Interfacial Catalysis during Amylolytic Degradation of Starch Granules: Current Understanding and Kinetic Approaches
Abstract
:1. Introduction
2. The Starch Granule
2.1. Nano-Level Structures
2.2. Blocklet Structures
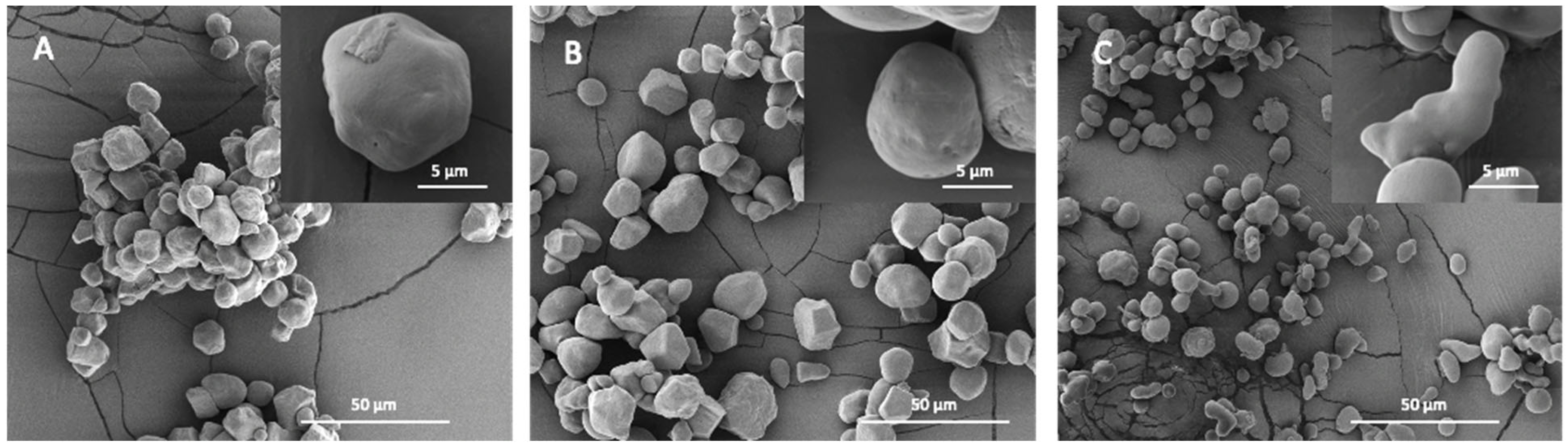
2.3. Starch Granular Topography and Morphology
3. Action of Hydrolase on Granular Substrates
3.1. Hydrolase Mechanisms on Granular Substrates
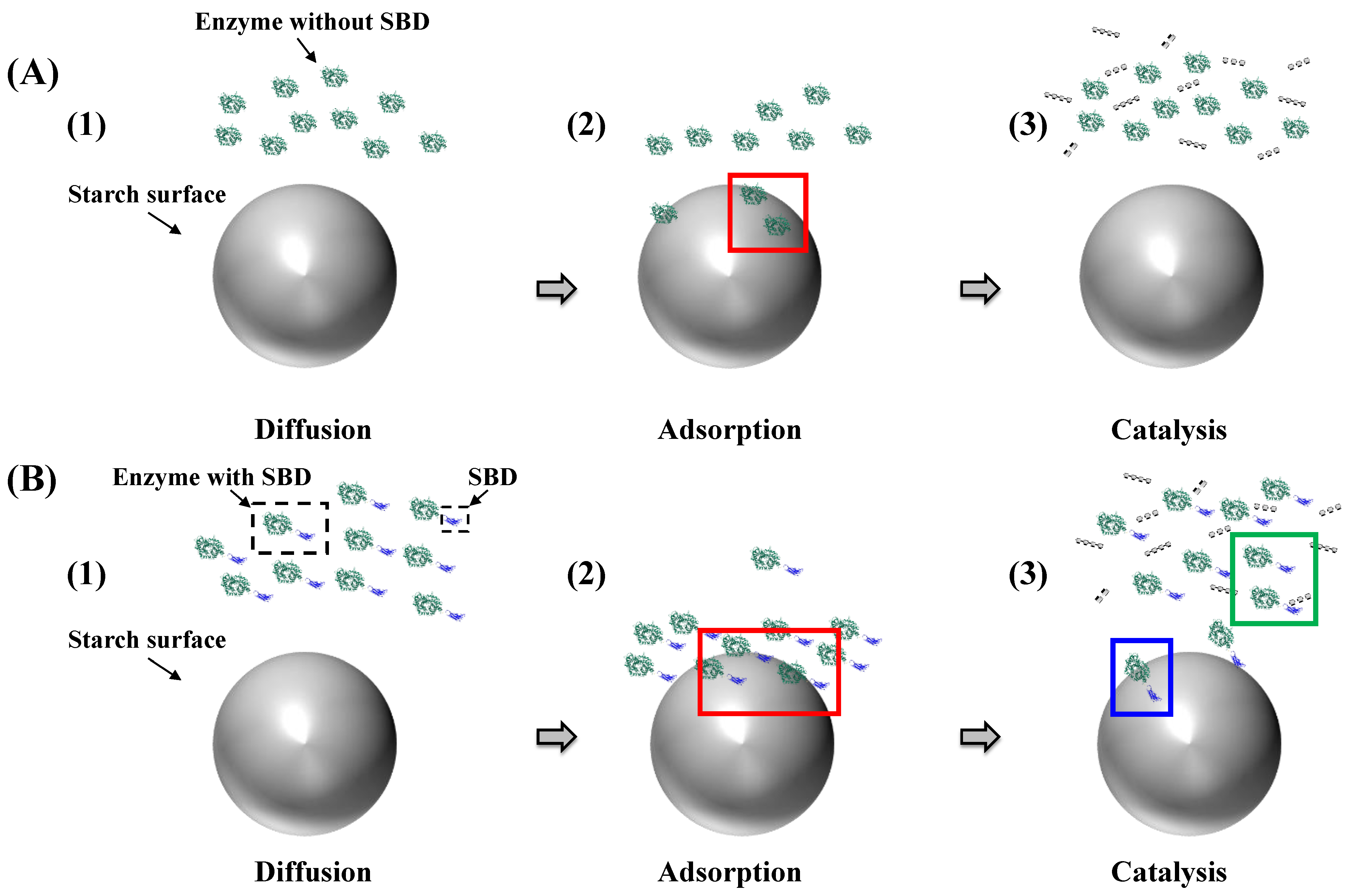
3.1.1. Diffusion Mechanism on the Granular Starch Surface
3.1.2. Starch Binding Ability
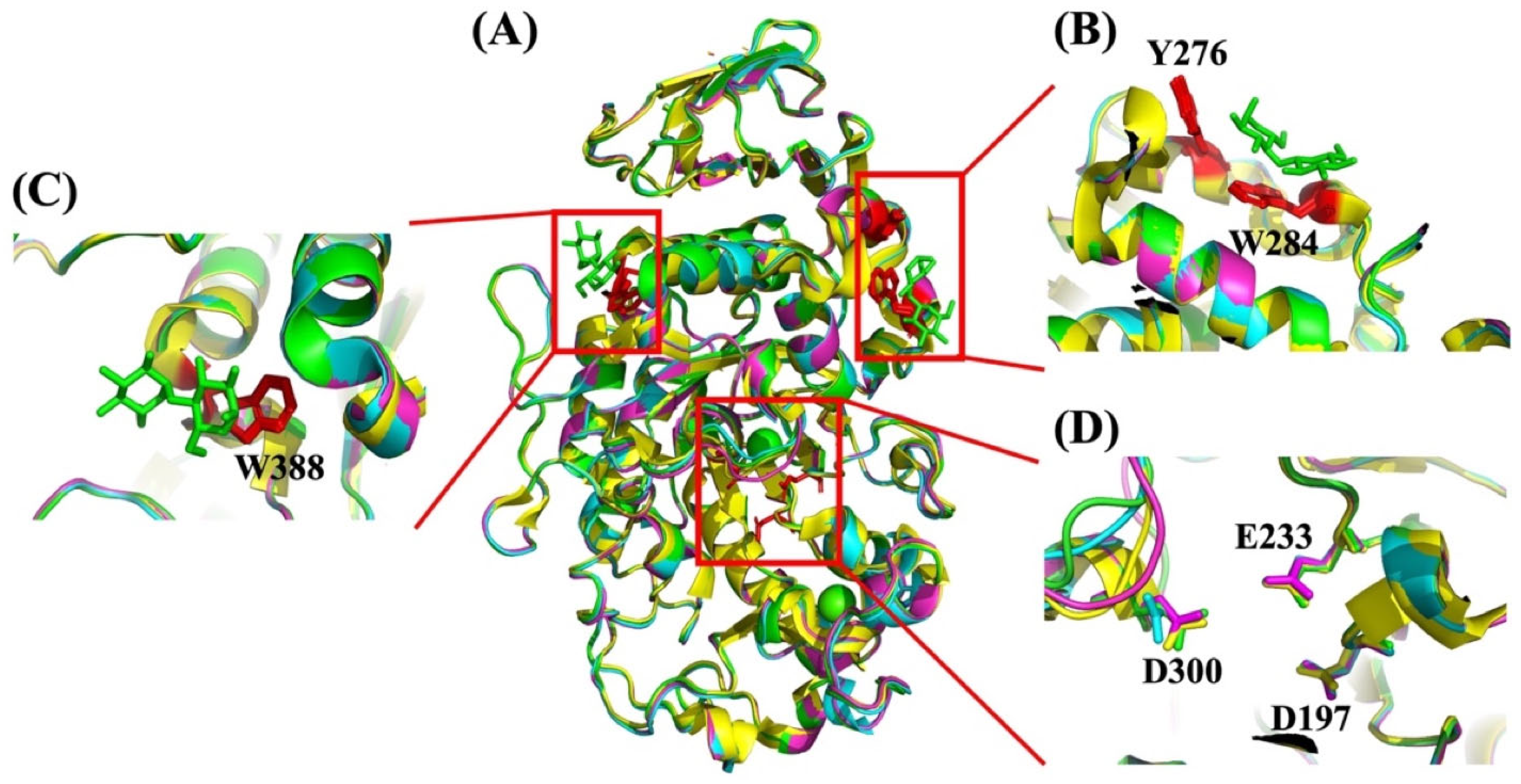
Starch Binding Domains
Surface Binding Sites
3.1.3. General Mechanism of Granular Starch Hydrolysis
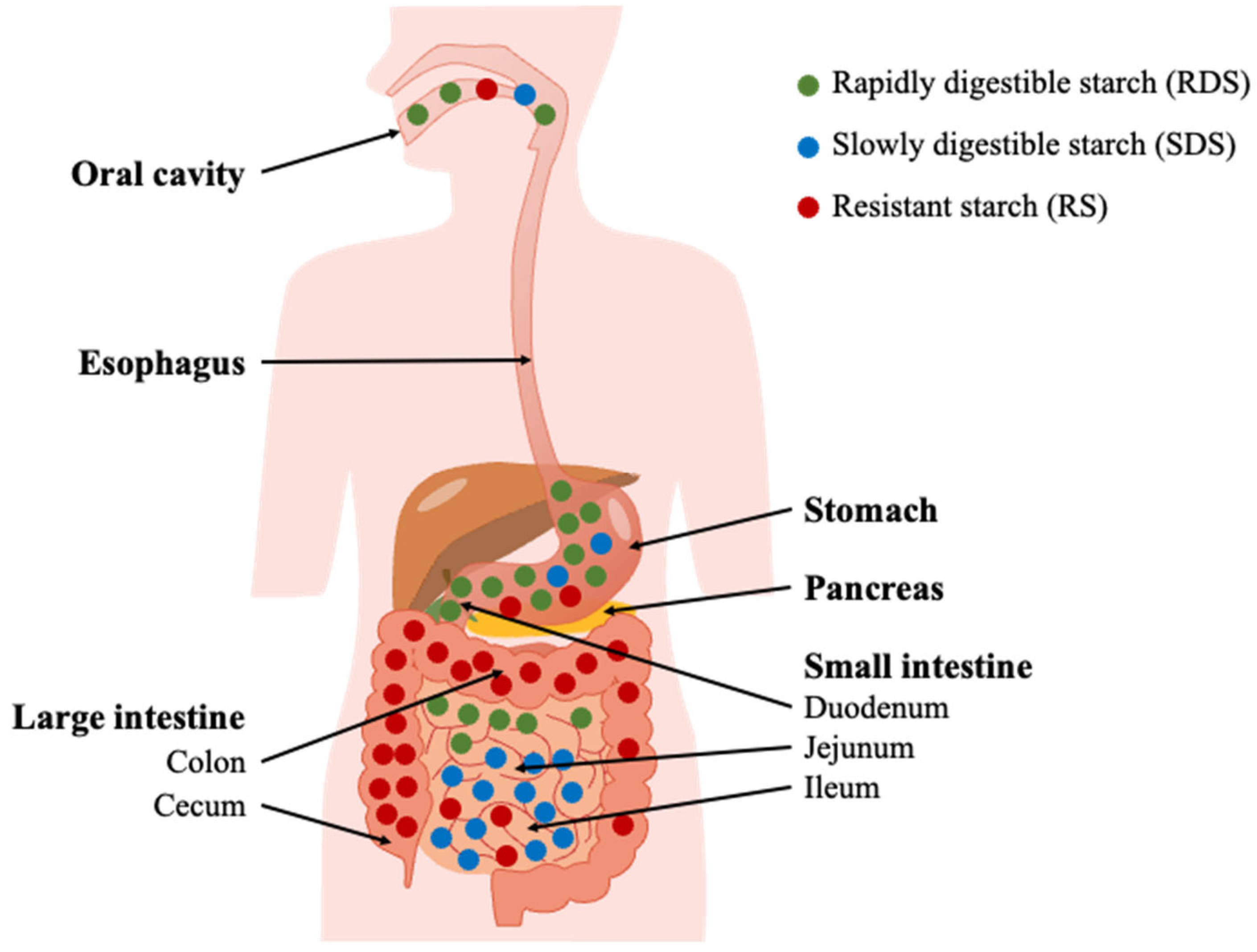
3.2. Dietary Starch Granule Digestion
3.2.1. Starch Hydrolysis in the Digestive System
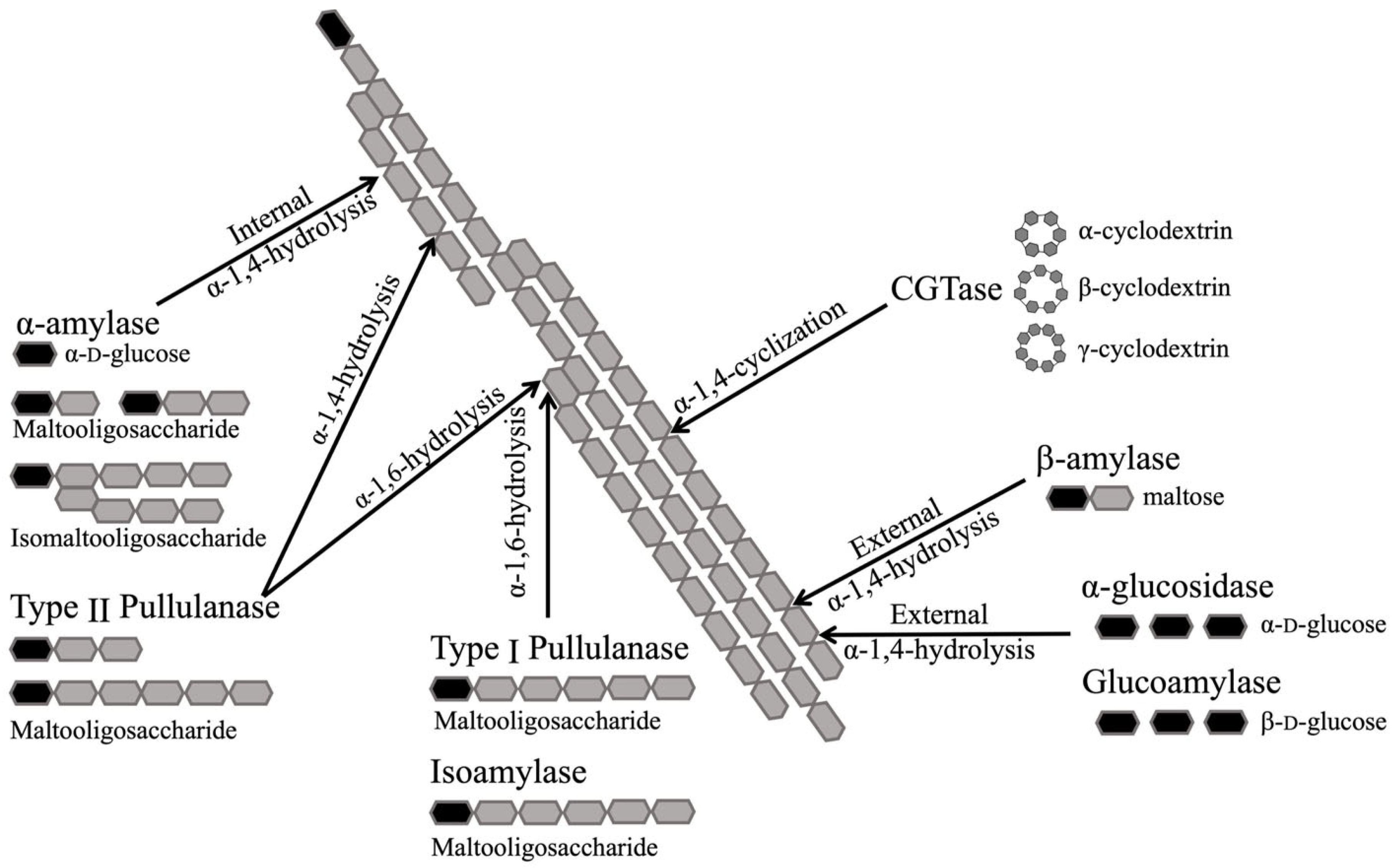
3.2.2. Glycoside Hydrolases Involved in Human Starch Digestion
α-Amylase
Glucoamylase
3.3. Starch Granule Structure and Digestion
4. Kinetics of Heterogeneous Systems
4.1. Analyzing the Digestive Rate and Efficiency of Granular Starch Degradation
4.2. Michaelis–Menten (M-M) Kinetics of Starch with Hydrolase
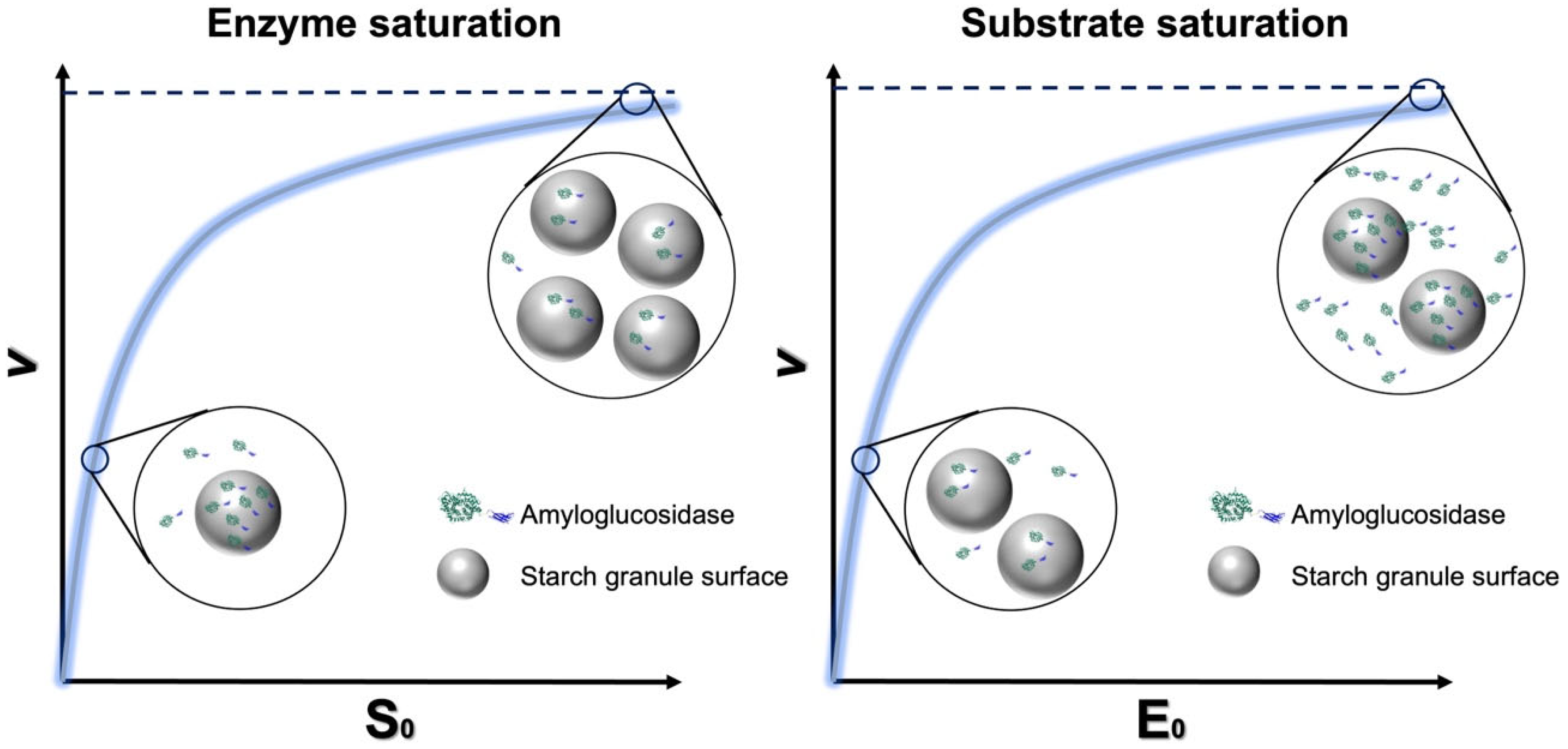
4.3. Handling Interfacial M-M Kinetics at High Amylase Concentrations
5. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cho, Y.G.; Kang, K.K. Functional analysis of starch metabolism in plants. Plants 2020, 9, 1152. [Google Scholar] [CrossRef] [PubMed]
- Blazek, J.; Gilbert, E.P. Effect of enzymatic hydrolysis on native starch granule structure. Biomacromolecules 2010, 11, 3275–3289. [Google Scholar] [CrossRef] [PubMed]
- Pietrzak, W.; Kawa-Rygielska, J. Ethanol fermentation of waste bread using granular starch hydrolyzing enzyme: Effect of raw material pretreatment. Fuel 2014, 134, 250–256. [Google Scholar] [CrossRef]
- Zhong, Y.; Xu, J.; Liu, X.; Ding, L.; Svensson, B.; Herburger, K.; Guo, K.; Pang, C.; Blennow, A. Recent advances in enzyme biotechnology on modifying gelatinized and granular starch. Trends Food Sci. Technol. 2022, 123, 343–354. [Google Scholar] [CrossRef]
- Kirk, O.; Borchert, T.V.; Fuglsang, C.C. Industrial enzyme applications. Curr. Opin. Biotechnol. 2002, 13, 345–351. [Google Scholar] [CrossRef]
- Cerqueira, F.M.; Photenhauer, A.L.; Pollet, R.M.; Brown, H.A.; Koropatkin, N.M. Starch digestion by gut bacteria: Crowdsourcing for carbs. Trends Microbiol. 2020, 28, 95–108. [Google Scholar] [CrossRef]
- Birt, D.F.; Boylston, T.; Hendrich, S.; Jane, J.-L.; Hollis, J.; Li, L.; McClelland, J.; Moore, S.; Phillips, G.J.; Rowling, M. Resistant starch: Promise for improving human health. Adv. Nutr. 2013, 4, 587–601. [Google Scholar] [CrossRef] [Green Version]
- Spinozzi, F.; Ferrero, C.; Perez, S. The architecture of starch blocklets follows phyllotaxic rules. Sci. Rep. 2020, 10, 20093. [Google Scholar] [CrossRef]
- Bertoft, E. Understanding Starch Structure: Recent Progress. Agronomy 2017, 7, 56. [Google Scholar] [CrossRef] [Green Version]
- Kari, J.; Olsen, J.P.; Jensen, K.; Badino, S.F.; Krogh, K.B.R.M.; Borch, K.; Westh, P. Sabatier principle for interfacial (heterogeneous) enzyme catalysis. ACS Catal. 2018, 8, 11966–11972. [Google Scholar] [CrossRef]
- Kari, J.; Andersen, M.; Borch, K.; Westh, P. An inverse Michaelis-Menten approach for interfacial enzyme kinetics. ACS Catal. 2017, 7, 4904–4914. [Google Scholar] [CrossRef]
- Smith, A.M.; Zeeman, S.C. Starch: A flexible, adaptable carbon store coupled to plant growth. Annu. Rev. Plant Biol. 2020, 71, 217–245. [Google Scholar] [CrossRef] [Green Version]
- Baghurst, P.A.; Baghurst, K.I.; Record, S.J. Dietary fibre, non-starch polysaccharides and resistant starch: A review. Food Aust. 1996, 48, S3–S35. [Google Scholar]
- Damager, I.; Engelsen, S.B.; Blennow, A.; Lindberg Møller, B.; Motawia, M.S. First principles insight into the α-glucan structures of starch: Their synthesis, conformation, and hydration. Chem. Rev. 2010, 110, 2049–2080. [Google Scholar] [CrossRef]
- Zhong, Y.; Li, Z.; Qu, J.; Bertoft, E.; Li, M.; Zhu, F.; Blennow, A.; Liu, X. Relationship between molecular structure and lamellar and crystalline structure of rice starch. Carbohydr. Polym. 2021, 258, 117616. [Google Scholar] [CrossRef]
- Nakamura, Y.; Kainuma, K. On the cluster structure of amylopectin. Plant Mol. Biol. 2022, 108, 291–306. [Google Scholar] [CrossRef]
- Jane, J. Current understanding on starch granule structures. J. Appl. Glycosci. 2006, 53, 205–213. [Google Scholar] [CrossRef] [Green Version]
- Jane, J.; Ao, Z.; Duvick, S.A.; Wiklund, M.; Yoo, S.-H.; Wong, K.-S.; Gardner, C. Structures of amylopectin and starch granules: How are they synthesized? J. Appl. Glycosci. 2003, 50, 167–172. [Google Scholar] [CrossRef]
- Alcázar-Alay, S.C.; Meireles, M.A.A. Physicochemical properties, modifications and applications of starches from different botanical sources. Food Sci. Technol. 2015, 35, 215–236. [Google Scholar] [CrossRef] [Green Version]
