Discovery of Pyrimidine- and Coumarin-Linked Hybrid Molecules as Inducers of JNK Phosphorylation through ROS Generation in Breast Cancer Cells
Abstract
1. Introduction
2. Results and Discussions
2.1. Chemical Synthesis of PCLs (5a–h)
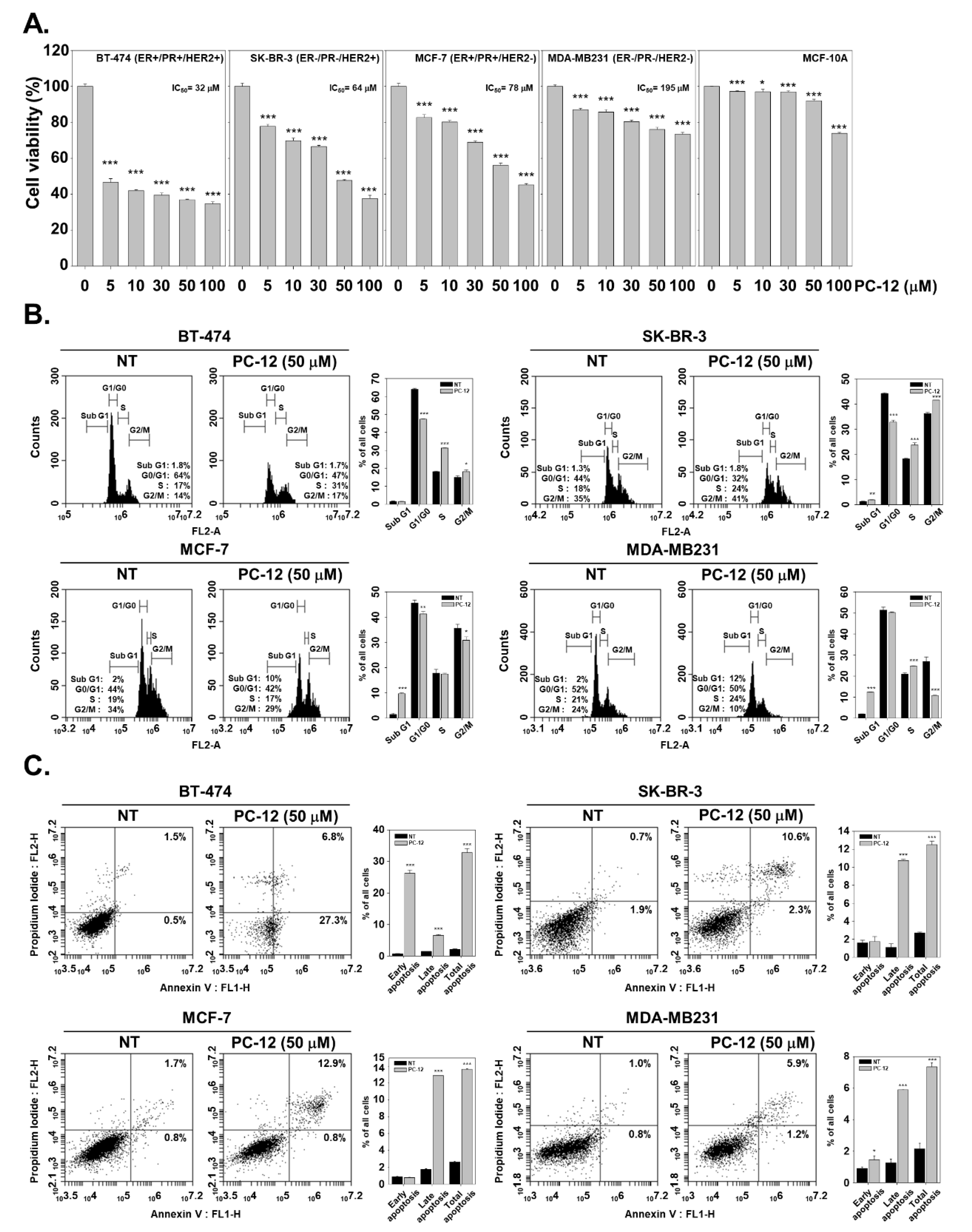
2.2. Efficacy of PCLs against a Variety of Breast Cancer Cells
2.3. PC-12 Induces Apoptosis in Human Breast Cancer Cells
2.4. PC-12 Promotes DNA Damage in Human Breast Cancer Cells
2.5. PC-12 Induces Cleavage of PARP and Regulated the Expression of Apoptotic Proteins
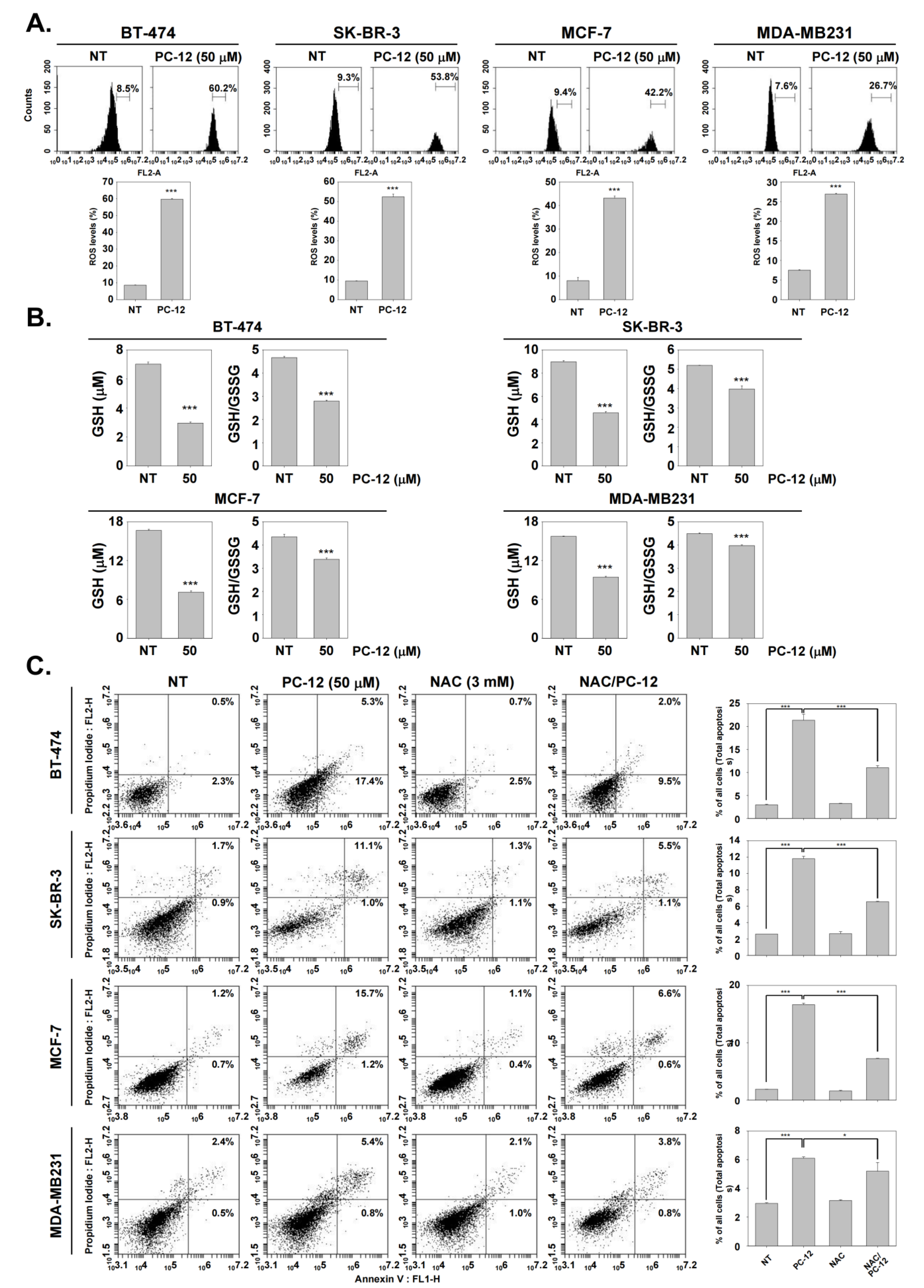
2.6. PC-12 Induces Cell Death through ROS Production
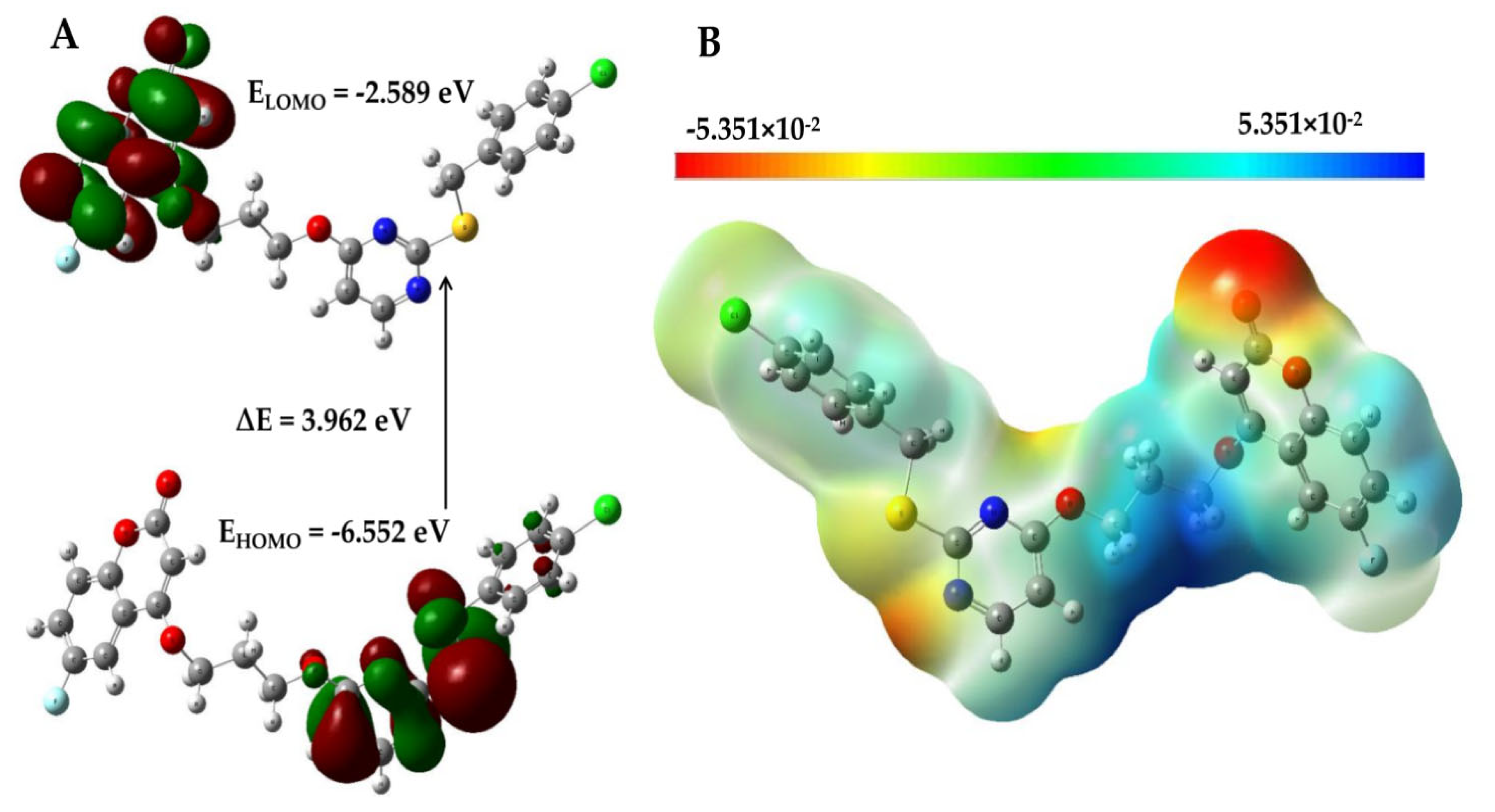
2.7. Mechanism for ROS Generation by PC-12 via Frontier Molecular Orbital (FMO) Analysis
2.8. PC-12 Interacts with JNK In Silico
2.9. PC-12 Induced Apoptosis via Up-Regulation of JNK Pathway
3. Materials and Methods
3.1. Experimental Materials and Method of Analysis
3.2. General Procedure for the Synthesis of Pyrimidine-Linked Coumarin Derivatives (PC-09 to PC-16) (5a–h)
3.2.1. 4-(2-((2-((4-Chlorobenzyl)thio)Pyrimidin-4-yl)oxy)Ethoxy)-2H-Chromen-2-One (PC-09) (5a)
3.2.2. 4-(3-((2-((4-Chlorobenzyl)thio)Pyrimidin-4-yl)oxy)Propoxy)-2H-Chromen-2-One (PC-10) (5b)
3.2.3. 4-(2-((2-((4-Chlorobenzyl)thio)Pyrimidin-4-yl)oxy)Ethoxy)-6-Fluoro-2H-Chromen-2-One (PC-11) (5c)
3.2.4. 4-(3-((2-((4-Chlorobenzyl)thio)Pyrimidin-4-yl)oxy)Propoxy)-6-Fluoro-2H-Chromen-2-One (PC-12) (5d)
3.2.5. 4-(2-((2-((4-Methoxybenzyl)thio)Pyrimidin-4-yl)oxy)Ethoxy)-2H-Chromen-2-One(PC-13) (5e)
3.2.6. 4-(3-((2-((4-Methoxybenzyl)thio)Pyrimidin-4-yl)oxy)Propoxy)-2H-Chromen-2-One (PC-14) (5f)
3.2.7. 6-Fluoro-4-(2-((2-((4-Methoxybenzyl)thio)Pyrimidin-4-yl)oxy)Ethoxy)-2H-Chromen-2-One (PC-15) (5g)
3.2.8. 6-Fluoro-4-(3-((2-((4-Methoxybenzyl)thio)Pyrimidin-4-yl)oxy)Propoxy)-2H-Chromen-2-One (PC-16) (5h)
3.3. Reagents
3.4. MTT Assay
3.5. Cell Cycle Analysis
3.6. Annexin/PI Staining Assay
3.7. Live and Dead Assay
3.8. Terminal Deoxynucleotidyl Transferase-Mediated dUTP Nick End Labeling (Tunel) Staining
3.9. Western Blot Analysis
3.10. ROS Measurement by H2DCF-DA
3.11. GSH/GSSG Assay for ROS Detection
3.12. In Silico DFT Calculations
3.13. Molecular Docking Analysis
3.14. Molecular Dynamics Simulations
3.15. Statistical Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
References
- Iqbal, N.; Iqbal, N. Human epidermal growth factor receptor 2 (HER2) in cancers: Overexpression and therapeutic implications. Mol. Biol. Int. 2014, 2014, 852748. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N. Insights into biology of luminal HER2 vs. enriched HER2 subtypes: Therapeutic implications. Breast 2015, 24 (Suppl. 2), S44–S48. [Google Scholar] [CrossRef] [PubMed]
