CSDR Coupling with Exo III for Ultrasensitive Electrochemistry Determination of miR-145
Abstract
:1. Introduction
2. Results and Discussions
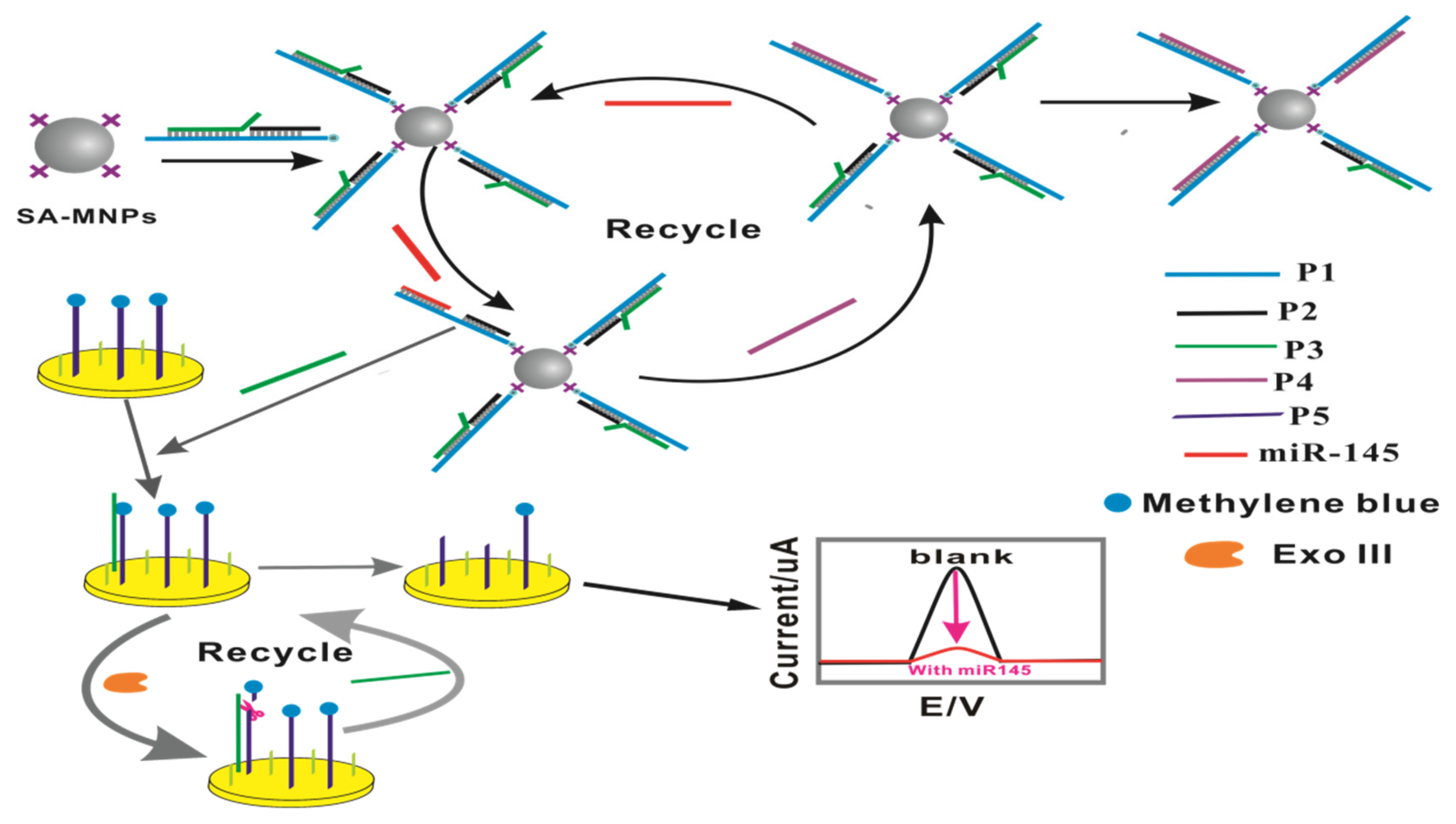
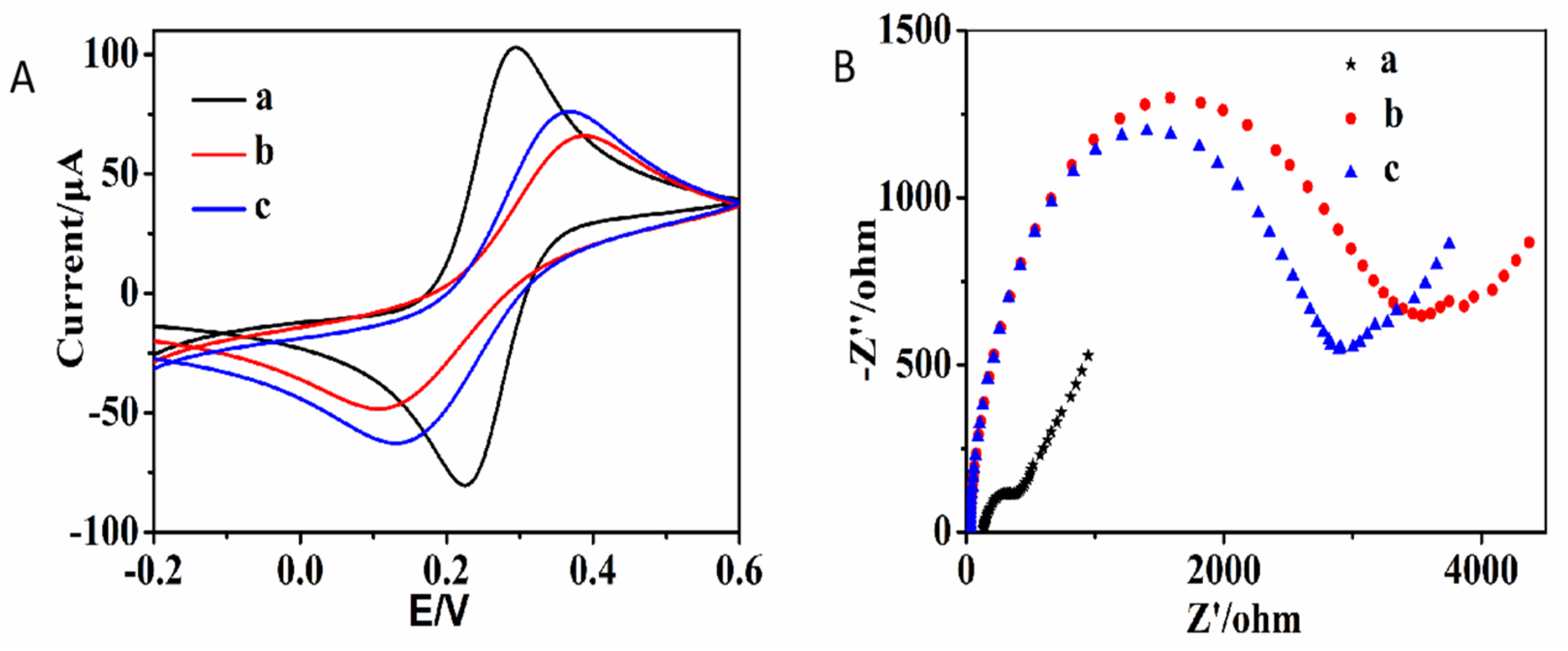
2.1. The Principle and Feasibility of This Biosensor
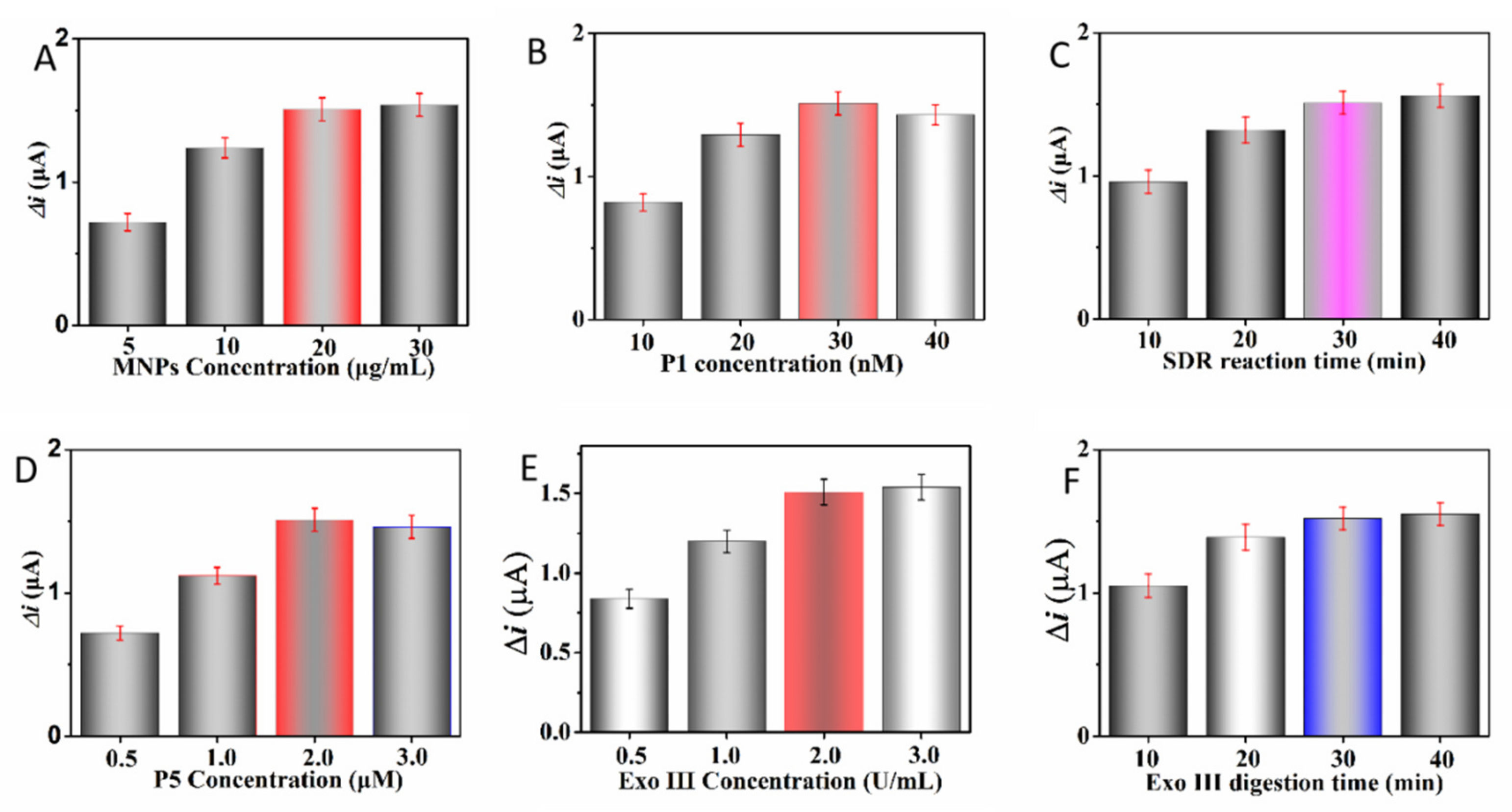
2.2. Optimization of the Assay Conditions
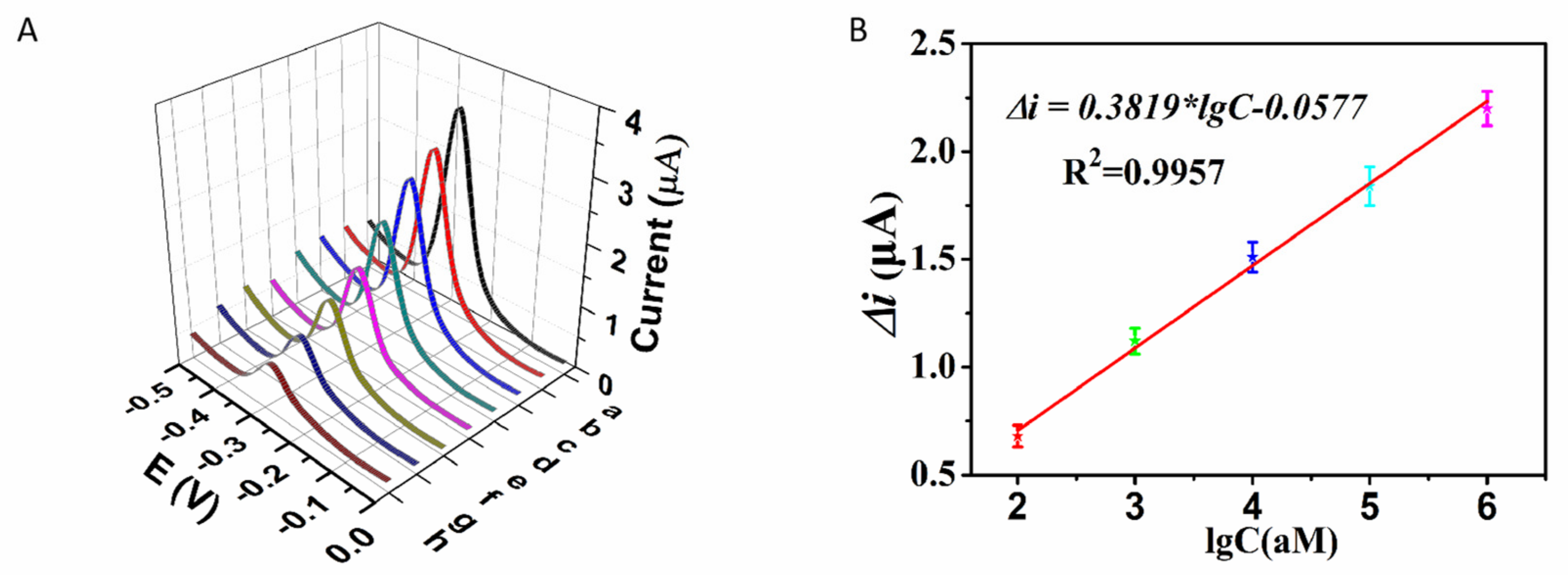
2.3. Sensitivity and Specificity for the Detection of miR-145
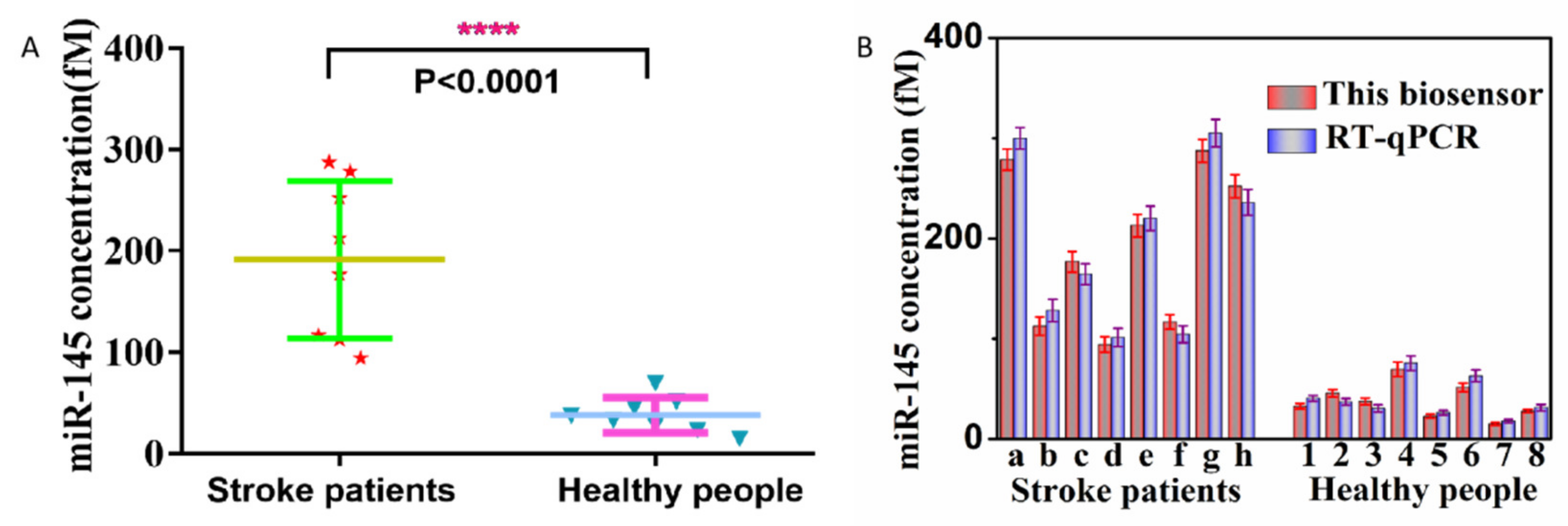
2.4. Clinical Samples
3. Experimental Procedure
3.1. Materials and Reagents
3.2. Preparation of the Modified Electrode
3.3. Determination of miR-145 and Clinical Samples
3.4. qRT-PCR Experiment Determination of miR-145
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Donnan, G.A.; Fisher, M.; Macleod, M.; Davis, S.M. Stroke. Lancet 2008, 371, 1612–1623. [Google Scholar] [CrossRef] [PubMed]
- Lopez, A.D.; Mathers, C.D.; Ezzati, M.; Jamison, D.T.; Murray, C.J. Global and regional burden of disease and risk factors, 2001: Systematic analysis of population health data. Lancet 2006, 367, 1747–1757. [Google Scholar] [CrossRef] [PubMed]
- López-Espuela, F.; González-Gil, T.; Jiménez-Gracia, M.A.; Bravo-Fernández, S.; Amarilla-Donoso, J. [Impact on quality of life in caregivers of stroke survivors]. Enferm Clin. 2015, 25, 49–56. [Google Scholar] [CrossRef] [PubMed]
