An Expedited Route to Optical and Electronic Properties at Finite Temperature via Unsupervised Learning
Abstract
1. Introduction
2. Results and Discussion
2.1. The TCNE::1ClN Case Study
2.2. The N34− Case Study
3. Materials and Methods
3.1. Ab Initio Molecular Dynamics
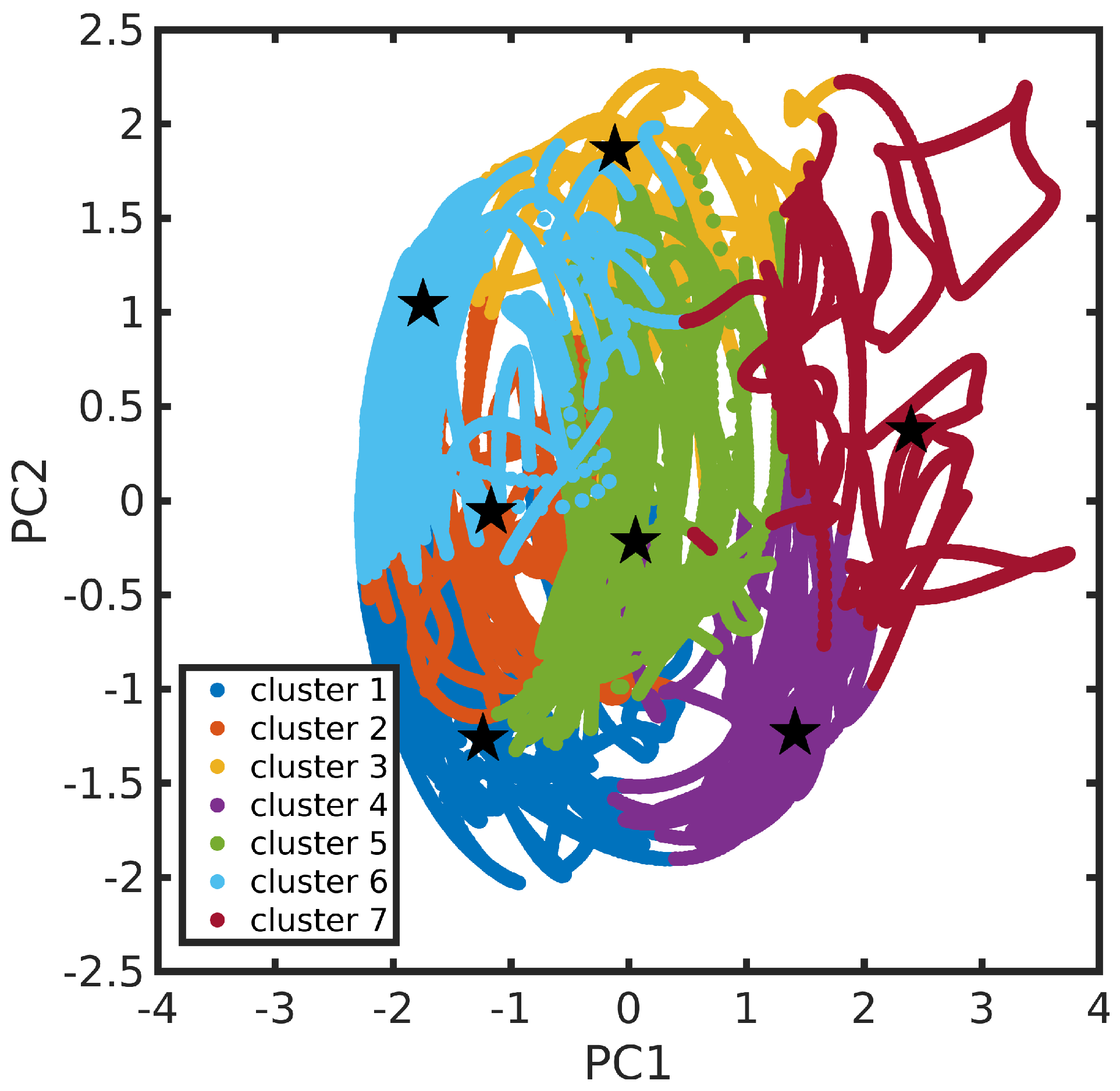
3.2. Feature Selection and Clustering of Molecular Dynamics Trajectories
3.3. Dimensionality Reduction for MD Data Visualization
3.4. Excited State Characterization and Spectra Simulations
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 1ClN | 1-chloronaphthalene |
| ADMP | Atom-centered density matrix propagation |
| C-PCM | Conductor-like polarizable continuum model |
| CT | Charge transfer |
| dcbpy | 4,4′-dicarboxy-2,2′-bipyridine |
| DCM | dichloromethane |
| DFT | Density functional theory |
| LMCT | Ligand-to-metal charge-transfer |
| MD | Molecular dynamics |
| ML | Machine learning |
| MLCT | Metal-to-ligand charge-transfer |
| MM | Molecular mechanics |
| N34− | [Ru(dcbpy)2(NCS)2]4− |
| PC | Principal component |
| PCA | Principal component analysis |
| PES | Potential energy surface |
| QM | Quantum mechanics |
| SDF | Spatial distribution function |
| TCNE | Tetracyanoethylene |
| TD-DFT | Time dependent density functional theory |
References
- Adamo, C.; Cossi, M.; Rega, N.; Barone, V. Chapter 12—New computational strategies for the quantum mechanical study of biological systems in condensed phases. In Theoretical Biochemistry; Eriksson, L.A., Ed.; Elsevier: Amsterdam, The Netherlands, 2001; Volume 9: Theoretical and Computational Chemistry; pp. 467–538. [Google Scholar] [CrossRef]
- Barone, V.; Improta, R.; Rega, N. Quantum Mechanical Computations and Spectroscopy: From Small Rigid Molecules in the Gas Phase to Large Flexible Molecules in Solution. Acc. Chem. Res. 2008, 41, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Reichardt, C. Solvatochromic Dyes as Solvent Polarity Indicators. Chem. Rev. 1994, 94, 2319–2358. [Google Scholar] [CrossRef]
- Barone, V.; Polimeno, A. Integrated computational strategies for UV/vis spectra of large molecules in solution. Chem. Soc. Rev. 2007, 36, 1724–1731. [Google Scholar] [CrossRef] [PubMed]
- Krystkowiak, E.; Dobek, K.; Maciejewski, A. Origin of the strong effect of protic solvents on the emission spectra, quantum yield of fluorescence and fluorescence lifetime of 4-aminophthalimide: Role of hydrogen bonds in deactivation of S1-4-aminophthalimide. J. Photochem. Photobiol. 2006, 184, 250–264. [Google Scholar] [CrossRef]
- Solntsev, K.M.; Huppert, D.; Agmon, N. Photochemistry of “Super”-Photoacids. Solvent Effects. J. Phys. Chem. A 1999, 103, 6984–6997. [Google Scholar] [CrossRef]
- Solntsev, K.M.; Huppert, D.; Tolbert, L.M.; Agmon, N. Solvatochromic shifts of “super” photoacids. J. Am. Chem. Soc. 1998, 120, 7981–7982. [Google Scholar] [CrossRef]
- Coppola, F.; Nucci, M.; Marazzi, M.; Rocca, D.; Pastore, M. Norbornadiene/Quadricyclane System in the Spotlight: The Role of Rydberg States and Dynamic Electronic Correlation in a Solar-Thermal Building Block. ChemPhotoChem 2023, e202200214. [Google Scholar] [CrossRef]
- Frank, H.A.; Bautista, J.A.; Josue, J.; Pendon, Z.; Hiller, R.G.; Sharples, F.P.; Gosztola, D.; Wasielewski, M.R. Effect of the Solvent Environment on the Spectroscopic Properties and Dynamics of the Lowest Excited States of Carotenoids. J. Phys. Chem. B 2000, 104, 4569–4577. [Google Scholar] [CrossRef]
- Raucci, U.; Perrella, F.; Donati, G.; Zoppi, M.; Petrone, A.; Rega, N. Ab-initio molecular dynamics and hybrid explicit-implicit solvation model for aqueous and nonaqueous solvents: GFP chromophore in water and methanol solution as case study. J. Comput. Chem. 2020, 41, 2228–2239. [Google Scholar] [CrossRef]
- Donati, G.; Petrone, A.; Rega, N. Multiresolution continuous wavelet transform for studying coupled solute–solvent vibrations via ab initio molecular dynamics. Phys. Chem. Chem. Phys. 2020, 22, 22645–22661. [Google Scholar] [CrossRef]
- Coppola, F.; Perrella, F.; Petrone, A.; Donati, G.; Rega, N. A not obvious correlation between the structure of green fluorescent protein chromophore pocket and hydrogen bond dynamics: A choreography from ab initio molecular dynamics. Front. Mol. Biosci. 2020, 7, 569990. [Google Scholar] [CrossRef]
- Raucci, U.; Savarese, M.; Adamo, C.; Ciofini, I.; Rega, N. Intrinsic and Dynamical Reaction Pathways of an Excited State Proton Transfer. J. Phys. Chem. B 2015, 119, 2650–2657. [Google Scholar] [CrossRef]
- Petrone, A.; Caruso, P.; Tenuta, S.; Rega, N. On the optical absorption of the anionic GFP chromophore in vacuum, solution, and protein. Phys. Chem. Chem. Phys. 2013, 15, 20536–20544. [Google Scholar] [CrossRef]
- Langella, E.; Rega, N.; Improta, R.; Crescenzi, O.; Barone, V. Conformational analysis of the tyrosine dipeptide analogue in the gas phase and in aqueous solution by a density functional/continuum solvent model. J. Comput. Chem. 2002, 23, 650–661. [Google Scholar] [CrossRef]