- Llombart-Cussac, A.; Cortés, J.; Paré, L.; Galván, P.; Bermejo, B.; Martínez, N.; Vidal, M.; Pernas, S.; López, R.; Muñoz, M.; et al. HER2-enriched subtype as a predictor of pathological complete response following trastuzumab and lapatinib without chemotherapy in early-stage HER2-positive breast cancer (PAMELA): An open-label, single-group, multicentre, phase 2 trial. Lancet Oncol. 2017, 18, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Varghese, E.; Samuel, S.; Abotaleb, M.; Cheema, S.; Mamtani, R.; Büsselberg, D. The “Yin and Yang” of Natural Compounds in Anticancer Therapy of Triple-Negative Breast Cancers. Cancers 2018, 10, 346. [Google Scholar] [CrossRef] [PubMed]
- Duong, M.T.H.; Lee, J.-H.; Ahn, H.-C. C-Jun N-terminal kinase inhibitors: Structural insight into kinase-inhibitor complexes. Comput. Struct. Biotechnol. J. 2020, 18, 1440–1457. [Google Scholar] [CrossRef]
- Kamenecka, T.; Jiang, R.; Song, X.; Duckett, D.; Chen, W.; Ling, Y.Y.; Habel, J.; Laughlin, J.D.; Chambers, J.; Figuera-Losada, M.; et al. Synthesis, Biological Evaluation, X-ray Structure, and Pharmacokinetics of Aminopyrimidine c-jun-N-terminal Kinase (JNK) Inhibitors. J. Med. Chem. 2010, 53, 419–431. [Google Scholar] [CrossRef]
- Fricker, M.; LoGrasso, P.; Ellis, S.; Wilkie, N.; Hunt, P.; Pollack, S.J. Substituting c-Jun N-terminal kinase-3 (JNK3) ATP-binding site amino acid residues with their p38 counterparts affects binding of JNK- and p38-selective inhibitors. Arch. Biochem. Biophys. 2005, 438, 195–205. [Google Scholar] [CrossRef]
- Palmer, W.S.; Alam, M.; Arzeno, H.B.; Chang, K.-C.; Dunn, J.P.; Goldstein, D.M.; Gong, L.; Goyal, B.; Hermann, J.C.; Hogg, J.H.; et al. Development of amino-pyrimidine inhibitors of c-Jun N-terminal kinase (JNK): Kinase profiling guided optimization of a 1,2,3-benzotriazole lead. Bioorg. Med. Chem. Lett. 2013, 23, 1486–1492. [Google Scholar] [CrossRef]
- Shuai, W.; Bu, F.; Zhu, Y.; Wu, Y.; Xiao, H.; Pan, X.; Zhang, J.; Sun, Q.; Wang, G.; Ouyang, L. Discovery of Novel Indazole Chemotypes as Isoform-Selective JNK3 Inhibitors for the Treatment of Parkinson’s Disease. J. Med. Chem. 2023, 66, 1273–1300. [Google Scholar] [CrossRef]
- Zheng, K.; Park, C.M.; Iqbal, S.; Hernandez, P.; Park, H.; LoGrasso, P.V.; Feng, Y. Pyridopyrimidinone Derivatives as Potent and Selective c-Jun N-Terminal Kinase (JNK) Inhibitors. ACS Med. Chem. Lett. 2015, 6, 413–418. [Google Scholar] [CrossRef]
- Liu, M.; Wang, S.; Clampit, J.E.; Gum, R.J.; Haasch, D.L.; Rondinone, C.M.; Trevillyan, J.M.; Abad-Zapatero, C.; Fry, E.H.; Sham, H.L.; et al. Discovery of a new class of 4-anilinopyrimidines as potent c-Jun N-terminal kinase inhibitors: Synthesis and SAR studies. Bioorg. Med. Chem. Lett. 2007, 17, 668–672. [Google Scholar] [CrossRef] [PubMed]
- Garver, L.; Oliveira, G.; Barillas-Mury, C. The JNK Pathway Is a Key Mediator of Anopheles gambiae Antiplasmodial Immunity. PLOS Pathog. 2013, 9, e1003622. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-A.; Liu, C.-K.; Hsu, M.-L.; Chi, C.-W.; Ko, C.-C.; Chen, J.-S.; Lai, C.-T.; Chang, H.-H.; Lee, T.-Y.; Lai, Y.-L.; et al. Daphnoretin modulates differentiation and maturation of human dendritic cells through down-regulation of c-Jun N-terminal kinase. Int. Immunopharmacol. 2017, 51, 25–30. [Google Scholar] [CrossRef]