- de Leciñana, M.A.; Egido, J.A.; Casado, I.; Ribó, M.; Dávalos, A.; Masjuan, J.; Caniego, J.L.; Martínez Vila, E.; Díez Tejedor, E.; ad hoc committee of the SEN Study Group for Cerebrovascular Diseases; et al. Guidelines for the treatment of acute ischaemic stroke. Neurologia 2014, 29, 102–122. [Google Scholar] [CrossRef]
- Novo, A.C.; Pinzón-Benavides, P.A.; Rozas-Fernández, P.; Martínez-Palicio, M.; García-Rúa, A.; Antón-González, C.; Casado-Menéndez, I.; Suárez-Moro, R.; Temprano-Fernández, M.T. [Ischaemic or transient attack? Magnetic resonance imaging in transient ischaemic attack: A review of 106 cases]. Rev. Neurol. 2022, 75, 333–339. [Google Scholar] [PubMed]
- Lehmonen, L.; Putaala, J.; Pöyhönen, P.; Kuusisto, J.; Pirinen, J.; Sinisalo, J.; Järvinen, V. MRI-derived cardiac washout is slowed in the left ventricle and associated with left ventricular non-compaction in young patients with cryptogenic ischemic stroke. Int. J. Cardiovasc. Imaging 2022, 38, 2395–2402. [Google Scholar] [CrossRef]
- Ferreira, B.; Caetano, J.; Barahona, F.; Lopes, R.; Carneiro, E.; Costa-Silva, B.; João, C. Liquid biopsies for multiple myeloma in a time of precision medicine. J. Mol. Med. 2020, 98, 513–525. [Google Scholar] [CrossRef] [Green Version]
- Xu, T.; Zhou, J.; Li, X.; Ke, W.; Liu, J.; Gao, H.; Dai, H. Electrochemical sensing technology for liquid biopsy of circulating tumor cells—A review. Bioelectrochemistry 2021, 140, 107823. [Google Scholar] [CrossRef]