- Carciofi, M.; Blennow, A.; Jensen, S.L.; Shaik, S.S.; Henriksen, A.; Buléon, A.; Holm, P.B.; Hebelstrup, K.H. Concerted suppression of all starch branching enzyme genes in barley produces amylose-only starch granules. BMC Plant Biol. 2012, 12, 223. [Google Scholar] [CrossRef] [Green Version]
- Gidley, M.J.; Bociek, S.M. Molecular organization in starches: A carbon 13 CP/MAS NMR study. J. Am. Chem. Soc. 1985, 107, 7040–7044. [Google Scholar] [CrossRef]
- Blennow, A.; Hansen, M.; Schulz, A.; Jørgensen, K.; Donald, A.M.; Sanderson, J. The molecular deposition of transgenically modified starch in the starch granule as imaged by functional microscopy. J. Struct. Biol. 2003, 143, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Tang, P.C.; Ash, J.S.; Bates, D.W.; Overhage, J.M.; Sands, D.Z. Personal health records: Definitions, benefits, and strategies for overcoming barriers to adoption. J. Am. Med. Inform. Assoc. 2006, 13, 121–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, H.; Mitsunaga, T.; Kawamura, Y. Molecular arrangement in blocklets and starch granule architecture. Carbohydr. Polym. 2006, 63, 555–560. [Google Scholar] [CrossRef]
- Baker, A.A.; Miles, M.J.; Helbert, W. Internal structure of the starch granule revealed by AFM. Carbohydr. Res. 2001, 330, 249–256. [Google Scholar] [CrossRef]
- Dang, J.M.C.; Copeland, L. Imaging rice grains using atomic force microscopy. J. Cereal Sci. 2003, 37, 165–170. [Google Scholar] [CrossRef]
- Barrera, G.N.; Calderón-Domínguez, G.; Chanona-Pérez, J.; Gutiérrez-López, G.F.; León, A.E.; Ribotta, P.D. Evaluation of the mechanical damage on wheat starch granules by SEM, ESEM, AFM and texture image analysis. Carbohydr. Polym. 2013, 98, 1449–1457. [Google Scholar] [CrossRef]
- Chen, L.; McClements, D.J.; Ma, Y.; Yang, T.; Ren, F.; Tian, Y.; Jin, Z. Analysis of porous structure of potato starch granules by low-field NMR cryoporometry and AFM. Int. J. Biol. Macromol. 2021, 173, 307–314. [Google Scholar] [CrossRef]
- Baldwin, P.M.; Davies, M.C.; Melia, C.D. Starch granule surface imaging using low-voltage scanning electron microscopy and atomic force microscopy. Int. J. Biol. Macromol. 1997, 21, 103–107. [Google Scholar] [CrossRef]
- Chen, L.; Ma, R.; Zhang, Z.; Huang, M.; Cai, C.; Zhang, R.; McClements, D.J.; Tian, Y.; Jin, Z. Comprehensive investigation and comparison of surface microstructure of fractionated potato starches. Food Hydrocoll. 2019, 89, 11–19. [Google Scholar] [CrossRef]
- Ridout, M.J.; Parker, M.L.; Hedley, C.L.; Bogracheva, T.Y.; Morris, V.J. Atomic force microscopy of pea starch granules: Granule architecture of wild-type parent, r and rb single mutants, and the rrb double mutant. Carbohydr. Res. 2003, 338, 2135–2147. [Google Scholar] [CrossRef]
- Jane, J.; Kasemsuwan, T.; Leas, S.; Zobel, H.; Robyt, J.F. Anthology of starch granule morphology by scanning electron microscopy. Starch-Stärke 1994, 46, 121–129. [Google Scholar] [CrossRef]
- Hall, D.M.; Sayre, J.G. A scanning electron-microscope study of starches: Part III: Miscellaneous starches. Text. Res. J. 1971, 41, 880–894. [Google Scholar] [CrossRef]
- Pérez, S.; Bertoft, E. The molecular structures of starch components and their contribution to the architecture of starch granules: A comprehensive review. Starch-Staerke 2010, 62, 389–420. [Google Scholar] [CrossRef]
- Hall, D.M.; Sayre, J.G. A scanning electron-microscope study of starches: Part II: Cereal starches. Text. Res. J. 1970, 40, 256–266. [Google Scholar] [CrossRef]
- French, D. Chemical and physical properties of starch. J. Anim. Sci. 1973, 37, 1048–1061. [Google Scholar] [CrossRef]
- Lindeboom, N.; Chang, P.R.; Tyler, R.T. Analytical, biochemical and physicochemical aspects of starch granule size, with emphasis on small granule starches: A review. Starch-Stärke 2004, 56, 89–99. [Google Scholar] [CrossRef]
- Zeng, J.; Gao, H.; Li, G. Functional properties of wheat starch with different particle size distribution. J. Sci. Food Agric. 2014, 94, 57–62. [Google Scholar] [CrossRef]
- Song, Y.; Jane, J.-L. Characterization of barley starches of waxy, normal, and high amylose varieties. Carbohydr. Polym. 2000, 41, 365–377. [Google Scholar] [CrossRef]
- Jiang, H.; Campbell, M.; Blanco, M.; Jane, J.-L. Characterization of maize amylose-extender (ae) mutant starches: Part II. Structures and properties of starch residues remaining after enzymatic hydrolysis at boiling-water temperature. Carbohydr. Polym. 2010, 80, 1–12. [Google Scholar] [CrossRef]
- Lin, L.; Guo, D.; Zhao, L.; Zhang, X.; Wang, J.; Zhang, F.; Wei, C. Comparative structure of starches from high-amylose maize inbred lines and their hybrids. Food Hydrocoll. 2016, 52, 19–28. [Google Scholar] [CrossRef]
- Cai, C.; Zhao, L.; Huang, J.; Chen, Y.; Wei, C. Morphology, structure and gelatinization properties of heterogeneous starch granules from high-amylose maize. Carbohydr. Polym. 2014, 102, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Waterschoot, J.; Gomand, S.V.; Fierens, E.; Delcour, J.A. Production, structure, physicochemical and functional properties of maize, cassava, wheat, potato and rice starches. Starch-Stärke 2015, 67, 14–29. [Google Scholar] [CrossRef]