- Deveshegowda, S.N.; Metri, P.K.; Shivakumar, R.; Yang, J.-R.; Rangappa, S.; Swamynayaka, A.; Shanmugam, M.K.; Nagaraja, O.; Madegowda, M.; Shubha, P.B.; et al. Development of 1-(4-(Substituted)piperazin-1-yl)-2-((2-((4-methoxybenzyl)thio)pyrimidin-4-yl)oxy)ethanones That Target Poly (ADP-Ribose) Polymerase in Human Breast Cancer Cells. Molecules 2022, 27, 2848. [Google Scholar] [CrossRef] [PubMed]
- Gilandoust, M.; Harsha, K.B.; Mohan, C.D.; Raquib, A.R.; Rangappa, S.; Pandey, V.; Lobie, P.E.; Basappa; Rangappa, K.S. Synthesis, characterization and cytotoxicity studies of 1,2,3-triazoles and 1,2,4-triazolo [1,5-a] pyrimidines in human breast cancer cells. Bioorg. Med. Chem. Lett. 2018, 28, 2314–2319. [Google Scholar] [CrossRef] [PubMed]
- Neelgundmath, M.; Dinesh, K.R.; Mohan, C.D.; Li, F.; Dai, X.; Siveen, K.S.; Paricharak, S.; Mason, D.J.; Fuchs, J.E.; Sethi, G.; et al. Novel synthetic coumarins that targets NF-κB in Hepatocellular carcinoma. Bioorg. Med. Chem. Lett. 2015, 25, 893–897. [Google Scholar] [CrossRef]
- Keerthy, H.K.; Mohan, C.D.; Siveen, K.S.; Fuchs, J.E.; Rangappa, S.; Sundaram, M.S.; Li, F.; Girish, K.S.; Sethi, G.; Basappa; et al. Novel Synthetic Biscoumarins Target Tumor Necrosis Factor-α in Hepatocellular Carcinoma in Vitro and in Vivo. J. Biol. Chem. 2014, 289, 31879–31890. [Google Scholar] [CrossRef]
- Bharathkumar, H.; Mohan, C.D.; Ananda, H.; Fuchs, J.E.; Li, F.; Rangappa, S.; Surender, M.; Bulusu, K.C.; Girish, K.S.; Sethi, G.; et al. Microwave-assisted synthesis, characterization and cytotoxic studies of novel estrogen receptor α ligands towards human breast cancer cells. Bioorg. Med. Chem. Lett. 2015, 25, 1804–1807. [Google Scholar] [CrossRef]
- Sadashiva, M.P.; Basappa, B.; NanjundaSwamy, S.; Li, F.; Manu, K.A.; Sengottuvelan, M.; Prasanna, D.S.; Anilkumar, N.C.; Sethi, G.; Sugahara, K.; et al. Anti-cancer activity of novel dibenzo[b,f]azepine tethered isoxazoline derivatives. BMC Chem. Biol. 2012, 12, 5–11. [Google Scholar] [CrossRef]
- Pandey, V.; Zhang, X.; Poh, H.-M.; Wang, B.; Dukanya, D.; Ma, L.; Yin, Z.; Bender, A.; Periyasamy, G.; Zhu, T.; et al. Monomerization of Homodimeric Trefoil Factor 3 (TFF3) by an Aminonitrile Compound Inhibits TFF3-Dependent Cancer Cell Survival. ACS Pharmacol. Transl. Sci. 2022, 5, 761–773. [Google Scholar] [CrossRef]
- Ko, H.; Lee, J.; Kim, H.; Kim, T.; Han, Y.; Suh, Y.-G.; Chun, J.; Kim, Y.; Ahn, K. Novel Galiellalactone Analogues Can Target STAT3 Phosphorylation and Cause Apoptosis in Triple-Negative Breast Cancer. Biomolecules 2019, 9, 170. [Google Scholar] [CrossRef]
- Anjali Rawat, A. Vijaya Bhaskar Reddy, Recent Advances on Anticancer Activity of Coumarin Derivatives. Eur. J. Med. Chem. Rep. 2022, 5, 100038. [Google Scholar] [CrossRef]