- Shi, J.; Lin, Y.; Qin, W.; Li, M.; Zhou, Y.; Wu, Y.; Luo, H.; Huang, K.-J.; Tan, X. Superior performance of a graphdiyne self-powered biosensor with exonuclease III-assisted signal amplification for sensitive detection of microRNAs. Analyst 2022, 147, 4991–4999. [Google Scholar] [CrossRef]
- Sun, A.L.; Zhang, Y.F.; Sun, G.P.; Wang, X.N.; Tang, D.P. Homogeneous electrochemical detection of ochratoxin A in foodstuff using aptamer–graphene oxide nanosheets and DNase I-based target recycling reaction. Biosens. Bioelectron. 2017, 89, 659–665. [Google Scholar] [CrossRef]
- Li, K.; Tu, J.; Zhang, Y.; Jin, D.; Li, T.; Li, J.; Ni, W.; Xiao, M.-M.; Zhang, Z.-Y.; Zhang, G.-J. Ultrasensitive detection of exosomal miRNA with PMO-graphene quantum dots-functionalized field-effect transistor biosensor. iScience 2022, 25, 104522. [Google Scholar] [CrossRef]
- Chen, X.; Xu, K.; Li, J.; Yang, M.; Li, X.; Chen, Q.; Lu, C.; Yang, H. Switch-conversional ratiometric fluorescence biosensor for miRNA detection. Biosens. Bioelectron. 2020, 155, 112104. [Google Scholar] [CrossRef]
- Li, D.; Xia, L.; Zhou, Q.; Wang, L.; Chen, D.; Gao, X.; Li, Y. Label-Free Detection of miRNA Using Surface-Enhanced Raman Spectroscopy. Anal. Chem. 2020, 92, 12769–12773. [Google Scholar] [CrossRef]