- Cerezo, J.; Petrone, A.; Ferrer, F.J.A.; Donati, G.; Santoro, F.; Improta, R.; Rega, N. Electronic spectroscopy of a solvatochromic dye in water: Comparison of static cluster/implicit and dynamical/explicit solvent models on structures and energies. Theor. Chem. Acc. 2016, 135, 263. [Google Scholar] [CrossRef]
- Cimino, P.; Raucci, U.; Donati, G.; Chiariello, M.G.; Schiazza, M.; Coppola, F.; Rega, N. On the different strength of photoacids. Theor. Chem. Acc. 2016, 135, 117. [Google Scholar] [CrossRef]
- Kim, P.; Valentine, A.J.S.; Roy, S.; Mills, A.W.; Chakraborty, A.; Castellano, F.N.; Li, X.; Chen, L.X. Ultrafast Excited-State Dynamics of Photoluminescent Pt(II) Dimers Probed by a Coherent Vibrational Wavepacket. J. Phys. Chem. Lett. 2021, 12, 6794–6803. [Google Scholar] [CrossRef]
- Lu, L.; Wildman, A.; Jenkins, A.J.; Young, L.; Clark, A.E.; Li, X. The “Hole” Story in Ionized Water from the Perspective of Ehrenfest Dynamics. J. Phys. Chem. Lett. 2020, 11, 9946–9951. [Google Scholar] [CrossRef]
- Leger, J.D.; Friedfeld, M.R.; Beck, R.A.; Gaynor, J.D.; Petrone, A.; Li, X.; Cossairt, B.M.; Khalil, M. Carboxylate Anchors Act as Exciton Reporters in 1.3 nm Indium Phosphide Nanoclusters. J. Phys. Chem. Lett. 2019, 10, 1833–1839. [Google Scholar] [CrossRef]
- Nascimento, D.R.; Zhang, Y.; Bergmann, U.; Govind, N. Near-Edge X-ray Absorption Fine Structure Spectroscopy of Heteroatomic Core-Hole States as a Probe for Nearly Indistinguishable Chemical Environments. J. Phys. Chem. Lett. 2020, 11, 556–561. [Google Scholar] [CrossRef]
- Alberto, M.E.; De Simone, B.C.; Mazzone, G.; Quartarolo, A.D.; Russo, N. Theoretical Determination of Electronic Spectra and Intersystem Spin–Orbit Coupling: The Case of Isoindole-BODIPY Dyes. J. Chem. Theory Comput. 2014, 10, 4006–4013. [Google Scholar] [CrossRef] [PubMed]
- Barone, V.; Alessandrini, S.; Biczysko, M.; Cheeseman, J.R.; Clary, D.C.; McCoy, A.B.; DiRisio, R.J.; Neese, F.; Melosso, M.; Puzzarini, C. Computational molecular spectroscopy. Nat. Rev. Methods Prim. 2021, 1, 38. [Google Scholar] [CrossRef]
- Barone, V.; Bloino, J.; Biczysko, M.; Santoro, F. Fully Integrated Approach to Compute Vibrationally Resolved Optical Spectra: From Small Molecules to Macrosystems. J. Chem. Theory Comput. 2009, 5, 540–554. [Google Scholar] [CrossRef] [PubMed]
- Santoro, F.; Lami, A.; Improta, R.; Barone, V. Effective method to compute vibrationally resolved optical spectra of large molecules at finite temperature in the gas phase and in solution. J. Chem. Phys. 2007, 126, 184102. [Google Scholar] [CrossRef]
- Avila Ferrer, F.J.; Cerezo, J.; Stendardo, E.; Improta, R.; Santoro, F. Insights for an Accurate Comparison of Computational Data to Experimental Absorption and Emission Spectra: Beyond the Vertical Transition Approximation. J. Chem. Theory Comput. 2013, 9, 2072–2082. [Google Scholar] [CrossRef]
- Dierksen, M.; Grimme, S. Density functional calculations of the vibronic structure of electronic absorption spectra. J. Chem. Phys. 2004, 120, 3544–3554. [Google Scholar] [CrossRef]
- Isborn, C.M.; Gotz, A.W.; Clark, M.A.; Walker, R.C.; Martínez, T.J. Electronic absorption spectra from MM and ab initio QM/MM molecular dynamics: Environmental effects on the absorption spectrum of photoactive yellow protein. J. Chem. Theory Comput. 2012, 8, 5092–5106. [Google Scholar] [CrossRef]
- Pagliai, M.; Mancini, G.; Carnimeo, I.; De Mitri, N.; Barone, V. Electronic absorption spectra of pyridine and nicotine in aqueous solution with a combined molecular dynamics and polarizable QM/MM approach. J. Chem. Theory Comput. 2017, 38, 319–335. [Google Scholar] [CrossRef]
- Mendanha, K.; Prado, R.C.; Oliveira, L.B.; Colherinhas, G. TD-DFT absorption spectrum of (poly) threonine in water: A study combining molecular dynamics and quantum mechanics calculations. Chem. Phys. Lett. 2021, 779, 138876. [Google Scholar] [CrossRef]
- Kasper, J.M.; Williams-Young, D.B.; Vecharynski, E.; Yang, C.; Li, X. A Well-Tempered Hybrid Method for Solving Challenging Time-Dependent Density Functional Theory (TDDFT) Systems. J. Chem. Theory Comput. 2018, 14, 2034–2041. [Google Scholar] [CrossRef]
- Van Beeumen, R.; Williams-Young, D.B.; Kasper, J.M.; Yang, C.; Ng, E.G.; Li, X. Model Order Reduction Algorithm for Estimating the Absorption Spectrum. J. Chem. Theory Comput. 2017, 13, 4950–4961. [Google Scholar] [CrossRef]
- Alberto, M.E.; Mazzone, G.; Quartarolo, A.D.; Sousa, F.F.R.; Sicilia, E.; Russo, N. Electronic spectra and intersystem spin-orbit coupling in 1,2- and 1,3-squaraines. J. Comput. Chem. 2014, 35, 2107–2113. [Google Scholar] [CrossRef]
- Petrone, A.; Perrella, F.; Coppola, F.; Crisci, L.; Donati, G.; Cimino, P.; Rega, N. Ultrafast photo-induced processes in complex environments: The role of accuracy in excited-state energy potentials and initial conditions. Chem. Phys. Rev. 2022, 3, 021307. [Google Scholar] [CrossRef]
- Coppola, F.; Cimino, P.; Perrella, F.; Crisci, L.; Petrone, A.; Rega, N. Electronic and Vibrational Manifold of Tetracyanoethylene–Chloronaphthalene Charge Transfer Complex in Solution: Insights from TD-DFT and Ab Initio Molecular Dynamics. J. Phys. Chem. A 2022, 126, 7179–7192. [Google Scholar] [CrossRef]