- Santacruz, S.; Koch, K.; Andersson, R.; Åman, P. Characterization of potato leaf starch. J. Agric. Food Chem. 2004, 52, 1985–1989. [Google Scholar] [CrossRef]
- Huber, K.C.; BeMiller, J.N. Channels of maize and sorghum starch granules. Carbohydr. Polym. 2000, 41, 269–276. [Google Scholar] [CrossRef]
- Baldwin, P.M.; Adler, J.; Davies, M.C.; Melia, C.D. Holes in starch granules: Confocal, SEM and light microscopy studies of starch granule structure. Starch-Stärke 1994, 46, 341–346. [Google Scholar] [CrossRef]
- Fannon, J.E.; Hauber, R.J.; BeMILLER, J.N. Surface pores of starch granules. Cereal Chem. 1992, 69, 284–288. [Google Scholar]
- Zhang, B.; Dhital, S.; Gidley, M.J. Synergistic and antagonistic effects of α-amylase and amyloglucosidase on starch digestion. Biomacromolecules 2013, 14, 1945–1954. [Google Scholar] [CrossRef]
- Benmoussa, M.; Hamaker, B.R.; Huang, C.P.; Sherman, D.M.; Weil, C.F.; BeMiller, J.N. Elucidation of maize endosperm starch granule channel proteins and evidence for plastoskeletal structures in maize endosperm amyloplasts. J. Cereal Sci. 2010, 52, 22–29. [Google Scholar] [CrossRef]
- Dhital, S.; Butardo, V.M., Jr.; Jobling, S.A.; Gidley, M.J. Rice starch granule amylolysis–Differentiating effects of particle size, morphology, thermal properties and crystalline polymorph. Carbohydr. Polym. 2015, 115, 305–316. [Google Scholar] [CrossRef]
- Huber, K.C.; BeMiller, J.N. Visualization of channels and cavities of corn and sorghum starch granules. Cereal Chem. 1997, 74, 537–541. [Google Scholar] [CrossRef]
- Fannon, J.E.; Gray, J.A.; Gunawan, N.; Huber, K.C.; BeMiller, J.N. Heterogeneity of starch granules and the effect of granule channelization on starch modification. Cellulose 2004, 11, 247–254. [Google Scholar] [CrossRef]
- Sujka, M.; Jamroz, J. Characteristics of pores in native and hydrolyzed starch granules. Starch-Stärke 2010, 62, 229–235. [Google Scholar] [CrossRef]
- Kuakpetoon, D.; Wang, Y.-J. Internal structure and physicochemical properties of corn starches as revealed by chemical surface gelatinization. Carbohydr. Res. 2007, 342, 2253–2263. [Google Scholar] [CrossRef]
- Glaring, M.A.; Koch, C.B.; Blennow, A. Genotype-specific spatial distribution of starch molecules in the starch granule: A combined CLSM and SEM approach. Biomacromolecules 2006, 7, 2310–2320. [Google Scholar] [CrossRef]
- Shaik, S.S.; Obata, T.; Hebelstrup, K.H.; Schwahn, K.; Fernie, A.R.; Mateiu, R.V.; Blennow, A. Starch granule re-structuring by starch branching enzyme and glucan water dikinase modulation affects caryopsis physiology and metabolism. PLoS ONE 2016, 11, e0149613. [Google Scholar] [CrossRef] [Green Version]
- Blennow, A.; Skryhan, K.; Tanackovic, V.; Krunic, S.L.; Shaik, S.S.; Andersen, M.S.; Kirk, H.G.; Nielsen, K.L. Non-GMO potato lines, synthesizing increased amylose and resistant starch, are mainly deficient in isoamylase debranching enzyme. Plant Biotechnol. J. 2020, 18, 2096. [Google Scholar] [CrossRef]
- Baldwin, A.J.; Egan, D.L.; Warren, F.J.; Barker, P.D.; Dobson, C.M.; Butterworth, P.J.; Ellis, P.R. Investigating the mechanisms of amylolysis of starch granules by solution-state NMR. Biomacromolecules 2015, 16, 1614–1621. [Google Scholar] [CrossRef]
- Ma, M.; Xu, Z.; Li, P.; Sui, Z.; Corke, H. Removal of starch granule-associated proteins affects amyloglucosidase hydrolysis of rice starch granules. Carbohydr. Polym. 2020, 247, 116674. [Google Scholar] [CrossRef]
- Schofield, J.D.; Greenwell, P. Wheat Starch Granule Proteins and Their Technological Significance. 1987. Available online: https://agris.fao.org/agris-search/search.do?recordID=US8907453 (accessed on 5 March 2023).
- Zhan, Q.; Ye, X.; Zhang, Y.; Kong, X.; Bao, J.; Corke, H.; Sui, Z. Starch granule-associated proteins affect the physicochemical properties of rice starch. Food Hydrocoll. 2020, 101, 105504. [Google Scholar] [CrossRef]
- Ye, X.; Zhang, Y.; Qiu, C.; Corke, H.; Sui, Z. Extraction and characterization of starch granule-associated proteins from rice that affect in vitro starch digestibility. Food Chem. 2019, 276, 754–760. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Ogawa, Y.; Shi, J.; Chen, S.; Zhang, H.; Liu, D.; Ye, X. The microstructure of starchy food modulates its digestibility. Crit. Rev. Food Sci. Nutr. 2019, 59, 3117–3128. [Google Scholar] [CrossRef] [PubMed]
- Gorodetskii, V.; Lauterbach, J.; Rotermund, H.-H.; Block, J.H.; Ertl, G. Coupling between adjacent crystal planes in heterogeneous catalysis by propagating reaction–diffusion waves. Nature 1994, 370, 276–279. [Google Scholar] [CrossRef]
- Juge, N.; Le Gal-Coëffet, M.F.; Furniss, C.S.M.; Gunning, A.P.; Kramhøft, B.; Morris, V.J.; Williamson, G.; Svensson, B. The starch binding domain of glucoamylase from Aspergillus niger: Overview of its structure, function, and role in raw-starch hydrolysis. Biol. Sect. Cell. Mol. Biol. 2002, 57, 239–245. [Google Scholar]