- Han, K.; Liu, H.; Cui, J.; Liu, Y.; Pan, P. Recent strategies for electrochemical sensing detection of miRNAs in lung cancer. Anal. Biochem. 2022, 661, 114986. [Google Scholar] [CrossRef]
- Liu, Q.; Liu, J.; He, N.; Zhang, M.; Wu, L.; Chen, X.; Zhu, J.; Ran, F.; Chen, Q.; Zhang, H. CRISPR/Cas12a Coupling with Magnetic Nanoparticles and Cascaded Strand Displacement Reaction for Ultrasensitive Fluorescence Determination of Exosomal miR-21. Molecules 2022, 27, 5338. [Google Scholar] [CrossRef]
- Fang, C.; Ouyang, P.; Yang, Y.; Qing, Y.; Han, J.; Shang, W.; Chen, Y.; Du, J. MiRNA Detection Using a Rolling Circle Amplification and RNA-Cutting Allosteric Deoxyribozyme Dual Signal Amplification Strategy. Biosensors 2021, 11, 222. [Google Scholar] [CrossRef]
- Yao, Y.; Zhang, H.; Tian, T.; Liu, Y.; Zhu, R.; Ji, J.; Liu, B. Iodide-modified Ag nanoparticles coupled with DSN-Assisted cycling amplification for label-free and ultrasensitive SERS detection of MicroRNA-21. Talanta 2021, 235, 122728. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, J.; Xie, H.; Liu, H.; Liu, S.; He, D.; Mi, P.; He, S.; Wang, J.; Sun, Y. NIR-to-Vis Handheld Platforms for Detecting miRNA Level and Mutation Based on Sub-10 nm Sulfide Nanodots and HCR Amplification. ACS Appl. Mater. Interfaces 2022, 14, 10212–10226. [Google Scholar] [CrossRef]
- Yan, B.; Zheng, X.; Shi, P. Electrochemical sensor propelled by exonuclease III for highly efficient microRNA-155 detection. Analyst 2022, 147, 4824–4828. [Google Scholar] [CrossRef]