- Segatta, F.; Nenov, A.; Nascimento, D.R.; Govind, N.; Mukamel, S.; Garavelli, M. iSPECTRON: A simulation interface for linear and nonlinear spectra with ab-initio quantum chemistry software. J. Comput. Chem. 2021, 42, 644–659. [Google Scholar] [CrossRef]
- Petrenko, T.; Neese, F. Analysis and prediction of absorption band shapes, fluorescence band shapes, resonance Raman intensities and excitation profiles using the time-dependent theory of electronic spectroscopy. J. Chem. Phys. 2007, 127, 164319. [Google Scholar] [CrossRef]
- Petrone, A.; Cerezo, J.; Ferrer, F.J.A.; Donati, G.; Improta, R.; Rega, N.; Santoro, F. Absorption and Emission Spectral Shapes of a Prototype Dye in Water by Combining Classical/Dynamical and Quantum/Static Approaches. J. Phys. Chem. A 2015, 119, 5426–5438. [Google Scholar] [CrossRef]
- De Mitri, N.; Monti, S.; Prampolini, G.; Barone, V. Absorption and emission spectra of a flexible dye in solution: A computational time-dependent approach. J. Chem. Theory Comput. 2013, 9, 4507–4516. [Google Scholar] [CrossRef]
- Hoffman, D.P.; Ellis, S.R.; Mathies, R.A. Characterization of a conical intersection in a charge-transfer dimer with two-dimensional time-resolved stimulated Raman spectroscopy. J. Phys. Chem. A 2014, 118, 4955–4965. [Google Scholar] [CrossRef]
- Dubinets, N.; Safonov, A.; Bagaturyants, A. Structures and binding energies of the naphthalene dimer in its ground and excited states. J. Phys. Chem. A 2016, 120, 2779–2782. [Google Scholar] [CrossRef]
- Hancock, A.C.; Goerigk, L. Noncovalently bound excited-state dimers: A perspective on current time-dependent density functional theory approaches applied to aromatic excimer models. RSC Adv. 2022, 12, 13014–13034. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.h.; Lischka, H.; Mueller, T.; Plasser, F.; Kertesz, M. Study of the Diradicaloid Character in a Prototypical Pancake-Bonded Dimer: The Stacked Tetracyanoethylene (TCNE) Anion Dimer and the Neutral K2TCNE2 Complex. ChemPhysChem 2014, 15, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Valente, D.C.A.; Do Casal, M.T.; Barbatti, M.; Niehaus, T.A.; Aquino, A.J.; Lischka, H.; Cardozo, T.M. Excitonic and charge transfer interactions in tetracene stacked and T-shaped dimers. J. Chem. Phys. 2021, 154, 044306. [Google Scholar] [CrossRef] [PubMed]
- Siddique, F.; Barbatti, M.; Cui, Z.; Lischka, H.; Aquino, A.J. Nonadiabatic dynamics of charge-transfer states using the anthracene–tetracyanoethylene complex as a prototype. J. Phys. Chem. A 2020, 124, 3347–3357. [Google Scholar] [CrossRef] [PubMed]
- Mauck, C.M.; Bae, Y.J.; Chen, M.; Powers-Riggs, N.; Wu, Y.L.; Wasielewski, M.R. Charge-transfer character in a covalent diketopyrrolopyrrole dimer: Implications for singlet fission. ChemPhotoChem 2018, 2, 223–233. [Google Scholar] [CrossRef]
- Cui, Z.h.; Aquino, A.J.; Sue, A.C.H.; Lischka, H. Analysis of charge transfer transitions in stacked π-electron donor–acceptor complexes. Phys. Chem. Chem. Phys. 2018, 20, 26957–26967. [Google Scholar] [CrossRef]
- Müller-Dethlefs, K.; Hobza, P. Noncovalent interactions: A challenge for experiment and theory. Chem. Rev. 2000, 100, 143–168. [Google Scholar] [CrossRef]
- Snyder, J.W.; Fales, B.S.; Hohenstein, E.G.; Levine, B.G.; Martínez, T.J. A direct-compatible formulation of the coupled perturbed complete active space self-consistent field equations on graphical processing units. J. Chem. Phys. 2017, 146, 174113. [Google Scholar] [CrossRef]
- Demel, O.; Pittner, J.; Neese, F. A Local Pair Natural Orbital-Based Multireference Mukherjee’s Coupled Cluster Method. J. Chem. Theory Comput. 2015, 11, 3104–3114. [Google Scholar] [CrossRef]
- Riplinger, C.; Sandhoefer, B.; Hansen, A.; Neese, F. Natural triple excitations in local coupled cluster calculations with pair natural orbitals. J. Chem. Phys. 2013, 139, 134101. [Google Scholar] [CrossRef]
- Chiariello, M.G.; Donati, G.; Raucci, U.; Perrella, F.; Rega, N. Structural Origin and Vibrational Fingerprints of the Ultrafast Excited State Proton Transfer of the Pyranine-Acetate Complex in Aqueous Solution. J. Phys. Chem. B 2021, 125, 10273–10281. [Google Scholar] [CrossRef]
- Chiariello, M.G.; Raucci, U.; Donati, G.; Rega, N. Water-mediated excited state proton transfer of pyranine–acetate in aqueous solution: Vibrational fingerprints from ab initio molecular dynamics. J. Phys. Chem. A 2021, 125, 3569–3578. [Google Scholar] [CrossRef]
- De Simone, B.C.; Alberto, M.E.; Marino, T.; Russo, N.; Toscano, M. The Contribution of Density Functional Theory to the Atomistic Knowledge of Electrochromic Processes. Molecules 2021, 26, 5793. [Google Scholar] [CrossRef]
- Raucci, U.; Chiariello, M.G.; Rega, N. Modeling excited-state proton transfer to solvent: A dynamics study of a super photoacid with a hybrid implicit/explicit solvent model. J. Chem. Theory Comput. 2020, 16, 7033–7043. [Google Scholar] [CrossRef]