- Kaneko, T.; Tahara, S.; Takabayashi, F. Inhibitory effect of natural coumarin compounds, esculetin and esculin, on oxidative DNA damage and formation of aberrant crypt foci and tumors induced by 1,2-dimethylhydrazine in rat colons. Biol. Pharm. Bull. 2007, 30, 2052–2057. [Google Scholar] [CrossRef] [PubMed]
- Gillan, L.; Evans, G.; Maxwell, W.M. Flow cytometric evaluation of sperm parameters in relation to fertility potential. Theriogenology 2005, 63, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Zhao, Y.; Gu, W.; Zhu, Y.; Wang, S. Novel camphor-based pyrimidine derivatives induced cancer cell death through a ROS-mediated mitochondrial apoptosis pathway. RSC Adv. 2019, 9, 29711–29720. [Google Scholar] [CrossRef]
- Jaiswal, P.K.; Goel, A.; Mittal, R.D. Survivin: A molecular biomarker in cancer. Indian J. Med. Res. 2015, 141, 389–397. [Google Scholar] [CrossRef]
- Baek, S.H.; Kim, C.; Lee, J.H.; Nam, D.; Lee, J.; Lee, S.-G.; Chung, W.-S.; Jang, H.-J.; Kim, S.-H.; Ahn, K.S. Cinobufagin exerts anti-proliferative and pro-apoptotic effects through the modulation ROS-mediated MAPKs signaling pathway. Immunopharmacol. Immunotoxicol. 2015, 37, 265–273. [Google Scholar] [CrossRef]
- Jagadish, S.; Rajeev, N.; NaveenKumar, S.K.; Kumar, K.S.S.; Paul, M.; Hegde, M.; Basappa; Sadashiva, M.P.; Girish, K.S.; Rangappa, K.S. Platelet protective efficacy of 3,4,5 trisubstituted isoxazole analogue by inhibiting ROS-mediated apoptosis and platelet aggregation. Mol. Cell. Biochem. 2016, 414, 137–151. [Google Scholar] [CrossRef]
- Dai, X.; Wang, L.; Deivasigamni, A.; Looi, C.Y.; Karthikeyan, C.; Trivedi, P.; Chinnathambi, A.; Alharbi, S.A.; Arfuso, F.; Dharmarajan, A.; et al. A novel benzimidazole derivative, MBIC inhibits tumor growth and promotes apoptosis via activation of ROS-dependent JNK signaling pathway in hepatocellular carcinoma. Oncotarget 2017, 8, 12831–12842. [Google Scholar] [CrossRef]
- Sulaiman, N.B.; Mohan, C.D.; Basappa, S.; Pandey, V.; Rangappa, S.; Bharathkumar, H.; Kumar, A.P.; Lobie, P.E.; Rangappa, K.S. An azaspirane derivative suppresses growth and induces apoptosis of ER-positive and ER-negative breast cancer cells through the modulation of JAK2/STAT3 signaling pathway. Int. J. Oncol. 2016, 49, 1221–1229. [Google Scholar] [CrossRef]
- Anusha, S.; Mohan, C.D.; Ananda, H.; Baburajeev, C.P.; Rangappa, S.; Mathai, J.; Fuchs, J.E.; Li, F.; Shanmugam, M.K.; Bender, A.; et al. Adamantyl-tethered-biphenylic compounds induce apoptosis in cancer cells by targeting Bcl homologs. Bioorg. Med. Chem. Lett. 2016, 26, 1056–1060. [Google Scholar] [CrossRef]
- Priya, B.S.; Swamy, S.N.; Tejesvi, M.V.; Basappa Sarala, G.; Gaonkar, S.L.; Naveen, S.; Prasad, J.S.; Rangappa, K.S. Synthesis, characterization, antimicrobial and single crystal X-ray crystallographic studies of some new sulfonyl, 4-chloro phenoxy benzene and dibenzoazepine substituted benzamides. Eur. J. Med. Chem. 2006, 41, 1262–1270. [Google Scholar] [CrossRef]