- Kadziola, A.; Søgaard, M.; Svensson, B.; Haser, R. Molecular structure of a barley α-amylase-inhibitor complex: Implications for starch binding and catalysis. J. Mol. Biol. 1998, 278, 205–217. [Google Scholar] [CrossRef]
- Robert, X.; Haser, R.; Gottschalk, T.E.; Ratajczak, F.; Driguez, H.; Svensson, B.; Aghajari, N. The structure of barley α-amylase isozyme 1 reveals a novel role of domain C in substrate recognition and binding: A pair of sugar tongs. Structure 2003, 11, 973–984. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, M.M.; Bozonnet, S.; Seo, E.N.; Mótyán, J.A.; Andersen, J.M.; Dilokpimol, A.; Hachem, M.A.; Gyémánt, G.; Næsted, H.; Kandra, L.; et al. Two secondary carbohydrate binding sites on the surface of barley α-amylase 1 have distinct functions and display synergy in hydrolysis of starch granules. Biochemistry 2009, 48, 7686–7697. [Google Scholar] [CrossRef]
- Christiansen, C.; Abou Hachem, M.; Glaring, M.A.; Viksø-Nielsen, A.; Sigurskjold, B.W.; Svensson, B.; Blennow, A. A CBM20 low-affinity starch-binding domain from glucan, water dikinase. FEBS Lett. 2009, 583, 1159–1163. [Google Scholar] [CrossRef] [Green Version]
- Giardina, T.; Gunning, A.P.; Juge, N.; Faulds, C.B.; Furniss, C.S.M.; Svensson, B.; Morris, V.J.; Williamson, G. Both binding sites of the starch-binding domain of Aspergillus niger glucoamylase are essential for inducing a conformational change in amylose. J. Mol. Biol. 2001, 313, 1149–1159. [Google Scholar] [CrossRef]
- Leloup, V.M.; Colonna, P.; Ring, S.G. α-Amylase adsorption on starch crystallites. Biotechnol. Bioeng. 1991, 38, 127–134. [Google Scholar] [CrossRef]
- Tian, Y.; Wang, Y.; Liu, X.; Herburger, K.; Westh, P.; Møller, M.S.; Svensson, B.; Zhong, Y.; Blennow, A. Interfacial enzyme kinetics reveals degradation mechanisms behind resistant starch. Food Hydrocoll. 2023, 140, 108621. [Google Scholar] [CrossRef]
- Arnling Bååth, J.; Jensen, K.; Borch, K.; Westh, P.; Kari, J. Sabatier principle for rationalizing enzymatic hydrolysis of a synthetic polyester. JACS Au 2022, 2, 1223–1231. [Google Scholar] [CrossRef]
- Feller, B.E.; Kellis, J.T.; Cascão-Pereira, L.G.; Robertson, C.R.; Frank, C.W. Interfacial biocatalysis on charged and immobilized substrates: The roles of enzyme and substrate surface charge. Langmuir 2011, 27, 250–263. [Google Scholar] [CrossRef]
- Warren, F.J.; Royall, P.G.; Gaisford, S.; Butterworth, P.J.; Ellis, P.R. Binding interactions of α-amylase with starch granules: The influence of supramolecular structure and surface area. Carbohydr. Polym. 2011, 86, 1038–1047. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- Janeček, Š.; Mareček, F.; MacGregor, E.A.; Svensson, B. Starch-binding domains as CBM families–history, occurrence, structure, function and evolution. Biotechnol. Adv. 2019, 37, 107451. [Google Scholar] [CrossRef]
- Ueda, S. Fungal glucoamylases and raw starch digestion. Trends Biochem. Sci. 1981, 6, 89–90. [Google Scholar] [CrossRef]
- Jespersen, H.M.; Macgregor, E.A.; Sierks, M.R.; Svensson, B. Comparison of the domain-level organization of starch hydrolases and related enzymes. Biochem. J 1991, 280, 51–55. [Google Scholar] [CrossRef] [Green Version]
- Valk, V.; Lammerts van Bueren, A.; van der Kaaij, R.M.; Dijkhuizen, L. Carbohydrate-binding module 74 is a novel starch-binding domain associated with large and multidomain α-amylase enzymes. FEBS J. 2016, 283, 2354–2368. [Google Scholar] [CrossRef] [Green Version]
- Southall, S.M.; Simpson, P.J.; Gilbert, H.J.; Williamson, G.; Williamson, M.P. The starch-binding domain from glucoamylase disrupts the structure of starch. FEBS Lett. 1999, 447, 58–60. [Google Scholar] [CrossRef] [Green Version]
- Armenta, S.; Moreno-Mendieta, S.; Sánchez-Cuapio, Z.; Sánchez, S.; Rodríguez-Sanoja, R. Advances in molecular engineering of carbohydrate-binding modules. Proteins Struct. Funct. Bioinform. 2017, 85, 1602–1617. [Google Scholar] [CrossRef] [PubMed]
- Sorimachi, K.; Le Gal-Coëffet, M.-F.; Williamson, G.; Archer, D.B.; Williamson, M.P. Solution structure of the granular starch binding domain of Aspergillus niger glucoamylase bound to β-cyclodextrin. Structure 1997, 5, 647–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cockburn, D.; Svensson, B. Surface binding sites in carbohydrate active enzymes: An emerging picture of structural and functional diversity. Carbohydr. Chem. 2013, 39, 204–221. [Google Scholar]
- Cockburn, D.; Nielsen, M.M.; Christiansen, C.; Andersen, J.M.; Rannes, J.B.; Blennow, A.; Svensson, B. Surface binding sites in amylase have distinct roles in recognition of starch structure motifs and degradation. Int. J. Biol. Macromol. 2015, 75, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Cockburn, D.; Wilkens, C.; Dilokpimol, A.; Nakai, H.; Lewińska, A.; Abou Hachem, M.; Svensson, B. Using carbohydrate interaction assays to reveal novel binding sites in carbohydrate active enzymes. PLoS ONE 2016, 11, e0160112. [Google Scholar] [CrossRef] [Green Version]