- Chiariello, M.G.; Donati, G.; Rega, N. Time-resolved vibrational analysis of excited state ab initio molecular dynamics to understand photorelaxation: The case of the pyranine photoacid in aqueous solution. J. Chem. Theory Comput. 2020, 16, 6007–6013. [Google Scholar] [CrossRef]
- Raucci, U.; Chiariello, M.G.; Coppola, F.; Perrella, F.; Savarese, M.; Ciofini, I.; Rega, N. An electron density based analysis to establish the electronic adiabaticity of proton coupled electron transfer reactions. J. Comput. Chem. 2020, 41, 1835–1841. [Google Scholar] [CrossRef]
- Chiariello, M.G.; Raucci, U.; Coppola, F.; Rega, N. Unveiling anharmonic coupling by means of excited state ab initio dynamics: Application to diarylethene photoreactivity. Phys. Chem. Chem. Phys. 2019, 21, 3606–3614. [Google Scholar] [CrossRef]
- Chiariello, M.G.; Rega, N. Exploring nuclear photorelaxation of pyranine in aqueous solution: An integrated ab-initio molecular dynamics and time resolved vibrational analysis approach. J. Phys. Chem. A 2018, 122, 2884–2893. [Google Scholar] [CrossRef]
- Perrella, F.; Raucci, U.; Chiariello, M.G.; Chino, M.; Maglio, O.; Lombardi, A.; Rega, N. Unveiling the structure of a novel artificial heme-enzyme with peroxidase-like activity: A theoretical investigation. Biopolymers 2018, 109, e23225. [Google Scholar] [CrossRef]
- Williams-Young, D.B.; Bagusetty, A.; de Jong, W.A.; Doerfler, D.; van Dam, H.J.; Vázquez-Mayagoitia, Á.; Windus, T.L.; Yang, C. Achieving performance portability in Gaussian basis set density functional theory on accelerator based architectures in NWChemEx. Parallel Comput. 2021, 108, 102829. [Google Scholar] [CrossRef]
- Williams-Young, D.B.; Petrone, A.; Sun, S.; Stetina, T.F.; Lestrange, P.; Hoyer, C.E.; Nascimento, D.R.; Koulias, L.; Wildman, A.; Kasper, J.; et al. The Chronus Quantum software package. WIREs Comput. Mol. Sci. 2020, 10, e1436. [Google Scholar] [CrossRef]
- Artrith, N.; Butler, K.T.; Coudert, F.X.; Han, S.; Isayev, O.; Jain, A.; Walsh, A. Best practices in machine learning for chemistry. Nat. Chem. 2021, 13, 505–508. [Google Scholar] [CrossRef] [PubMed]
- Butler, K.T.; Davies, D.W.; Cartwright, H.; Isayev, O.; Walsh, A. Machine learning for molecular and materials science. Nature 2018, 559, 547–555. [Google Scholar] [CrossRef]
- Pflüger, P.M.; Glorius, F. Molecular Machine Learning: The Future of Synthetic Chemistry? Angew. Chem. 2020, 59, 18860–18865. [Google Scholar] [CrossRef] [PubMed]
- Stocker, S.; Csányi, G.; Reuter, K.; Margraf, J.T. Machine learning in chemical reaction space. Nat. Chem. 2020, 11, 5505. [Google Scholar] [CrossRef]
- Sanchez-Lengeling, B.; Aspuru-Guzik, A. Inverse molecular design using machine learning: Generative models for matter engineering. Science 2018, 361, 360–365. [Google Scholar] [CrossRef]
- Dral, P.O. Quantum Chemistry in the Age of Machine Learning. J. Phys. Chem. Lett. 2020, 11, 2336–2347. [Google Scholar] [CrossRef]
- Keith, J.A.; Vassilev-Galindo, V.; Cheng, B.; Chmiela, S.; Gastegger, M.; Müller, K.R.; Tkatchenko, A. Combining Machine Learning and Computational Chemistry for Predictive Insights Into Chemical Systems. Chem. Rev. 2021, 121, 9816–9872. [Google Scholar] [CrossRef]
- Ramakrishnan, R.; Dral, P.O.; Rupp, M.; von Lilienfeld, O.A. Big Data Meets Quantum Chemistry Approximations: The Δ-Machine Learning Approach. J. Chem. Theory Comput. 2015, 11, 2087–2096. [Google Scholar] [CrossRef]
- von Lilienfeld, O.A.; Müller, K.R.; Tkatchenko, A. Exploring chemical compound space with quantum-based machine learning. Nat. Rev. Chem. 2020, 4, 347–358. [Google Scholar] [CrossRef]
- Häse, F.; Roch, L.M.; Friederich, P.; Aspuru-Guzik, A. Designing and understanding light-harvesting devices with machine learning. Nat. Chem. 2020, 11, 4587. [Google Scholar] [CrossRef]
- Rosen, A.S.; Iyer, S.M.; Ray, D.; Yao, Z.; Aspuru-Guzik, A.; Gagliardi, L.; Notestein, J.M.; Snurr, R.Q. Machine learning the quantum-chemical properties of metal–organic frameworks for accelerated materials discovery. Matter 2021, 4, 1578–1597. [Google Scholar] [CrossRef]
- Häse, F.; Galván, I.F.; Aspuru-Guzik, A.; Lindh, R.; Vacher, M. How machine learning can assist the interpretation of ab initio molecular dynamics simulations and conceptual understanding of chemistry. Chem. Sci. 2019, 10, 2298–2307. [Google Scholar] [CrossRef]
- Häse, F.; Valleau, S.; Pyzer-Knapp, E.; Aspuru-Guzik, A. Machine learning exciton dynamics. Chem. Sci. 2016, 7, 5139–5147. [Google Scholar] [CrossRef]
- Schriber, J.B.; Nascimento, D.R.; Koutsoukas, A.; Spronk, S.A.; Cheney, D.L.; Sherrill, C.D. CLIFF: A component-based, machine-learned, intermolecular force field. J. Chem. Phys. 2021, 154, 184110. [Google Scholar] [CrossRef]
- Glielmo, A.; Husic, B.E.; Rodriguez, A.; Clementi, C.; Noé, F.; Laio, A. Unsupervised Learning Methods for Molecular Simulation Data. Chem. Rev. 2021, 121, 9722–9758. [Google Scholar] [CrossRef]
- Falbo, E.; Fusè, M.; Lazzari, F.; Mancini, G.; Barone, V. Integration of Quantum Chemistry, Statistical Mechanics and Artificial Intelligence for Computational Spectroscopy: The UV–Vis Spectrum of TEMPO Radical in Different Solvents. J. Chem. Theory Comput. 2022, 18, 6203–6216. [Google Scholar] [CrossRef]