- Paul, M.; Hemshekhar, M.; Thushara, R.M.; Sundaram, M.S.; NaveenKumar, S.K.; Naveen, S.; Devaraja, S.; Somyajit, K.; West, R.; Basappa Nayaka, S.C.; et al. Methotrexate Promotes Platelet Apoptosis via JNK-Mediated Mitochondrial Damage: Alleviation by N-Acetylcysteine and N-Acetylcysteine Amide. PLoS ONE. 2015, 10, e0127558. [Google Scholar] [CrossRef]
- Bhat, A.; Tan, V.; Heng, B.; Chow, S.; Basappa, S.; Essa, M.M.; Chidambaram, S.B.; Guillemin, G.J. Papaverine, a Phosphodiesterase 10A Inhibitor, Ameliorates Quinolinic Acid-Induced Synaptotoxicity in Human Cortical Neurons. Neurotox. Res. 2021, 39, 1238–1250. [Google Scholar] [CrossRef] [PubMed]
- Swamynayaka, A.; Srinivas, M.S.; Vahini, M.V.; Khamees, H.A.; Madegowda, M.; Hegde, V.N.; Hegde, T.A.; Vinitha, G. Third-order nonlinear optical studies of Bis(4-methylbenzylammonium) tetrachloridocuprate metal-organic crystal with optical limiting behavior: Experimental and theoretical investigations. J. Mol. Struct. 2022, 1269, 1333827. [Google Scholar] [CrossRef]
- Oliveros, E.; Infelta, P.P.; Ramsteiner, K. Singlet Oxygen and Superoxide: Experimental Differentiation and Analysis. Helvetica Chim. Acta 1983, 66, 722–733. [Google Scholar] [CrossRef]
- Lin, A. Activation of the JNK signaling pathway: Breaking the brake on apoptosis. Bioessays 2002, 25, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Yin, X.; Huang, X.; Guo, Q.; Ma, M.; Guo, L. Astragaloside IV ameliorates sepsis-induced myocardial dysfunction by regulating NOX4/JNK/BAX pathway. Life Sci. 2022, 310, 121123. [Google Scholar] [CrossRef]
- Yang, M.H.; Baek, S.H.; Hwang, S.T.; Um, J.; Ahn, K.S. Corilagin exhibits differential anticancer effects through the modulation of STAT3 /5 and MAPKs in human gastric cancer cells. Phytotherapy Res. 2022, 36, 2449–2462. [Google Scholar] [CrossRef]
- Jung, Y.Y.; Baek, S.H.; Ha, I.J.; Ahn, K.S. Regulation of apoptosis and autophagy by albendazole in human colon adenocarcinoma cells. Biochimie 2022, 198, 155–166. [Google Scholar] [CrossRef]
- Jung, Y.Y.; Ha, I.J.; Um, J.-Y.; Sethi, G.; Ahn, K.S. Fangchinoline diminishes STAT3 activation by stimulating oxidative stress and targeting SHP-1 protein in multiple myeloma model. J. Adv. Res. 2022, 35, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.Y.; Jung, Y.Y.; Yang, M.H.; Um, J.-Y.; Sethi, G.; Ahn, K.S. Isoimperatorin down-regulates epithelial mesenchymal transition through modulating NF-κB signaling and CXCR4 expression in colorectal and hepatocellular carcinoma cells. Cell. Signal. 2022, 99, 110433. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.Y.; Mohan, C.D.; Chinnathambi, A.; Alharbi, S.A.; Sethi, G.; Rangappa, K.S.; Ahn, K.S. Euphorbiasteroid Abrogates EGFR and Wnt/β-Catenin Signaling in Non-Small-Cell Lung Cancer Cells to Impart Anticancer Activity. Molecules 2022, 27, 3824. [Google Scholar] [CrossRef] [PubMed]