- Hoque, S.; Gonçales, V.R.; Bakthavathsalam, P.; Tilley, R.D.; Gooding, J.J. A calibration-free approach to detecting microRNA with DNA-modified gold coated magnetic nanoparticles as dispersible electrodes. Anal. Methods 2022, 14, 4861–4866. [Google Scholar] [CrossRef]
- Beyrampour-Basmenj, H.; Pourhassan-Moghamddam, M.; Nakhjavani, S.A.; Faraji, N.; Alivand, M.; Zarghami, N.; Talebi, M.; Rahmati, M.; Ebrahimi-Kalan, A. Sensitive and convenient detection of miRNA-145 using a gold nanoparticle-HCR coupled system: Computational and in vitro validations. IEEE Trans. Nanobiosci. 2022. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.X.; Xie, T.J.; Li, C.H.; Ye, Q.C.; Tian, L.L.; Li, Y.F.; Huang, C.Z.; Zhen, S.J. A crosslinked submicro-hydrogel formed by DNA circuit-driven protein aggregation amplified fluorescence anisotropy for biomolecules detection. Anal. Chim. Acta 2021, 1154, 338319. [Google Scholar] [CrossRef] [PubMed]
- Aviñó, A.; Huertas, C.S.; Lechuga, L.M.; Eritja, R. Sensitive and label-free detection of miRNA-145 by triplex formation. Anal. Bioanal. Chem. 2016, 408, 885–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, J.A.; Sales MG, F.; Pereira, C.M. Electrochemistry-assisted surface plasmon resonance detection of miRNA-145 at femtomolar level. Sens. Actuators B Chem. 2020, 316, 128129. [Google Scholar] [CrossRef]