- Mancini, G.; Fusè, M.; Lipparini, F.; Nottoli, M.; Scalmani, G.; Barone, V. Molecular Dynamics Simulations Enforcing Nonperiodic Boundary Conditions: New Developments and Application to the Solvent Shifts of Nitroxide Magnetic Parameters. J. Chem. Theory Comput. 2022, 18, 2479–2493. [Google Scholar] [CrossRef]
- Mancini, G.; Fusè, M.; Lazzari, F.; Barone, V. Fast exploration of potential energy surfaces with a joint venture of quantum chemistry, evolutionary algorithms and unsupervised learning. Digit. Discov. 2022, 1, 790–805. [Google Scholar] [CrossRef]
- Barone, V.; Puzzarini, C.; Mancini, G. Integration of theory, simulation, artificial intelligence and virtual reality: A four-pillar approach for reconciling accuracy and interpretability in computational spectroscopy. Phys. Chem. Chem. Phys. 2021, 23, 17079–17096. [Google Scholar] [CrossRef]
- Mancini, G.; Del Galdo, S.; Chandramouli, B.; Pagliai, M.; Barone, V. Computational Spectroscopy in Solution by Integration of Variational and Perturbative Approaches on Top of Clusterized Molecular Dynamics. J. Chem. Theory Comput. 2020, 16, 5747–5761. [Google Scholar] [CrossRef] [PubMed]
- Del Galdo, S.; Chandramouli, B.; Mancini, G.; Barone, V. Assessment of Multi-Scale Approaches for Computing UV–Vis Spectra in Condensed Phases: Toward an Effective yet Reliable Integration of Variational and Perturbative QM/MM Approaches. J. Chem. Theory Comput. 2019, 15, 3170–3184. [Google Scholar] [CrossRef] [PubMed]
- Troyer, J.M.; Cohen, F.E. Protein conformational landscapes: Energy minimization and clustering of a long molecular dynamics trajectory. Proteins 1995, 23, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Wolf, A.; Kirschner, K.N. Principal component and clustering analysis on molecular dynamics data of the ribosomal L11·23S subdomain. J. Mol. Model. 2013, 19, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Papaleo, E.; Mereghetti, P.; Fantucci, P.; Grandori, R.; De Gioia, L. Free-energy landscape, principal component analysis and structural clustering to identify representative conformations from molecular dynamics simulations: The myoglobin case. J. Mol. Graph. Model. 2009, 27, 889–899. [Google Scholar] [CrossRef]
- Shao, J.; Tanner, S.W.; Thompson, N.; Cheatham, T.E. Clustering Molecular Dynamics Trajectories: 1. Characterizing the Performance of Different Clustering Algorithms. J. Chem. Theory Comput. 2007, 3, 2312–2334. [Google Scholar] [CrossRef]
- Torda, A.E.; van Gunsteren, W.F. Algorithms for clustering molecular dynamics configurations. J. Comput. Chem. 1994, 15, 1331–1340. [Google Scholar] [CrossRef]
- Phillips, J.L.; Colvin, M.E.; Newsam, S. Validating clustering of molecular dynamics simulations using polymer models. BMC Bioinform. 2011, 12, 445. [Google Scholar] [CrossRef]
- Karpen, M.E.; Tobias, D.J.; Brooks, C.L.I. Statistical clustering techniques for the analysis of long molecular dynamics trajectories: Analysis of 2.2-ns trajectories of YPGDV. Biochemistry 1993, 32, 412–420. [Google Scholar] [CrossRef]
- Peng, J.H.; Wang, W.; Yu, Y.Q.; Gu, H.L.; Huang, X. Clustering algorithms to analyze molecular dynamics simulation trajectories for complex chemical and biological systems. Chin. J. Chem. Phys. 2018, 31, 404–420. [Google Scholar] [CrossRef]
- González-Alemán, R.; Hernández-Castillo, D.; Rodríguez-Serradet, A.; Caballero, J.; Hernández-Rodríguez, E.W.; Montero-Cabrera, L. BitClust: Fast Geometrical Clustering of Long Molecular Dynamics Simulations. J. Chem. Inf. Model. 2020, 60, 444–448. [Google Scholar] [CrossRef]
- Ellis, S.R.; Hoffman, D.P.; Park, M.; Mathies, R.A. Difference bands in time-resolved femtosecond stimulated Raman spectra of photoexcited intermolecular electron transfer from chloronaphthalene to tetracyanoethylene. J. Phys. Chem. A 2018, 122, 3594–3605. [Google Scholar] [CrossRef]
- Coppola, F.; Cimino, P.; Raucci, U.; Chiariello, M.G.; Petrone, A.; Rega, N. Exploring the Franck–Condon region of a photoexcited charge transfer complex in solution to interpret femtosecond stimulated Raman spectroscopy: Excited state electronic structure methods to unveil non-radiative pathways. Chem. Sci. 2021, 12, 8058–8072. [Google Scholar] [CrossRef]
- Hagfeldt, A.; Boschloo, G.; Sun, L.; Kloo, L.; Pettersson, H. Dye-Sensitized Solar Cells. Chem. Rev. 2010, 110, 6595–6663. [Google Scholar] [CrossRef]
- Grätzel, M. Dye-sensitized solar cells. J. Photochem. Photobiol. C 2003, 4, 145–153. [Google Scholar] [CrossRef]
- Grätzel, M. Solar Energy Conversion by Dye-Sensitized Photovoltaic Cells. Inorg. Chem. 2005, 44, 6841–6851. [Google Scholar] [CrossRef]
- McCusker, J.K.; Vlcek, A., Jr. Ultrafast Excited-State Processes in Inorganic Systems. Acc. Chem. Res. 2015, 48, 1207–1208. [Google Scholar] [CrossRef]
- Chergui, M. Ultrafast Photophysics of Transition Metal Complexes. Acc. Chem. Res. 2015, 48, 801–808. [Google Scholar] [CrossRef]