- Mohan, C.D.; Anilkumar, N.C.; Rangappa, S.; Shanmugam, M.K.; Mishra, S.; Chinnathambi, A.; Alharbi, S.A.; Bhattacharjee, A.; Sethi, G.; Kumar, A.P.; et al. Novel 1,3,4-Oxadiazole Induces Anticancer Activity by Targeting NF-κB in Hepatocellular Carcinoma Cells. Front. Oncol. 2018, 8, 42. [Google Scholar] [CrossRef]
- Kumar, C.A.; Jayarama, S.; Basappa Salimath, B.P.; Rangappa, K.S. Pro-apoptotic activity of imidazole derivatives mediated by up-regulation of Bax and activation of CAD in Ehrlich Ascites Tumor cells. Investig. New Drugs 2007, 25, 343–350. [Google Scholar] [CrossRef]
- Li, P.; Zhao, Q.-L.; Rehman, M.U.; Jawaid, P.; Cui, Z.-G.; Ahmed, K.; Kondo, T.; Saitoh, J.-I.; Noguchi, K. Isofraxidin enhances hyperthermia-induced apoptosis via redox modification in acute monocytic leukemia U937 cells. Mol. Med. Rep. 2023, 27, 41. [Google Scholar] [CrossRef]
- Jung, Y.Y.; Um, J.-Y.; Chinnathambi, A.; Govindasamy, C.; Sethi, G.; Ahn, K.S. Leelamine Modulates STAT5 Pathway Causing Both Autophagy and Apoptosis in Chronic Myelogenous Leukemia Cells. Biology 2022, 11, 366. [Google Scholar] [CrossRef]
- Kim, N.Y.; Jung, Y.Y.; Yang, M.H.; Chinnathambi, A.; Govindasamy, C.; Narula, A.S.; Namjoshi, O.A.; Blough, B.E.; Ahn, K.S. Tanshinone IIA exerts autophagic cell death through down-regulation of β-catenin in renal cell carcinoma cells. Biochimie 2022, 200, 119–130. [Google Scholar] [CrossRef]
- Lee, J.; Kim, C.; Lee, S.-G.; Sethi, G.; Ahn, K. Ophiopogonin D, a Steroidal Glycoside Abrogates STAT3 Signaling Cascade and Exhibits Anti-Cancer Activity by Causing GSH/GSSG Imbalance in Lung Carcinoma. Cancers 2018, 10, 427. [Google Scholar] [CrossRef]
- Somu, C.; Mohan, C.D.; Ambekar, S.; Dukanya Rangappa, S.; Baburajeev, C.P.; Sukhorukov, A.; Mishra, S.; Shanmugam, M.K.; Chinnathambi, A.; Awad Alahmadi, T.; et al. Identification of a novel 1,2 oxazine that can induce apoptosis by targeting NF-κB in hepatocellular carcinoma cells. Biotechnol. Rep. 2020, 19, e00438. [Google Scholar] [CrossRef]
- Lee, J.H.; Mohan, C.D.; Shanmugam, M.K.; Rangappa, S.; Sethi, G.; Siveen, K.S.; Chinnathambi, A.; Alahmadi, T.A.; Alharbi, S.A.; Basappa, S.; et al. Vitexin abrogates invasion and survival of hepatocellular carcinoma cells through targeting STAT3 signaling pathway. Biochimie 2020, 175, 58–68. [Google Scholar] [CrossRef]
- Malojirao, V.H.; Girimanchanaika, S.S.; Shanmugam, M.K.; Sherapura, A.; Dukanya Metri, P.K.; Vigneshwaran, V.; Chinnathambi, A.; Alharbi, S.A.; Rangappa, S.; Mohan, C.D.; et al. Novel 1,3,4-oxadiazole Targets STAT3 Signaling to Induce Antitumor Effect in Lung Cancer. Biomedicines 2020, 8, 368. [Google Scholar] [CrossRef]