- Cockburn, D.; Wilkens, C.; Ruzanski, C.; Andersen, S.; Willum Nielsen, J.; Smith, A.M.; Field, R.A.; Willemoës, M.; Abou Hachem, M.; Svensson, B. Analysis of surface binding sites (SBSs) in carbohydrate active enzymes with focus on glycoside hydrolase families 13 and 77—A mini-review. Biol. 2014, 69, 705–712. [Google Scholar] [CrossRef]
- Gašperík, J.; Hostinová, E.; Ševočík, J. Acarbose binding at the surface of Saccharomycopsis fibuligera glucoamylase suggests the presence of a raw starch-binding site. Biol. Sect. Cell. Mol. Biol. 2005, 60, 167–170. [Google Scholar]
- Malle, D.; Itoh, T.; Hashimoto, W.; Murata, K.; Utsumi, S.; Mikami, B. Overexpression, purification and preliminary X-ray analysis of pullulanase from Bacillus subtilis strain 168. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2006, 62, 381–384. [Google Scholar] [CrossRef] [Green Version]
- Åkerberg, C.; Zacchi, G.; Torto, N.; Gorton, L. A kinetic model for enzymatic wheat starch saccharification. J. Chem. Technol. Biotechnol. 2000, 75, 306–314. [Google Scholar] [CrossRef]
- Apar, D.K.; Özbek, B. Estimation of kinetic parameters for rice starch hydrolysis inhibited by added materials. Chem. Eng. Commun. 2007, 194, 334–344. [Google Scholar] [CrossRef]
- Wang, J.P.; Zeng, A.W.; Liu, Z.; Yuan, X.G. Kinetics of glucoamylase hydrolysis of corn starch. J. Chem. Technol. Biotechnol. 2006, 81, 727–729. [Google Scholar] [CrossRef]
- Uitdehaag, J.; Mosi, R.; Kalk, K.H.; van der Veen, B.A.; Dijkhuizen, L.; Withers, S.G.; Dijkstra, B.W. X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the α-amylase family. Nat. Struct. Biol. 1999, 6, 432–436. [Google Scholar] [CrossRef] [Green Version]
- Ardèvol, A.; Rovira, C. Reaction mechanisms in carbohydrate-active enzymes: Glycoside hydrolases and glycosyltransferases. Insights from ab initio quantum mechanics/molecular mechanics dynamic simulations. J. Am. Chem. Soc. 2015, 137, 7528–7547. [Google Scholar] [CrossRef]
- Janeček, Š.; Svensson, B.; MacGregor, E.A. α-Amylase: An enzyme specificity found in various families of glycoside hydrolases. Cell. Mol. Life Sci. 2014, 71, 1149–1170. [Google Scholar] [CrossRef]
- Roman, L.; Sahagun, M.; Gomez, M.; Martinez, M.M. Nutritional and physical characterization of sugar-snap cookies: Effect of banana starch in native and molten states. Food Funct. 2019, 10, 616–624. [Google Scholar] [CrossRef]
- Weurding, R.E.; Enting, H.; Verstegen, M.W.A. The effect of site of starch digestion on performance of broiler chickens. Anim. Feed Sci. Technol. 2003, 110, 175–184. [Google Scholar] [CrossRef]
- Sorndech, W.; Tongta, S.; Blennow, A. Slowly digestible-and non-digestible α-glucans: An enzymatic approach to starch modification and nutritional effects. Starch-Stärke 2018, 70, 1700145. [Google Scholar] [CrossRef]
- Dhital, S.; Warren, F.J.; Butterworth, P.J.; Ellis, P.R.; Gidley, M.J. Mechanisms of starch digestion by α-amylase—Structural basis for kinetic properties. Crit. Rev. Food Sci. Nutr. 2017, 57, 875–892. [Google Scholar] [CrossRef]
- Li, Z.; Hu, G.; Zhu, L.; Zhao, Z.; Jiang, Y.; Gao, M.; Zhan, X. In vitro digestion and fecal fermentation of highly resistant starch rice and its effect on the gut microbiota. Food Chem. 2021, 361, 130095. [Google Scholar] [CrossRef]
- Teichmann, J.; Cockburn, D.W. In vitro fermentation reveals changes in butyrate production dependent on resistant starch source and microbiome composition. Front. Microbiol. 2021, 12, 640253. [Google Scholar] [CrossRef]
- Li, C.; Hu, Y. New definition of resistant starch types from the gut microbiota perspectives—A review. Crit. Rev. Food Sci. Nutr. 2022, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ze, X.; Ben David, Y.; Laverde-Gomez, J.A.; Dassa, B.; Sheridan, P.O.; Duncan, S.H.; Louis, P.; Henrissat, B.; Juge, N.; Koropatkin, N.M.; et al. Unique organization of extracellular amylases into amylosomes in the resistant starch-utilizing human colonic firmicutes bacterium Ruminococcus bromii. MBio 2015, 6, e01058-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, D.H.; Seo, D.H.; Kim, Y.J.; Chung, W.H.; Nam, Y.D.; Park, C.S. The presence of resistant starch-degrading amylases in Bifidobacterium adolescentis of the human gut. Int. J. Biol. Macromol. 2020, 161, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Raigond, P.; Ezekiel, R.; Raigond, B. Resistant starch in food: A review. J. Sci. Food Agric. 2015, 95, 1968–1978. [Google Scholar] [CrossRef]
- Lopez-Rubio, A.; Flanagan, B.M.; Shrestha, A.K.; Gidley, M.J.; Gilbert, E.P. Molecular rearrangement of starch during in vitro digestion: Toward a better understanding of enzyme resistant starch formation in processed starches. Biomacromolecules 2008, 9, 1951–1958. [Google Scholar] [CrossRef]
- Møller, M.S.; Svensson, B. Structural biology of starch-degrading enzymes and their regulation. Curr. Opin. Struct. Biol. 2016, 40, 33–42. [Google Scholar] [CrossRef] [Green Version]