- Pettersson Rimgard, B.; Föhlinger, J.; Petersson, J.; Lundberg, M.; Zietz, B.; Woys, A.M.; Miller, S.A.; Wasielewski, M.R.; Hammarström, L. Ultrafast interligand electron transfer in cis-[Ru(4,4′-dicarboxylate-2,2′-bipyridine)2(NCS)2]4− and implications for electron injection limitations in dye sensitized solar cells. Chem. Sci. 2018, 9, 7958–7967. [Google Scholar] [CrossRef]
- Waterland, M.R.; Kelley, D.F. Photophysics and Relaxation Dynamics of Ru(4,4′-Dicarboxy-2,2′-bipyridine)2cis(NCS)2 in Solution. J. Phys. Chem. A 2001, 105, 4019–4028. [Google Scholar] [CrossRef]
- Atkins, A.J.; González, L. Trajectory Surface-Hopping Dynamics Including Intersystem Crossing in [Ru(bpy)3]2+. J. Phys. Chem. Lett. 2017, 8, 3840–3845. [Google Scholar] [CrossRef] [PubMed]
- Zobel, J.P.; González, L. The Quest to Simulate Excited-State Dynamics of Transition Metal Complexes. JACS Au 2021, 1, 1116–1140. [Google Scholar] [CrossRef] [PubMed]
- Perrella, F.; Li, X.; Petrone, A.; Rega, N. Nature of the Ultrafast Interligands Electron Transfers in Dye-Sensitized Solar Cells. JACS Au 2023, 3, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Perrella, F.; Petrone, A.; Rega, N. Understanding Charge Dynamics in Dense Electronic Manifolds in Complex Environments. J. Chem. Theory Comput. 2023, 19, 626–639. [Google Scholar] [CrossRef] [PubMed]
- Baldini, E.; Palmieri, T.; Rossi, T.; Oppermann, M.; Pomarico, E.; Auböck, G.; Chergui, M. Interfacial Electron Injection Probed by a Substrate-Specific Excitonic Signature. J. Am. Chem. Soc. 2017, 139, 11584–11589. [Google Scholar] [CrossRef]
- Wei, H.; Luo, J.W.; Li, S.S.; Wang, L.W. Revealing the Origin of Fast Electron Transfer in TiO2-Based Dye-Sensitized Solar Cells. J. Am. Chem. Soc. 2016, 138, 8165–8174. [Google Scholar] [CrossRef]
- Tiwana, P.; Docampo, P.; Johnston, M.B.; Snaith, H.J.; Herz, L.M. Electron Mobility and Injection Dynamics in Mesoporous ZnO, SnO2 and TiO2 Films Used in Dye-Sensitized Solar Cells. ACS Nano 2011, 5, 5158–5166. [Google Scholar] [CrossRef]
- Katoh, R.; Furube, A.; Yoshihara, T.; Hara, K.; Fujihashi, G.; Takano, S.; Murata, S.; Arakawa, H.; Tachiya, M. Efficiencies of Electron Injection from Excited N3 Dye into Nanocrystalline Semiconductor (ZrO2, TiO2, ZnO, Nb2O2, SnO2, In2O2) Films. J. Phys. Chem. B 2004, 108, 4818–4822. [Google Scholar] [CrossRef]
- Asbury, J.B.; Ellingson, R.J.; Ghosh, H.N.; Ferrere, S.; Nozik, A.J.; Lian, T. Femtosecond IR Study of Excited-State Relaxation and Electron-Injection Dynamics of Ru(dcbpy)2(NCS)2 in Solution and on Nanocrystalline TiO2 and Al2O2 Thin Films. J. Phys. Chem. B 1999, 103, 3110–3119. [Google Scholar] [CrossRef]
- Perrella, F.; Petrone, A.; Rega, N. Direct observation of the solvent organization and nuclear vibrations of [Ru(dcbpy)2(NCS)2]4−, [dcbpy = (4,4′-dicarboxy-2,2′-bipyridine)], via ab initio molecular dynamics. Phys. Chem. Chem. Phys. 2021, 23, 22885–22896. [Google Scholar] [CrossRef]
- Brehm, M.; Thomas, M.; Gehrke, S.; Kirchner, B. TRAVIS—A free analyzer for trajectories from molecular simulation. J. Chem. Phys. 2020, 152, 164105. [Google Scholar] [CrossRef]
- Brehm, M.; Kirchner, B. TRAVIS—A free analyzer and visualizer for Monte Carlo and molecular dynamics trajectories. J. Chem. Inf. Model. 2011, 51, 8. [Google Scholar] [CrossRef]
- De Angelis, F.; Fantacci, S.; Selloni, A.; Nazeeruddin, M.K. Time dependent density functional theory study of the absorption spectrum of the [Ru(4,4′-COO−-2,2′-bpy)2(X)2]4− (X=NCS, Cl) dyes in water solution. Chem. Phys. Lett. 2005, 415, 115–120. [Google Scholar] [CrossRef]
- Schlegel, H.B.; Millam, J.M.; Iyengar, S.S.; Voth, G.A.; Daniels, A.D.; Scuseria, G.E.; Frisch, M.J. Ab initio molecular dynamics: Propagating the density matrix with Gaussian orbitals. J. Chem. Phys. 2001, 114, 9758–9763. [Google Scholar] [CrossRef]
- Iyengar, S.S.; Schlegel, H.B.; Millam, J.M.; Voth, G.A.; Scuseria, G.E.; Frisch, M.J. Ab initio molecular dynamics: Propagating the density matrix with Gaussian orbitals. II. Generalizations based on mass-weighting, idempotency, energy conservation and choice of initial conditions. J. Chem. Phys. 2001, 115, 10291–10302. [Google Scholar] [CrossRef]
- Schlegel, H.B.; Iyengar, S.S.; Li, X.; Millam, J.M.; Voth, G.A.; Scuseria, G.E.; Frisch, M.J. Ab initio molecular dynamics: Propagating the density matrix with Gaussian orbitals. III. Comparison with Born–Oppenheimer dynamics. J. Chem. Phys. 2002, 117, 8694–8704. [Google Scholar] [CrossRef]
- Iyengar, S.S.; Schlegel, H.B.; Voth, G.A.; Millam, J.M.; Scuseria, G.E.; Frisch, M.J. Ab initio molecular dynamics: Propagating the density matrix with Gaussian orbitals. IV. Formal analysis of the deviations from born-oppenheimer dynamics. Isr. J. Chem. 2002, 42, 191–202. [Google Scholar] [CrossRef]
- Rega, N.; Iyengar, S.S.; Voth, G.A.; Schlegel, H.B.; Vreven, T.; Frisch, M.J. Hybrid Ab-Initio/Empirical Molecular Dynamics: Combining the ONIOM Scheme with the Atom-Centered Density Matrix Propagation (ADMP) Approach. J. Phys. Chem. B 2004, 108, 4210–4220. [Google Scholar] [CrossRef]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef]