- Bhuvanalakshmi, G.; Basappa Rangappa, K.S.; Dharmarajan, A.; Sethi, G.; Kumar, A.P.; Warrier, S. Breast Cancer Stem-Like Cells Are Inhibited by Diosgenin, a Steroidal Saponin, by the Attenuation of the Wnt β-Catenin Signaling via the Wnt Antagonist Secreted Frizzled Related Protein-4. Front. Pharmacol. 2017, 8, 124. [Google Scholar] [CrossRef]
- Ananda, S.; Khamees, H.A.; Mahendra, M.; Kumara, C.; Prasad, D.J.; Hegde, T.A.; Vinitha, G. Structural, thermal, dielectric, nonlinear optical properties and DFT investigations of a novel material 2-(6-chloropyridin-3-yl)-N’-(2,3-dihydro-1,4-benzodioxin-6-ylmethylidene)acetohydrazide for optoelectronic applications. J. Mater. Sci. Mater. Electron. 2021, 32, 14677–14702. [Google Scholar] [CrossRef]
- Hiscocks, J.; Frisch, M.J. Gaussian 09: IOps Reference, 2nd ed.; Caricato, M., Frisch, M.J., Eds.; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Demirpolat, A.; Akman, F.; Kazachenko, A.S. An Experimental and Theoretical Study on Essential Oil of Aethionema sancakense: Characterization, Molecular Properties and RDG Analysis. Molecules 2022, 27, 6129. [Google Scholar] [CrossRef] [PubMed]
- Sanner, M.F. Python: A programming language for software integration and development. J. Mol. Graph. Model. 1999, 17, 57–61. [Google Scholar] [PubMed]
- Bowers, K.J.; Chow, D.E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable algorithms for molecular dynamics simulations on commodity clusters. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, FL, USA, 11–17 November 2006; p. 43. [Google Scholar]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Munshi, A.; Ramesh, R. Mitogen-Activated Protein Kinases and Their Role in Radiation Response. Genes Cancer 2013, 4, 401–408. [Google Scholar] [CrossRef]
- Ha, J.; Kang, E.; Seo, J.; Cho, S. Phosphorylation Dynamics of JNK Signaling: Effects of Dual-Specificity Phosphatases (DUSPs) on the JNK Pathway. Int. J. Mol. Sci. 2019, 20, 6157. [Google Scholar] [CrossRef] [PubMed]
- Dhanasekaran, D.N.; Reddy, E.P. JNK signaling in apoptosis. Oncogene 2008, 27, 6245–6251. [Google Scholar] [CrossRef] [PubMed]
- Ray, P.D.; Huang, B.-W.; Tsuji, Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell. Signal. 2012, 24, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, S.; Carter, C.; Lynch, M.; Lowinger, T.; Dumas, J.; Smith, R.A.; Schwartz, B.; Simantov, R.; Kelley, S. Discovery and development of sorafenib: A multikinase inhibitor for treating cancer. Nat. Rev. Drug Discov. 2006, 5, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; An, G.; Kuo, M.T. C-Jun N-terminal kinase signalling pathway in response to cisplatin. J. Cell Mol. Med. 2016, 20, 2013–2019. [Google Scholar] [CrossRef] [PubMed]
- Bharathkumar, H.; Mohan, C.D.; Rangappa, S.; Kang, T.; Keerthy, H.K.; Fuchs, J.E.; Kwon, N.H.; Bender, A.; Kim, S.; Mohan, C.D.; et al. Screening of quinoline, 1,3-benzoxazine, and 1,3-oxazine-based small molecules against isolated methionyl-tRNA synthetase and A549 and HCT116 cancer cells including an in silico binding mode analysis. Org Biomol Chem. 2015, 28, 9381–9387. [Google Scholar] [CrossRef]