- Mohamadi, M.; Mostafavi, A.; Torkzadeh-Mahani, M. Design of a Sensitive and Selective Electrochemical Aptasensor for the Determination of the Complementary cDNA of miRNA-145 Based on the Intercalation and Electrochemical Reduction of Doxorubicin. J. AOAC Int. 2017, 100, 1754–1760. [Google Scholar] [CrossRef]
- Chen, H.; Li, Z.-Y.; Chen, J.; Yu, H.; Zhou, W.; Shen, F.; Chen, Q.; Wu, L. CRISPR/Cas12a-based electrochemical biosensor for highly sensitive detection of cTnI. Bioelectrochemistry 2022, 146, 108167. [Google Scholar] [CrossRef]
| Analytical Methods | Detection Limit | Linear Range | Ref. |
|---|---|---|---|
| Gold nanoparticle-HCR coupled system | 0.519 nM | 1 pM–1 nM | [21] |
| Fluorescence anisotropy (FA) amplifier methods | 2.5 nM | 6.0 nM to 21.0 nM | [22] |
| Modified parallel tail-clamps methods | 2.43 nM | 2 nM to 30 nM | [23] |
| eSPR methods | 0.56 fM | 1.0 fM to 10 nM | [24] |
| DNA electrochemical aptasensor | 0.27 nM | 2.0 to 80.0 nM | [25] |
| CSDR coupling with Exo III | 100 aM | 1 × 102 to 1 × 106 aM | This method |
| Name | Sequences Description |
|---|---|
| P1 | 5′-biotin-TTTTTTTTGAA A TG G TG GAA AGG AGG G AG GGA TTC CTG GGA AAA CTGGAC-3′ |
| P2 | 5′-CCTTTCCACCATTTC-3′ |
| P3 | 5′-TTT TCC CAG GAA TCC CTC CCT AAC TGC-3′ |
| P4 | 5′-TT TCC CAG TCC CCT CC TTT CCA CCA TTT-3′ |
| MiR-145 | 5′-G UCCAG UUU UCC CAG GAA UCC CU-3′ |