- Miehlich, B.; Savin, A.; Stoll, H.; Preuss, H. Results obtained with the correlation energy density functionals of becke and Lee, Yang and Parr. Chem. Phys. Lett. 1989, 157, 200–206. [Google Scholar] [CrossRef]
- Tomasi, J.; Mennucci, B.; Cammi, R. Quantum Mechanical Continuum Solvation Models. Chem. Rev. 2005, 105, 2999–3094. [Google Scholar] [CrossRef] [PubMed]
- Brancato, G.; Rega, N.; Barone, V. A hybrid explicit/implicit solvation method for first-principle molecular dynamics simulations. J. Chem. Phys. 2008, 128, 144501. [Google Scholar] [CrossRef] [PubMed]
- Cossi, M.; Barone, V.; Cammi, R.; Tomasi, J. Ab initio study of solvated molecules: A new implementation of the polarizable continuum model. Chem. Phys. Lett. 1996, 255, 327–335. [Google Scholar] [CrossRef]
- Cossi, M.; Scalmani, G.; Rega, N.; Barone, V. New developments in the polarizable continuum model for quantum mechanical and classical calculations on molecules in solution. J. Chem. Phys. 2002, 117, 43–54. [Google Scholar] [CrossRef]
- Mennucci, B. Polarizable continuum model. WIREs Comput. Mol. Sci. 2012, 2, 386–404. [Google Scholar] [CrossRef]
- Cossi, M.; Barone, V. Solvent effect on vertical electronic transitions by the polarizable continuum model. J. Chem. Phys. 2000, 112, 2427–2435. [Google Scholar] [CrossRef]
- Grimme, S.; Ehrlich, S.; Goerigk, L. Effect of the damping function in dispersion corrected density functional theory. J. Comput. Chem. 2011, 32, 1456–1465. [Google Scholar] [CrossRef]
- Grimme, S. Density functional theory with London dispersion corrections. WIREs Comput. Mol. Sci. 2011, 1, 211–228. [Google Scholar] [CrossRef]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010, 132, 154104. [Google Scholar] [CrossRef]
- Ehrlich, S.; Moellmann, J.; Grimme, S. Dispersion-Corrected Density Functional Theory for Aromatic Interactions in Complex Systems. Acc. Chem. Res. 2013, 46, 916–926. [Google Scholar] [CrossRef] [PubMed]
- Risthaus, T.; Grimme, S. Benchmarking of London Dispersion-Accounting Density Functional Theory Methods on Very Large Molecular Complexes. J. Chem. Theory Comput. 2013, 9, 1580–1591. [Google Scholar] [CrossRef] [PubMed]
- Grimme, S. Do Special Noncovalent π–π Stacking Interactions Really Exist? Angew. Chem. 2008, 47, 3430–3434. [Google Scholar] [CrossRef] [PubMed]
- Weigend, F.; Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys. Chem. Chem. Phys. 2005, 7, 3297–3305. [Google Scholar] [CrossRef] [PubMed]
- Andrae, D.; Haeussermann, U.; Dolg, M.; Stoll, H.; Preuss, H. Energy-adjusted ab initio pseudopotentials for the second and third row transition elements. Theor. Chem. Acc. 1990, 77, 123–141. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Svensson, M.; Humbel, S.; Froese, R.D.J.; Matsubara, T.; Sieber, S.; Morokuma, K. ONIOM: A Multilayered Integrated MO + MM Method for Geometry Optimizations and Single Point Energy Predictions. A Test for Diels-Alder Reactions and Pt(P(t-Bu)3)2 + H2 Oxidative Addition. J. Phys. Chem. 1996, 100, 19357–19363. [Google Scholar] [CrossRef]
- Vreven, T.; Byun, K.S.; Komáromi, I.; Dapprich, S.; Montgomery, J.A.J.; Morokuma, K.; Frisch, M.J. Combining Quantum Mechanics Methods with Molecular Mechanics Methods in ONIOM. J. Chem. Theory Comput. 2006, 2, 815–826. [Google Scholar] [CrossRef]
- Chung, L.W.; Sameera, W.M.C.; Ramozzi, R.; Page, A.J.; Hatanaka, M.; Petrova, G.P.; Harris, T.V.; Li, X.; Ke, Z.; Liu, F.; et al. The ONIOM Method and Its Applications. Chem. Rev. 2015, 115, 5678–5796. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Brancato, G.; Rega, N.; Barone, V. Molecular dynamics simulations in a NpT ensemble using non-periodic boundary conditions. Chem. Phys. Lett. 2009, 483, 177–181. [Google Scholar] [CrossRef]
- Rega, N.; Brancato, G.; Barone, V. Non-periodic boundary conditions for ab initio molecular dynamics in condensed phase using localized basis functions. Chem. Phys. Lett. 2006, 422, 367–371. [Google Scholar] [CrossRef]
- Brancato, G.; Barone, V.; Rega, N. Theoretical modeling of spectroscopic properties of molecules in solution: Toward an effective dynamical discrete/continuum approach. Theor. Chem. Acc. 2007, 117, 1001–1015. [Google Scholar] [CrossRef]
- Zabrodsky, H.; Peleg, S.; Avnir, D. Continuous symmetry measures. J. Am. Chem. Soc. 1992, 114, 7843–7851. [Google Scholar] [CrossRef]
- Pinsky, M.; Casanova, D.; Alemany, P.; Alvarez, S.; Avnir, D.; Dryzun, C.; Kizner, Z.; Sterkin, A. Symmetry operation measures. J. Comput. Chem. 2008, 29, 190–197. [Google Scholar] [CrossRef]
- Pinsky, M.; Dryzun, C.; Casanova, D.; Alemany, P.; Avnir, D. Analytical methods for calculating Continuous Symmetry Measures and the Chirality Measure. J. Comput. Chem. 2008, 29, 2712–2721. [Google Scholar] [CrossRef]
- Mu, Y.; Nguyen, P.H.; Stock, G. Energy landscape of a small peptide revealed by dihedral angle principal component analysis. Proteins 2005, 58, 45–52. [Google Scholar] [CrossRef]
- Géron, A. Hands-On Machine Learning with Scikit-Learn, Keras and TensorFlow: Concepts, Tools and Techniques to Build Intelligent Systems; O’Reilly Media: Sebastopol, CA, USA, 2019. [Google Scholar]