- Kanchugarakoppal, S.R.; Basappa. New cholinesterase inhibitors: Synthesis and structure-activity relationship studies of 1,2-benzisoxazole series and novel imidazolyl-d 2-isoxazolines. J. Phys. Organ. Chem. 2005, 18, 773–778. [Google Scholar]
- Basappa; Kavitha, C.V.; Rangappa, K.S. Simple and an efficient method for the synthesis of 1-[2-dimethylamino-1-(4-methoxy-phenyl)-ethyl]-cyclohexanol hydrochloride: (+/−) venlafaxine racemic mixtures. Bioorg Med. Chem Lett. 2004, 21, 3279–3281. [Google Scholar] [CrossRef]
- Blanchard, V.; Chevalier, F.; Imberty, A.; Leeflang, B.R.; Basappa; Sugahara, K.; Kamerling, J.P. Conformational studies on five octasaccharides isolated from chondroitin sulfate using NMR spectroscopy and molecular modeling. Biochemistry. 2007, 46, 1167–1175. [Google Scholar] [CrossRef]
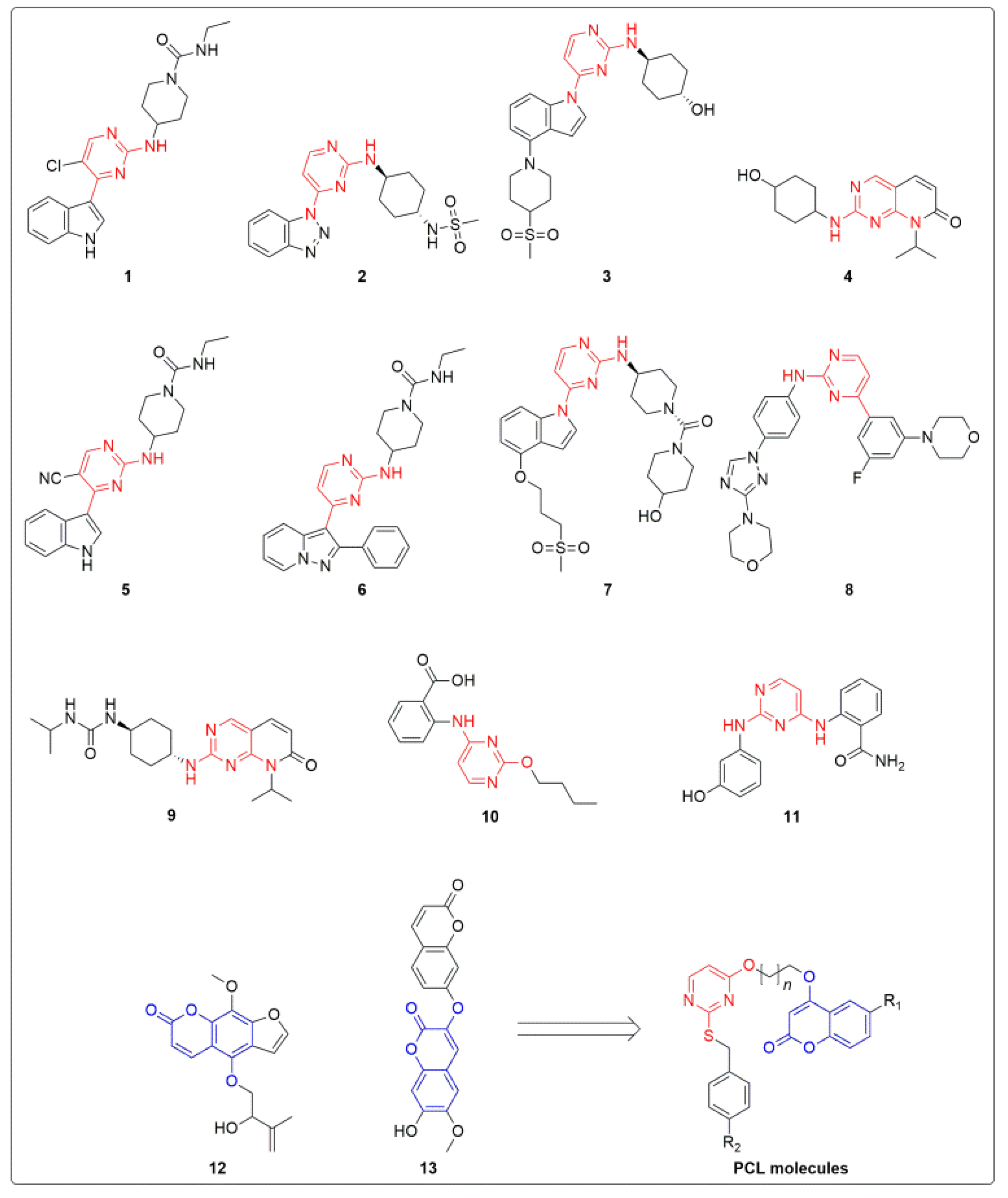


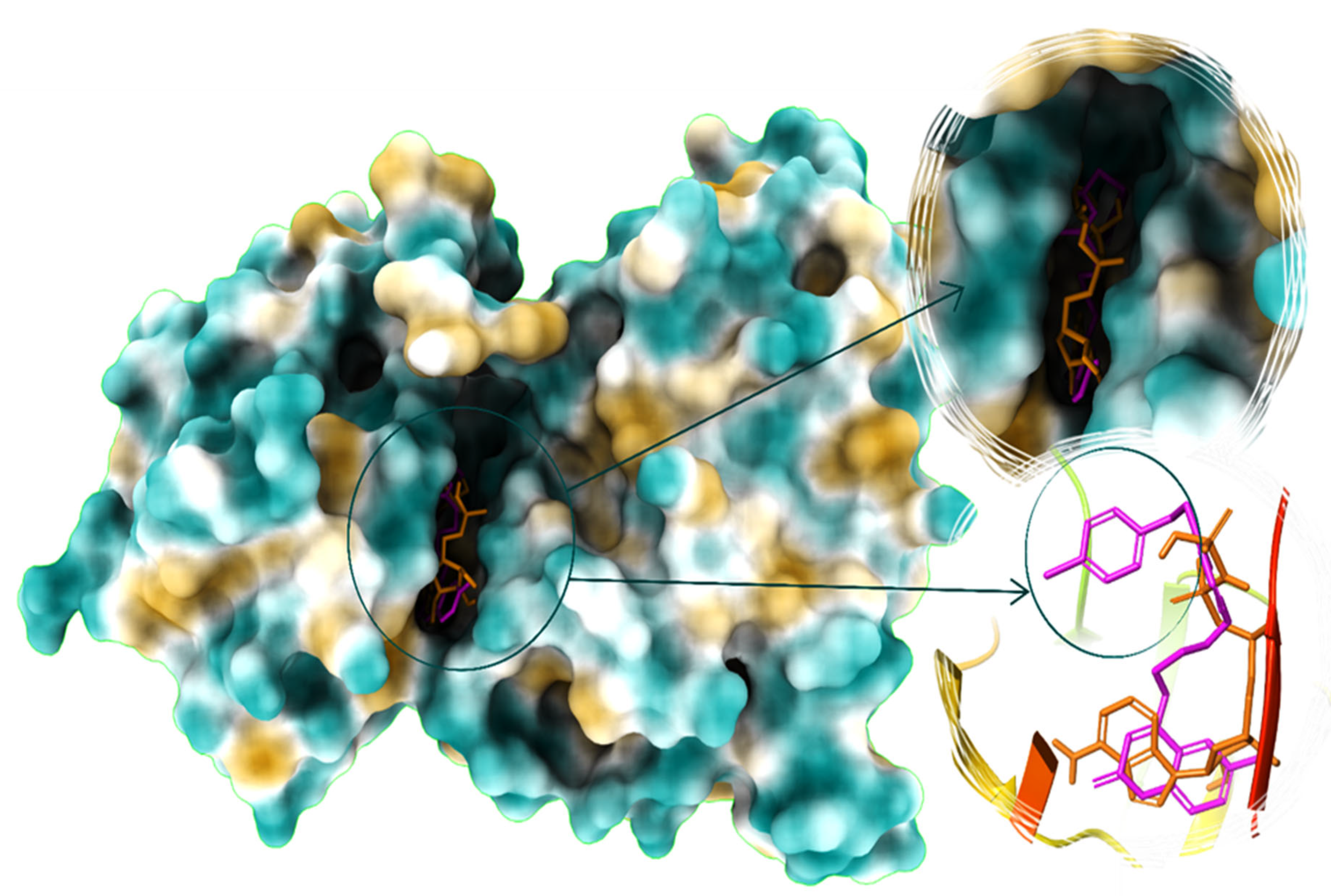
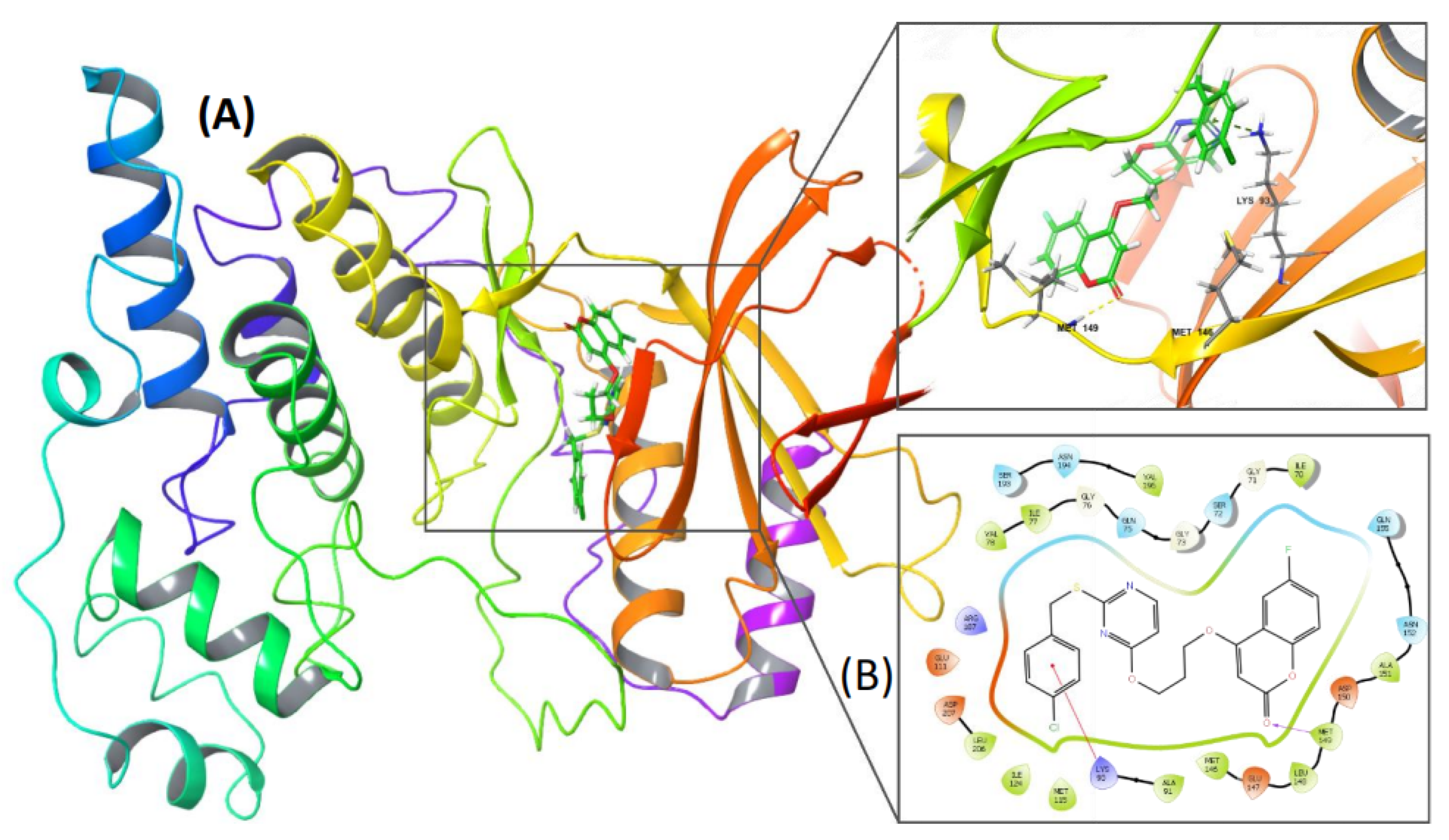

 | ||||
| Compound Code | R1 | R2 | n | MCF-7 (IC50 in μM) |
| PC-09 (5a) | H | Cl | 1 | 31.29 |
| PC-10 (5b) | H | Cl | 2 | 16.42 |
| PC-11 (5c) | F | Cl | 1 | 47.22 |
| PC-12 (5d) | F | Cl | 2 | 8.00 |
| PC-13 (5e) | H | OCH3 | 1 | >100 |
| PC-14 (5f) | H | OCH3 | 2 | 13.33 |
| PC-15 (5g) | F | OCH3 | 1 | >100 |
| PC-16 (5h) | F | OCH3 | 2 | >100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, N.Y.; Vishwanath, D.; Xi, Z.; Nagaraja, O.; Swamynayaka, A.; Kumar Harish, K.; Basappa, S.; Madegowda, M.; Pandey, V.; Sethi, G.; et al. Discovery of Pyrimidine- and Coumarin-Linked Hybrid Molecules as Inducers of JNK Phosphorylation through ROS Generation in Breast Cancer Cells. Molecules 2023, 28, 3450. https://doi.org/10.3390/molecules28083450
Kim NY, Vishwanath D, Xi Z, Nagaraja O, Swamynayaka A, Kumar Harish K, Basappa S, Madegowda M, Pandey V, Sethi G, et al. Discovery of Pyrimidine- and Coumarin-Linked Hybrid Molecules as Inducers of JNK Phosphorylation through ROS Generation in Breast Cancer Cells. Molecules. 2023; 28(8):3450. https://doi.org/10.3390/molecules28083450
Chicago/Turabian StyleKim, Na Young, Divakar Vishwanath, Zhang Xi, Omantheswara Nagaraja, Ananda Swamynayaka, Keshav Kumar Harish, Shreeja Basappa, Mahendra Madegowda, Vijay Pandey, Gautam Sethi, and et al. 2023. "Discovery of Pyrimidine- and Coumarin-Linked Hybrid Molecules as Inducers of JNK Phosphorylation through ROS Generation in Breast Cancer Cells" Molecules 28, no. 8: 3450. https://doi.org/10.3390/molecules28083450
APA StyleKim, N. Y., Vishwanath, D., Xi, Z., Nagaraja, O., Swamynayaka, A., Kumar Harish, K., Basappa, S., Madegowda, M., Pandey, V., Sethi, G., Lobie, P. E., Ahn, K. S., & Basappa, B. (2023). Discovery of Pyrimidine- and Coumarin-Linked Hybrid Molecules as Inducers of JNK Phosphorylation through ROS Generation in Breast Cancer Cells. Molecules, 28(8), 3450. https://doi.org/10.3390/molecules28083450