- Kaoutari, A.E.; Armougom, F.; Gordon, J.I.; Raoult, D.; Henrissat, B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 2013, 11, 497–504. [Google Scholar] [CrossRef]
- MacGregor, E.A.; Janeček, Š.; Svensson, B. Relationship of sequence and structure to specificity in the α-amylase family of enzymes. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 2001, 1546, 1–20. [Google Scholar] [CrossRef]
- Janeek, Š.; Svensson, B. How many α-amylase GH families are there in the CAZy database? Amylase 2022, 6, 1–10. [Google Scholar] [CrossRef]
- Krishnan, T.; Chandra, A.K. Purification and characterization of α-amylase from Bacillus licheniformis CUMC305. Appl. Environ. Microbiol. 1983, 46, 430–437. [Google Scholar] [CrossRef] [Green Version]
- Janeček, Š. α-amylases from archaea: Sequences, structures and evolution. In Biotechnology of Extremophiles: Advances and Challenges; Springer: Cham, Switzerland, 2016; pp. 505–524. [Google Scholar]
- Janíčková, Z.; Janeček, Š. Fungal α-amylases from three GH13 subfamilies: Their sequence-structural features and evolutionary relationships. Int. J. Biol. Macromol. 2020, 159, 763–772. [Google Scholar] [CrossRef]
- Janeček, Š. Amylolytic enzymes—Focus on the alpha-amylases from archaea and plants. Nov. Biotechnol. 2009, 9, 5–25. [Google Scholar] [CrossRef]
- Ramasubbu, N.; Paloth, V.; Luo, Y.; Brayer, G.D.; Levine, M.J. Structure of human salivary α-amylase at 1.6 Å resolution: Implications for its role in the oral cavity. Acta Crystallogr. Sect. D Biol. Crystallogr. 1996, 52, 435–446. [Google Scholar] [CrossRef]
- Brayer, G.D.; Luo, Y.; Withers, S.G. The structure of human pancreatic α-amylase at 1.8 Å resolution and comparisons with related enzymes. Protein Sci. 1995, 4, 1730–1742. [Google Scholar] [CrossRef]
- Butterworth, P.J.; Warren, F.J.; Ellis, P.R. Human α-amylase and starch digestion: An interesting marriage. Starch-Stärke 2011, 63, 395–405. [Google Scholar] [CrossRef]
- Kandra, L.; Gyémánt, G.; Farkas, E.; Lipták, A. Action pattern of porcine pancreatic alpha-amylase on three different series of β-maltooligosaccharide glycosides. Carbohydr. Res. 1997, 298, 237–242. [Google Scholar] [CrossRef]
- Whitcomb, D.C.; Lowe, M.E. Human pancreatic digestive enzymes. Dig. Dis. Sci. 2007, 52, 1–17. [Google Scholar] [CrossRef]
- Wang, S.; Guo, P. Botanical Sources of Starch. In Starch Structure, Functionality and Application in Foods; Springer: Singapore, 2020; ISBN 9789811506215. [Google Scholar]
- Mandel, A.L.; Peyrot des Gachons, C.; Plank, K.L.; Alarcon, S.; Breslin, P.A.S. Individual differences in AMY1 gene copy number, salivary α-amylase levels, and the perception of oral starch. PLoS ONE 2010, 5, e13352. [Google Scholar] [CrossRef] [Green Version]
- Lapis, T.J.; Penner, M.H.; Balto, A.S.; Lim, J. Oral digestion and perception of starch: Effects of cooking, tasting time, and salivary α-amylase activity. Chem. Sens. 2017, 42, 635–645. [Google Scholar] [CrossRef] [Green Version]
- Champ, M.; Martin, L.; Noah, L.; Gratas, M. Analytical methods for resistant starch. In Complex Carbohydrates in Foods; CRC Press: Boca Raton, FL, USA, 1999; pp. 173–193. ISBN 0203909577. [Google Scholar]
- McCleary, B.V.; Monaghan, D.A. Measurement of resistant starch. J. AOAC Int. 2002, 85, 665–675. [Google Scholar] [CrossRef] [Green Version]
- Kondo, H.; Nakatani, H.; Hiromi, K. In vitro action of human and porcine α-amylases on cyclomalto-oligosaccharides. Carbohydr. Res. 1990, 204, 207–213. [Google Scholar] [CrossRef]
- Zhang, X.; Caner, S.; Kwan, E.; Li, C.; Brayer, G.D.; Withers, S.G. Evaluation of the significance of starch surface binding sites on human pancreatic α-amylase. Biochemistry 2016, 55, 6000–6009. [Google Scholar] [CrossRef]
- Machius, M.; Vértesy, L.; Huber, R.; Wiegand, G. Carbohydrate and protein-based inhibitors of porcine pancreatic α-amylase: Structure analysis and comparison of their binding characteristics. J. Mol. Biol. 1996, 260, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Al Kazaz, M.; Desseaux, V.; Marchis-Mouren, G.; Prodanov, E.; Santimone, M. The mechanism of porcine pancreatic α-amylase. Inhibition of maltopentaose hydrolysis by acarbose, maltose and maltotriose. Eur. J. Biochem. 1998, 252, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Ragunath, C.; Manuel, S.G.A.; Venkataraman, V.; Sait, H.B.R.; Kasinathan, C.; Ramasubbu, N. Probing the role of aromatic residues at the secondary saccharide-binding sites of human salivary α-amylase in substrate hydrolysis and bacterial binding. J. Mol. Biol. 2008, 384, 1232–1248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sim, L.; Quezada-Calvillo, R.; Sterchi, E.E.; Nichols, B.L.; Rose, D.R. Human intestinal maltase-glucoamylase: Crystal structure of the N-terminal catalytic subunit and basis of inhibition and substrate specificity. J. Mol. Biol. 2008, 375, 782–792. [Google Scholar] [CrossRef]
- Gericke, B.; Schecker, N.; Amiri, M.; Naim, H.Y. Structure-function analysis of human sucrase-isomaltase identifies key residues required for catalytic activity. J. Biol. Chem. 2017, 292, 11070–11078. [Google Scholar] [CrossRef] [Green Version]