| SM | 5′-CUCCAG UUU UCC CAG GAA UCC CU-3′ |
| DM | 5′-CACCAG UUU UCC CAG GAA UCC CU-3′ |
| TM | 5′-CAGAG UUU UCC CAG GAA UCC CU-3′ |
| Random | 5′-UUAGCUUAUCAGACUGAUGUUGA-3′ |
| P5 | 5′-SH-TTTTTT GTT AGG GAG GGA TTC CTG GGA-MB-3′ |
| primer | 5′-GTCCAGTTTTCC CAG GAA TCC CT-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Yang, Y.; Xin, L.; Zhang, H.; Wu, L.; Zhu, J.; Zhu, J.; Liu, S.; Wang, Z.; Chen, Q.; et al. CSDR Coupling with Exo III for Ultrasensitive Electrochemistry Determination of miR-145. Molecules 2023, 28, 2208. https://doi.org/10.3390/molecules28052208
Zhang M, Yang Y, Xin L, Zhang H, Wu L, Zhu J, Zhu J, Liu S, Wang Z, Chen Q, et al. CSDR Coupling with Exo III for Ultrasensitive Electrochemistry Determination of miR-145. Molecules. 2023; 28(5):2208. https://doi.org/10.3390/molecules28052208
Chicago/Turabian StyleZhang, Moli, Yang Yang, Lingyi Xin, Hua Zhang, Lun Wu, Jun Zhu, Jing Zhu, Shiyun Liu, Zhaohui Wang, Qinhua Chen, and et al. 2023. "CSDR Coupling with Exo III for Ultrasensitive Electrochemistry Determination of miR-145" Molecules 28, no. 5: 2208. https://doi.org/10.3390/molecules28052208
APA StyleZhang, M., Yang, Y., Xin, L., Zhang, H., Wu, L., Zhu, J., Zhu, J., Liu, S., Wang, Z., Chen, Q., & Yang, G. (2023). CSDR Coupling with Exo III for Ultrasensitive Electrochemistry Determination of miR-145. Molecules, 28(5), 2208. https://doi.org/10.3390/molecules28052208








