; Zhang, Y.; Zhang, H. Bioactive Pyridone Alkaloids from a Deep-Sea-Derived Fungus Arthrinium sp. UJNMF0008. Mar. Drugs 2018, 16, 174. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Umeokoli, B.O.; Eze, P.; Heering, C.; Janiak, C.; Müller, W.E.G.; Orfali, R.S.; Hartmann, R.; Dai, H.; Lin, W.; et al. Secondary metabolites of the lichen-associated fungus Apiospora montagnei. Tetrahedron Lett. 2017, 58, 1702–1705. [Google Scholar] [CrossRef]
- Bockstahler, E.R.; Weaver, L.C.; Wright, D.L. 7-Oxabicyclo [2.2.1] heptane-2,3-dicarboximides with anticonvulsant activity. J. Med. Chem. 1968, 11, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Misra, R.N.; Brown, B.R.; Sher, P.M.; Patel, M.M.; Hall, S.E.; Han, W.C.; Barrish, J.C.; Kocy, O.; Harris, D.N.; Goldenberg, H.J.; et al. Interphenylene 7-oxabicyclo [2.2.1] heptane oxazoles. Highly potent, selective, and long-acting thromboxane A2 receptor antagonists. J. Med. Chem. 1993, 36, 1401–1417. [Google Scholar] [CrossRef]
- Walter, W.G. Antitumor Imide Derivatives of 7-Oxabicyclo [2.2.1] heptane-2,3-dimethyl-2,3-dicarboxylic Acid. J. Pharm. Sci. 1989, 78, 66–67. [Google Scholar] [CrossRef]
- Li, Y.; Zeng, Y.; Wang, Z.; Fan, Y.; Yang, X.; Wang, Z.; Yu, S.; Pang, Q.; Cao, A. Deoxymikanolide adversely altered physiology and ultrastructure of Ralstonia solanacearum. Pestic. Biochem. Phys. 2021, 174, 104803. [Google Scholar] [CrossRef]
- Dockray, G.J. Cholecystokinins in rat cerebral cortex: Identification, purification and characterization by immunochemical methods. Brain Res. 1980, 188, 155–165. [Google Scholar] [CrossRef]
- Cai, L.; Qin, X.; Xu, Z.; Song, Y.; Jiang, H.; Wu, Y.; Ruan, H.; Chen, J. Comparison of Cytotoxicity Evaluation of Anticancer Drugs between Real-Time Cell Analysis and CCK-8 Method. ACS Omega 2019, 4, 12036–12042. [Google Scholar] [CrossRef]
- Wufuer, H.; Xu, Y.; Wu, D.; He, W.; Wang, D.; Zhu, W.; Wang, L. Liglaurates A–E, cytotoxic bis (lauric acid-12yl) lignanoates from the rhizomes of Drynaria roosii Nakaike. Phytochemistry 2022, 198, 113143. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