- Lloyd, S. Least squares quantization in PCM. IEEE Trans. Inf. Theory 1982, 28, 129–137. [Google Scholar] [CrossRef]
- Kaufman, L.; Rousseeuw, P.J. Finding Groups in Data: An Introduction to Cluster Analysis; John Wiley & Sons: Hoboken, NJ, USA, 2009. [Google Scholar]
- Schubert, E.; Lenssen, L. Fast k-medoids Clustering in Rust and Python. J. Open Source Softw. 2022, 7, 4183. [Google Scholar] [CrossRef]
- Schubert, E.; Rousseeuw, P.J. Fast and eager k-medoids clustering: O(k) runtime improvement of the PAM, CLARA and CLARANS algorithms. Inform. Syst. 2021, 101, 101804. [Google Scholar] [CrossRef]
- Caliński, T.; Harabasz, J. A dendrite method for cluster analysis. Commun. Stat. 1974, 3, 1–27. [Google Scholar] [CrossRef]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Phil. Trans. R. Soc. A 2016, 374, 20150202. [Google Scholar] [CrossRef] [PubMed]
- Plasser, F. TheoDORE: A toolbox for a detailed and automated analysis of electronic excited state computations. J. Chem. Phys. 2020, 152, 084108. [Google Scholar] [CrossRef] [PubMed]
- Plasser, F.; Lischka, H. Analysis of Excitonic and Charge Transfer Interactions from Quantum Chemical Calculations. J. Chem. Theory Comput. 2012, 8, 2777–2789. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16 Revision C.01; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]

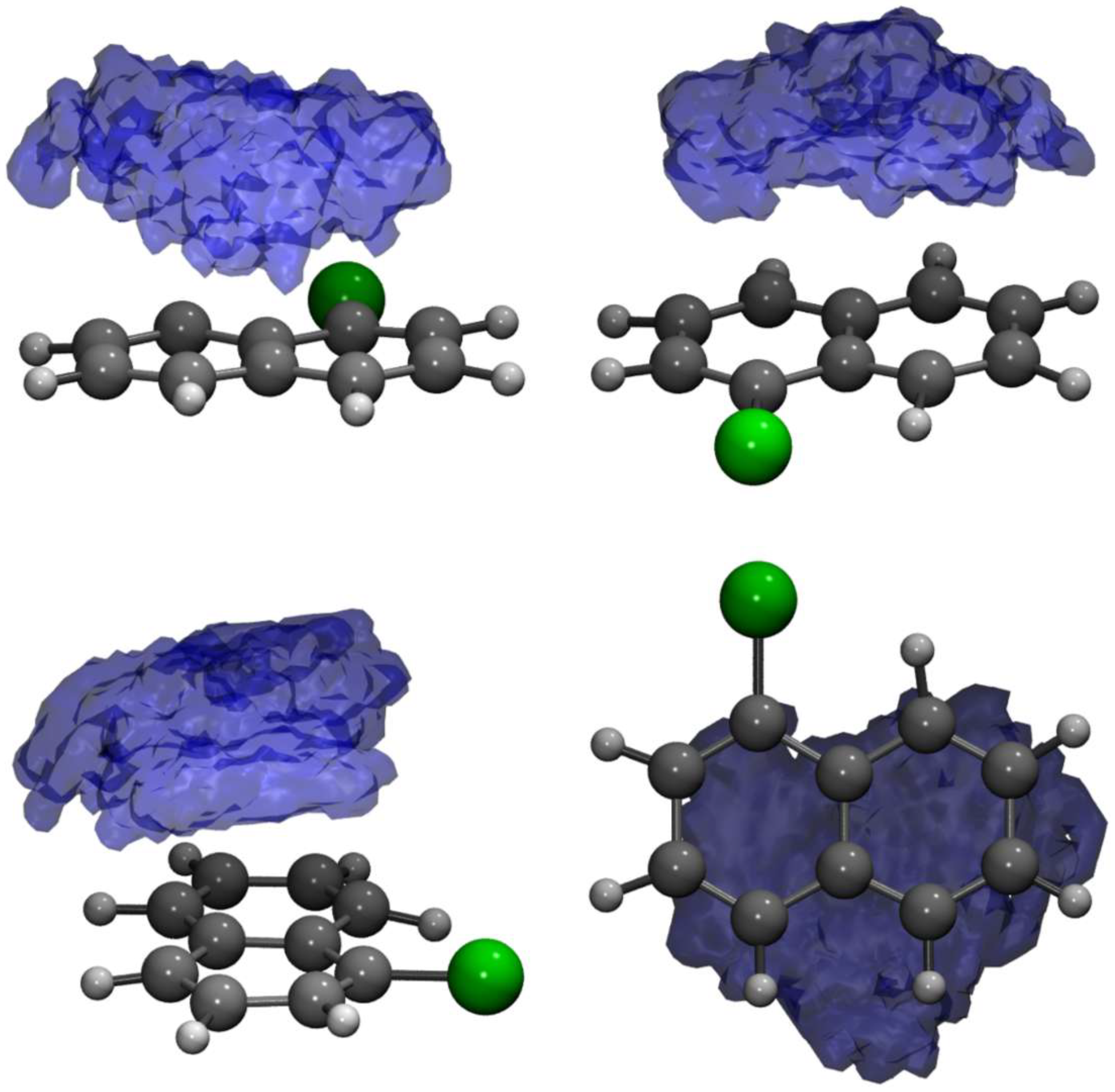
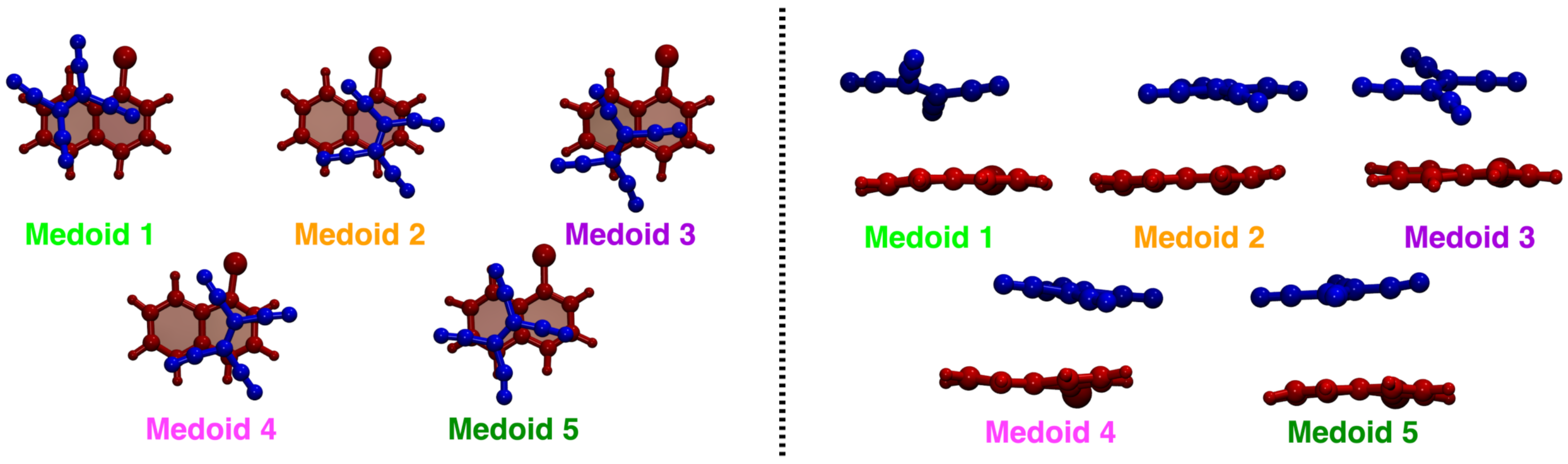

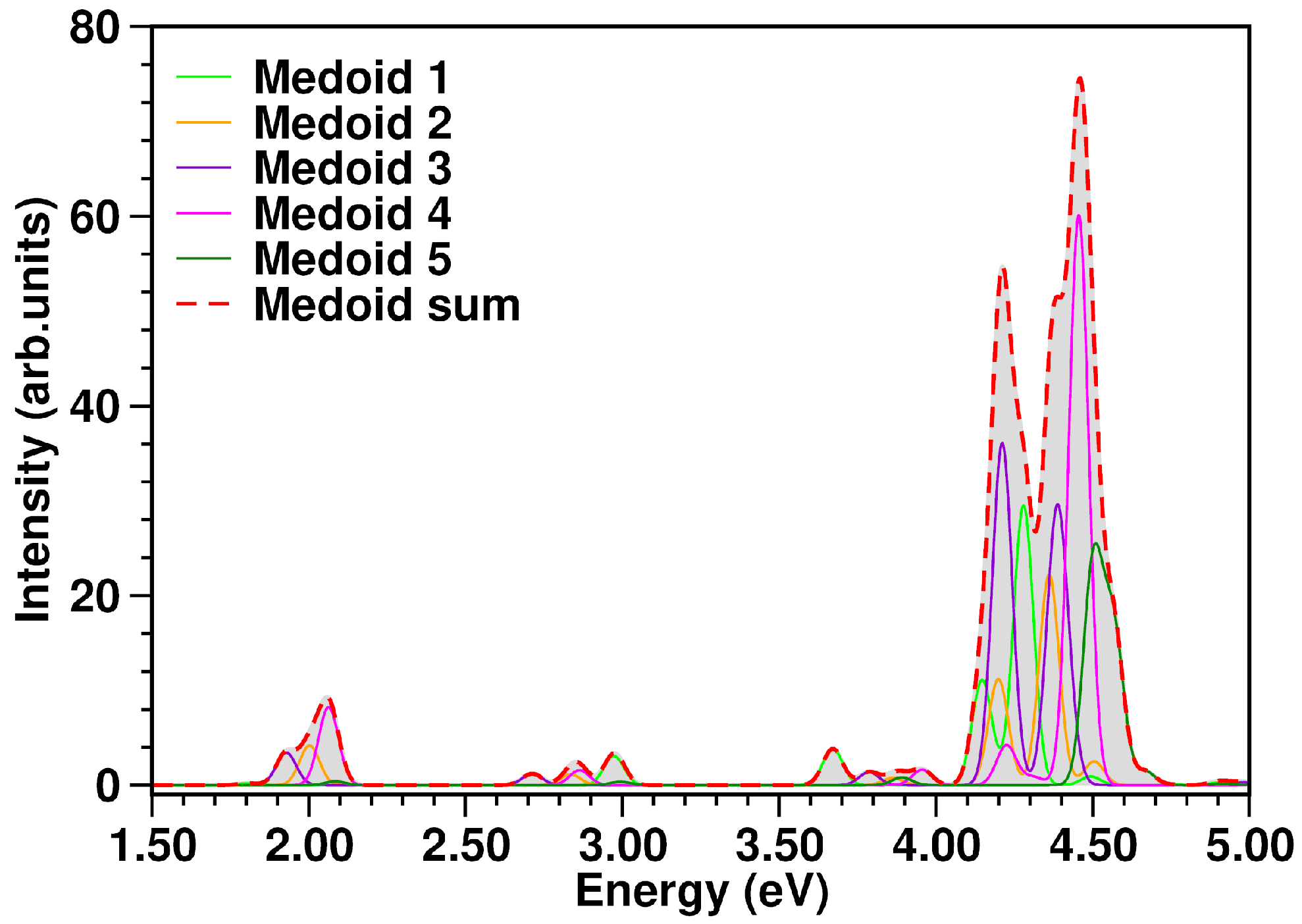

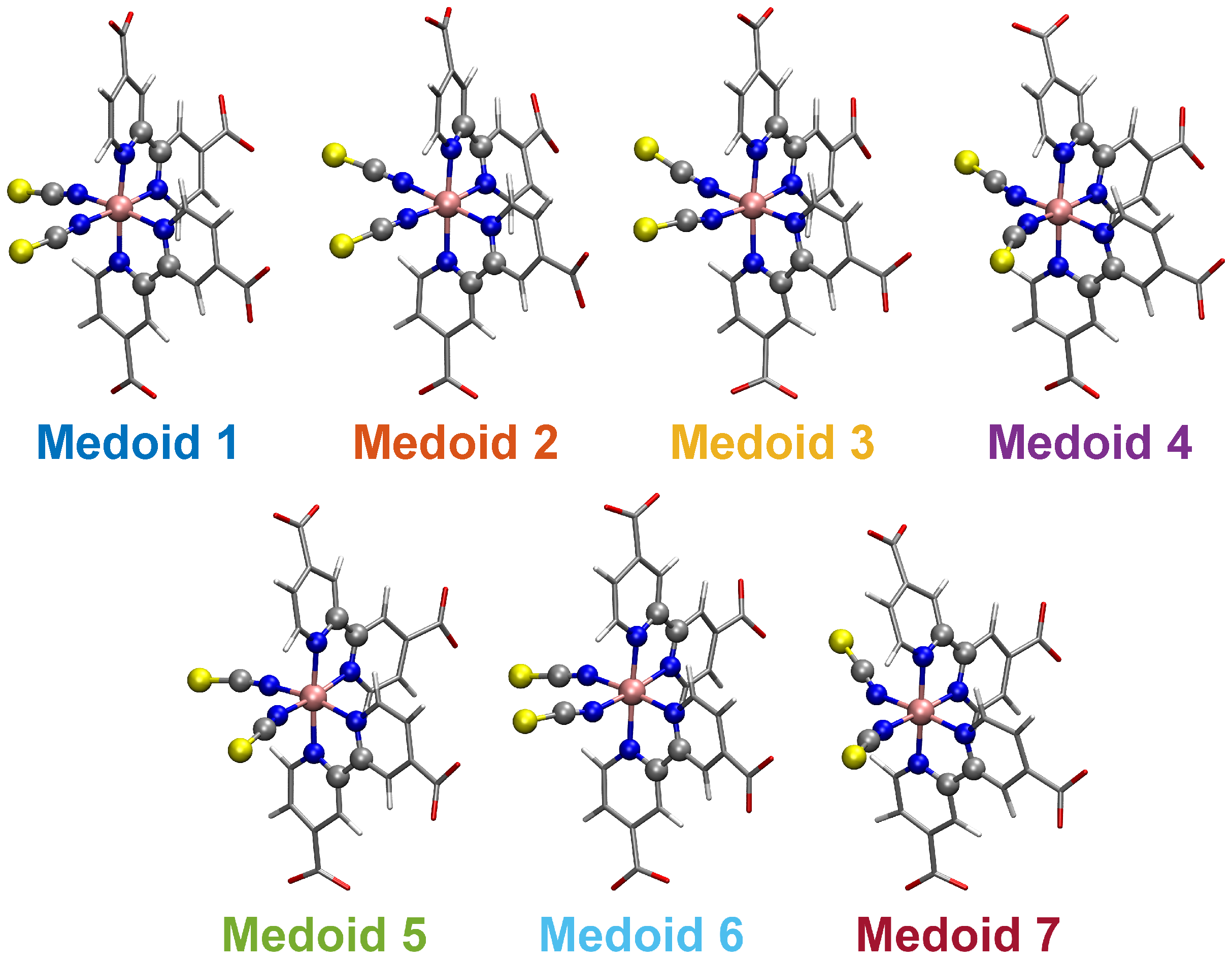
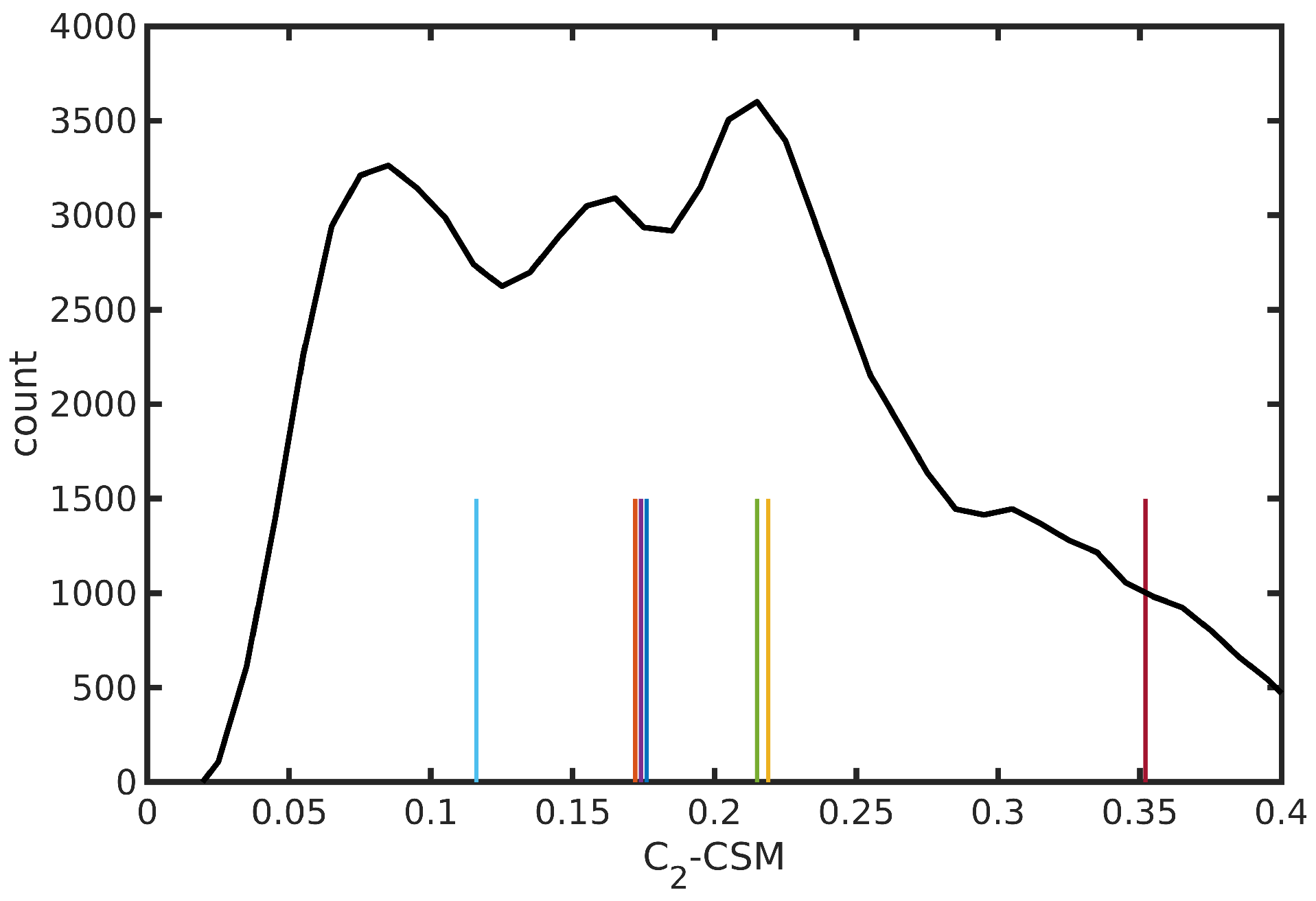

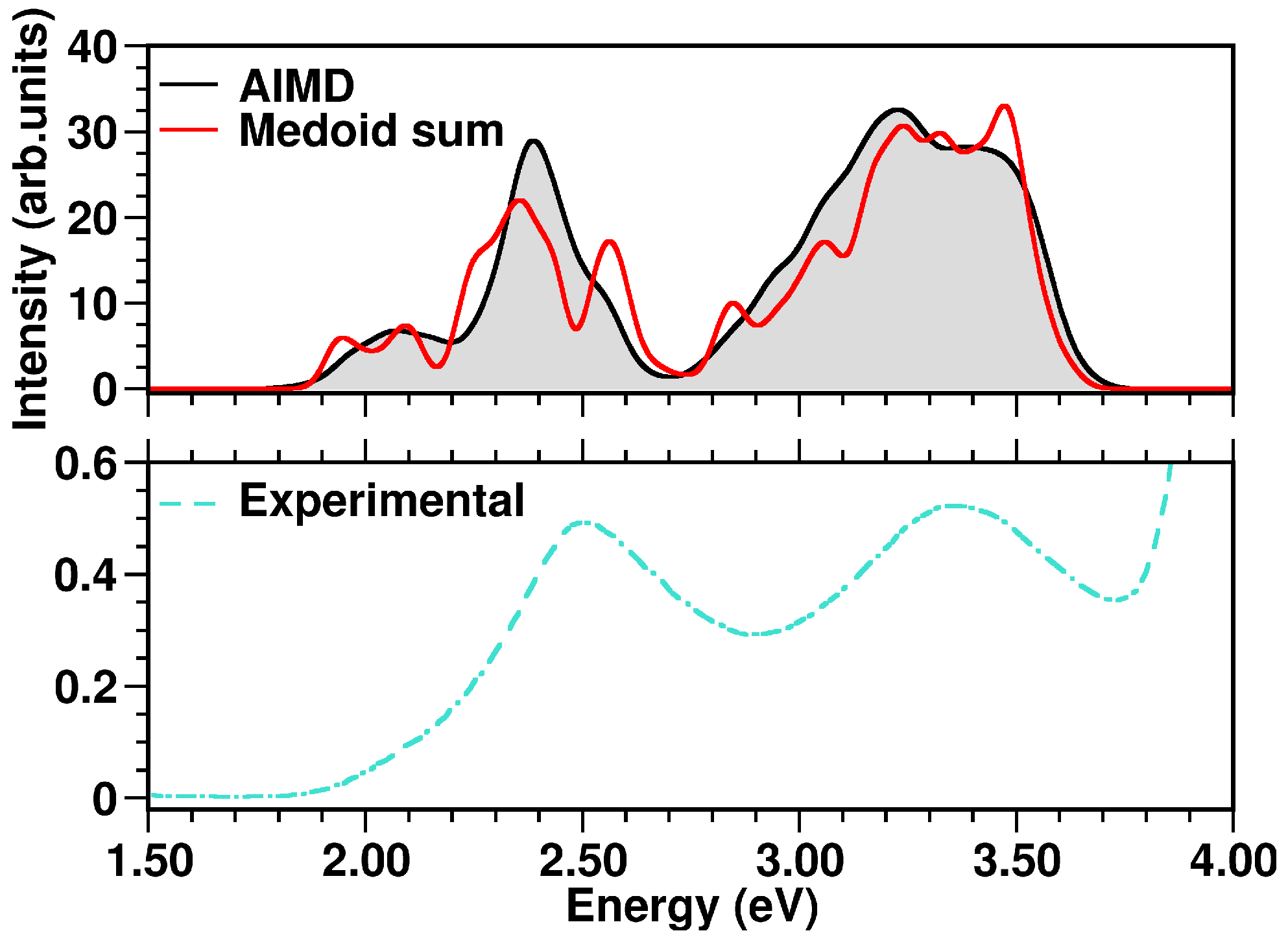
| Medoid | |||||||
|---|---|---|---|---|---|---|---|
| 1 | 16.68 | −0.660 | 0.398 | 0.638 | 1.513 | 2.847 | 2.190 |
| 2 | 14.60 | −0.816 | 0.141 | 0.561 | 2.604 | 1.673 | 1.852 |
| 3 | 19.36 | 0.723 | −0.407 | −0.558 | 1.543 | 2.444 | 2.276 |
| 4 | 15.19 | 0.714 | 0.021 | −0.699 | 2.666 | 1.773 | 1.736 |
| 5 | 16.27 | 0.112 | −0.736 | 0.668 | 1.833 | 2.401 | 2.436 |
| Medoid | ||||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 1.807 | 0.002 | 0.014 | 0.000 | 0.968 | 0.018 | 0.968 | |
| 2.973 | 0.035 | 0.016 | 0.000 | 0.965 | 0.018 | 0.966 | ||
| 2 | 2.003 | 0.052 | 0.031 | 0.001 | 0.943 | 0.025 | 0.944 | |
| 2.835 | 0.014 | 0.018 | 0.001 | 0.955 | 0.027 | 0.955 | ||
| 3 | 1.928 | 0.026 | 0.019 | 0.000 | 0.963 | 0.017 | 0.963 | |
| 2.713 | 0.009 | 0.018 | 0.000 | 0.963 | 0.018 | 0.964 | ||
| 4 | 2.063 | 0.067 | 0.031 | 0.001 | 0.938 | 0.030 | 0.939 | |
| 2.863 | 0.013 | 0.014 | 0.000 | 0.953 | 0.032 | 0.954 | ||
| 5 | 2.084 | 0.005 | 0.009 | 0.000 | 0.980 | 0.012 | 0.980 | |
| 2.994 | 0.005 | 0.017 | 0.000 | 0.971 | 0.012 | 0.971 |
| Medoid | -CSM | ||
|---|---|---|---|
| 1 | −30.77 | 5.57 | 0.176 |
| 2 | −54.10 | 107.01 | 0.172 |
| 3 | −143.43 | 140.61 | 0.219 |
| 4 | 52.26 | −131.41 | 0.174 |
| 5 | 69.09 | 83.05 | 0.215 |
| 6 | −127.10 | 40.02 | 0.116 |
| 7 | 107.85 | −137.07 | 0.352 |
| Medoid | |||||||
|---|---|---|---|---|---|---|---|
| 1 | 2.097 | 0.029 | 0.269 | 0.566 | 0.113 | 0.858 | |
| 2.342 | 0.086 | 0.284 | 0.542 | 0.119 | 0.851 | ||
| 3.556 | 0.056 | 0.578 | 0.235 | 0.127 | 0.851 | ||
| 2 | 2.083 | 0.026 | 0.235 | 0.580 | 0.111 | 0.846 | |
| 2.569 | 0.079 | 0.323 | 0.507 | 0.103 | 0.862 | ||
| 2.840 | 0.055 | 0.275 | 0.536 | 0.114 | 0.843 | ||
| 3.237 | 0.046 | 0.265 | 0.422 | 0.271 | 0.713 | ||
| 3.498 | 0.063 | 0.401 | 0.219 | 0.310 | 0.662 | ||
| 3 | 2.536 | 0.118 | 0.294 | 0.545 | 0.108 | 0.862 | |
| 3.389 | 0.043 | 0.133 | 0.215 | 0.583 | 0.391 | ||
| 4 | 2.983 | 0.056 | 0.366 | 0.511 | 0.087 | 0.894 | |
| 5 | 2.114 | 0.022 | 0.276 | 0.572 | 0.086 | 0.876 | |
| 3.454 | 0.057 | 0.373 | 0.277 | 0.289 | 0.693 | ||
| 6 | 2.041 | 0.025 | 0.242 | 0.627 | 0.093 | 0.884 | |
| 2.433 | 0.141 | 0.267 | 0.570 | 0.109 | 0.859 | ||
| 3.472 | 0.030 | 0.335 | 0.099 | 0.521 | 0.469 | ||
| 7 | 1.935 | 0.029 | 0.249 | 0.575 | 0.136 | 0.842 | |
| 2.316 | 0.101 | 0.267 | 0.551 | 0.121 | 0.844 | ||
| 3.022 | 0.056 | 0.278 | 0.553 | 0.148 | 0.840 | ||
| 3.165 | 0.059 | 0.427 | 0.312 | 0.212 | 0.773 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perrella, F.; Coppola, F.; Rega, N.; Petrone, A. An Expedited Route to Optical and Electronic Properties at Finite Temperature via Unsupervised Learning. Molecules 2023, 28, 3411. https://doi.org/10.3390/molecules28083411
Perrella F, Coppola F, Rega N, Petrone A. An Expedited Route to Optical and Electronic Properties at Finite Temperature via Unsupervised Learning. Molecules. 2023; 28(8):3411. https://doi.org/10.3390/molecules28083411
Chicago/Turabian StylePerrella, Fulvio, Federico Coppola, Nadia Rega, and Alessio Petrone. 2023. "An Expedited Route to Optical and Electronic Properties at Finite Temperature via Unsupervised Learning" Molecules 28, no. 8: 3411. https://doi.org/10.3390/molecules28083411
APA StylePerrella, F., Coppola, F., Rega, N., & Petrone, A. (2023). An Expedited Route to Optical and Electronic Properties at Finite Temperature via Unsupervised Learning. Molecules, 28(8), 3411. https://doi.org/10.3390/molecules28083411