- Quezada-Calvillo, R.; Robayo-Torres, C.C.; Opekun, A.R.; Sen, P.; Ao, Z.; Hamaker, B.R.; Quaroni, A.; Brayer, G.D.; Wattler, S.; Nehls, M.C.; et al. Contribution of mucosal maltase-glucoamylase activities to mouse small intestinal starch α-glucogenesis. J. Nutr. 2007, 137, 1725–1733. [Google Scholar] [CrossRef] [Green Version]
- Dhital, S.; Lin, A.H.-M.; Hamaker, B.R.; Gidley, M.J.; Muniandy, A. Mammalian mucosal α-glucosidases coordinate with α-amylase in the initial starch hydrolysis stage to have a role in starch digestion beyond glucogenesis. PLoS ONE 2013, 8, e62546. [Google Scholar] [CrossRef] [Green Version]
- Muir, J.G.; O’Dea, K. Validation of an in vitro assay for predicting the amount of starch that escapes digestion in the small intestine of humans. Am. J. Clin. Nutr. 1993, 57, 540–546. [Google Scholar] [CrossRef]
- Warren, F.J.; Zhang, B.; Waltzer, G.; Gidley, M.J.; Dhital, S. The interplay of α-amylase and amyloglucosidase activities on the digestion of starch in in vitro enzymic systems. Carbohydr. Polym. 2015, 117, 192–200. [Google Scholar] [CrossRef] [Green Version]
- Sierks, M.R.; Ford, C.; Reilly, P.J.; Svensson, B. Catalytic mechanism of fungal glucoamylase as defined by mutagenesis of Asp176, Glu179 and Glu180 in the enzyme from Aspergillus awamori. Protein Eng. Des. Sel. 1990, 3, 193–198. [Google Scholar] [CrossRef]
- Svensson, B.; Larsen, K.; Svendsen, I.; Boel, E. The complete amino acid sequence of the glycoprotein, glucoamylase G1, from Aspergillus niger. Carlsberg Res. Commun. 1983, 48, 529–544. [Google Scholar] [CrossRef] [Green Version]
- Svensson, B.; Jespersen, H.; Sierks, M.R.; MacGregor, E.A. Sequence homology between putative raw-starch binding domains from different starch-degrading enzymes. Biochem. J. 1989, 264, 309–311. [Google Scholar] [CrossRef] [Green Version]
- Bertoldo, C.; Antranikian, G. Starch-hydrolyzing enzymes from thermophilic archaea and bacteria. Curr. Opin. Chem. Biol. 2002, 6, 151–160. [Google Scholar] [CrossRef]
- Sauer, J.; Sigurskjold, B.W.; Christensen, U.; Frandsen, T.P.; Mirgorodskaya, E.; Harrison, M.; Roepstorff, P.; Svensson, B. Glucoamylase: Structure/function relationships, and protein engineering. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 2000, 1543, 275–293. [Google Scholar] [CrossRef]
- Tung, J.Y.; Chang, M.D.T.; Chou, W.I.; Liu, Y.Y.; Yeh, Y.H.; Chang, F.Y.; Lin, S.C.; Qiu, Z.L.; Sun, Y.J. Crystal structures of the starch-binding domain from Rhizopus oryzae glucoamylase reveal a polysaccharide-binding path. Biochem. J. 2008, 416, 27–36. [Google Scholar] [CrossRef] [Green Version]
- Gous, P.W.; Fox, G.P. Amylopectin synthesis and hydrolysis-Understanding isoamylase and limit dextrinase and their impact on starch structure on barley (Hordeum vulgare) quality. Trends Food Sci. Technol. 2017, 62, 23–32. [Google Scholar] [CrossRef] [Green Version]
- Mikami, B.; Degano, M.; Hehre, E.J.; Sacchettini, J.C. Crystal structures of soybean. beta-amylase reacted with. beta-maltose and maltal: Active site components and their apparent roles in catalysis. Biochemistry 1994, 33, 7779–7787. [Google Scholar] [CrossRef]
- Lundgard, R.; Svensson, B. The four major forms of barley β-amylase. Purification, characterization and structural relationship. Carlsberg Res. Commun. 1987, 52, 313–326. [Google Scholar] [CrossRef] [Green Version]
- Hii, S.L.; Tan, J.S.; Ling, T.C.; Ariff, A. Bin Pullulanase: Role in starch hydrolysis and potential industrial applications. Enzyme Res. 2012, 2012, 921362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szerman, N.; Schroh, I.; Rossi, A.L.; Rosso, A.M.; Krymkiewicz, N.; Ferrarotti, S.A. Cyclodextrin production by cyclodextrin glycosyltransferase from Bacillus circulans DF 9R. Bioresour. Technol. 2007, 98, 2886–2891. [Google Scholar] [CrossRef] [PubMed]
- Kong, B.W.; Kim, J.I.; Kim, M.J.; Kim, J.C. Porcine pancreatic α-amylase hydrolysis of native starch granules as a function of granule surface area. Biotechnol. Prog. 2003, 19, 1162–1166. [Google Scholar] [CrossRef] [PubMed]
- Chi, C.; Li, X.; Huang, S.; Chen, L.; Zhang, Y.; Li, L.; Miao, S. Basic principles in starch multi-scale structuration to mitigate digestibility: A review. Trends Food Sci. Technol. 2021, 109, 154–168. [Google Scholar] [CrossRef]
- Zhu, L.-J.; Liu, Q.-Q.; Wilson, J.D.; Gu, M.-H.; Shi, Y.-C. Digestibility and physicochemical properties of rice (Oryza sativa L.) flours and starches differing in amylose content. Carbohydr. Polym. 2011, 86, 1751–1759. [Google Scholar] [CrossRef]
- Gerard, C.; Colonna, P.; Buleon, A.; Planchot, V. Amylolysis of maize mutant starches. J. Sci. Food Agric. 2001, 81, 1281–1287. [Google Scholar] [CrossRef]
- Huang, J.; Shang, Z.; Man, J.; Liu, Q.; Zhu, C.; Wei, C. Comparison of molecular structures and functional properties of high-amylose starches from rice transgenic line and commercial maize. Food Hydrocoll. 2015, 46, 172–179. [Google Scholar] [CrossRef]
- Zhang, B.; Dhital, S.; Gidley, M.J. Densely packed matrices as rate determining features in starch hydrolysis. Trends Food Sci. Technol. 2015, 43, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Gong, B.; Hu, Y.; Liu, X.; Guan, X.; Zhang, B. Combined crystalline, lamellar and granular structural insights into in vitro digestion rate of native starches. Food Hydrocoll. 2020, 105, 105823. [Google Scholar] [CrossRef]
- Warren, F.J.; Butterworth, P.J.; Ellis, P.R. The surface structure of a complex substrate revealed by enzyme kinetics and Freundlich constants for α-amylase interaction with the surface of starch. Biochim. Biophys. Acta Gen. Subj. 2013, 1830, 3095–3101. [Google Scholar] [CrossRef]
- Jamme, F.; Bourquin, D.; Tawil, G.; Viksø-Nielsen, A.; Buléon, A.; Réfrégiers, M. 3D imaging of enzymes working in situ. Anal. Chem. 2014, 86, 5265–5270. [Google Scholar] [CrossRef]
- Dhital, S.; Warren, F.J.; Zhang, B.; Gidley, M.J. Amylase binding to starch granules under hydrolysing and non-hydrolysing conditions. Carbohydr. Polym. 2014, 113, 97–107. [Google Scholar] [CrossRef] [Green Version]
- Ma, M.; Xu, Y.; Liu, Z.; Sui, Z.; Corke, H. Removal of starch granule-associated proteins promotes α-amylase hydrolysis of rice starch granule. Food Chem. 2020, 330, 127313. [Google Scholar] [CrossRef]
- Hu, P.; Fan, X.; Lin, L.; Wang, J.; Zhang, L.; Wei, C. Effects of surface proteins and lipids on molecular structure, thermal properties, and enzymatic hydrolysis of rice starch. Food Sci. Technol. 2017, 38, 84–90. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Hu, X.; Luo, S.; McClements, D.J.; Liang, L.; Liu, C. Effect of endogenous proteins and lipids on starch digestibility in rice flour. Food Res. Int. 2018, 106, 404–409. [Google Scholar] [CrossRef]
- Butterworth, P.J.; Ellis, P.R. Kinetics of α-amylase action on starch. In Interdisciplinary Approaches to Food Digestion; Springer: Cham, Switzerland, 2019; pp. 291–302. [Google Scholar]
- Dona, A.C.; Pages, G.; Gilbert, R.G.; Kuchel, P.W. Digestion of starch: In vivo and in vitro kinetic models used to characterise oligosaccharide or glucose release. Carbohydr. Polym. 2010, 80, 599–617. [Google Scholar] [CrossRef]
- Zhang, B.; Li, H.; Wang, S.; Junejo, S.A.; Liu, X.; Huang, Q. In Vitro Starch Digestion: Mechanisms and Kinetic Models. In Starch Structure, Functionality and Application in Foods; Springer: Singapore, 2020; pp. 151–167. [Google Scholar]
- Benavent-Gil, Y.; Rosell, C.M. Performance of granular starch with controlled pore size during hydrolysis with digestive enzymes. Plant Foods Hum. Nutr. 2017, 72, 353–359. [Google Scholar] [CrossRef]
- Li, P.; Dhital, S.; Zhang, B.; He, X.; Fu, X.; Huang, Q. Surface structural features control in vitro digestion kinetics of bean starches. Food Hydrocoll. 2018, 85, 343–351. [Google Scholar] [CrossRef] [Green Version]
- Tahir, R.; Ellis, P.R.; Butterworth, P.J. The relation of physical properties of native starch granules to the kinetics of amylolysis catalysed by porcine pancreatic α-amylase. Carbohydr. Polym. 2010, 81, 57–62. [Google Scholar] [CrossRef]
- Slaughter, S.L.; Ellis, P.R.; Butterworth, P.J. An investigation of the action of porcine pancreatic α-amylase on native and gelatinised starches. Biochim. Biophys. Acta (BBA) Gen. Subj. 2001, 1525, 29–36. [Google Scholar] [CrossRef]
- Kim, M.-J.; Jung, Y.-J.; Lee, S.H.; Lee, H.; Kim, J.C. Kinetic analysis and enzyme concentration effect relevant to dependence of amylolysis of starch granules on specific surface area concentration. Food Sci. Biotechnol. 2014, 23, 475–481. [Google Scholar] [CrossRef]
- Takagi, H.; Kubo, A.; Inoue, M.; Nakaya, M.; Suzuki, S.; Kitamura, S. Binding interaction of porcine pancreatic α-amylase with waxy/amylose extender double-mutant rice starch granules does not determine their susceptibility to hydrolysis. Food Sci. Technol. Res. 2018, 24, 363–368. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Cai, L.; Gu, Z.; Shi, Y.-C. Effects of granule swelling on starch saccharification by granular starch hydrolyzing enzyme. J. Agric. Food Chem. 2014, 62, 8114–8119. [Google Scholar] [CrossRef] [PubMed]
- Patel, H.; Royall, P.G.; Gaisford, S.; Williams, G.R.; Edwards, C.H.; Warren, F.J.; Flanagan, B.M.; Ellis, P.R.; Butterworth, P.J. Structural and enzyme kinetic studies of retrograded starch: Inhibition of α-amylase and consequences for intestinal digestion of starch. Carbohydr. Polym. 2017, 164, 154–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhital, S.; Gidley, M.J.; Warren, F.J. Inhibition of α-amylase activity by cellulose: Kinetic analysis and nutritional implications. Carbohydr. Polym. 2015, 123, 305–312. [Google Scholar] [CrossRef]
- Li, H.T.; Sartika, R.S.; Kerr, E.D.; Schulz, B.L.; Gidley, M.J.; Dhital, S. Starch granular protein of high-amylose wheat gives innate resistance to amylolysis. Food Chem. 2020, 330, 127328. [Google Scholar] [CrossRef]
- Wang, S.; Wang, S.; Liu, L.; Wang, S.; Copeland, L. Structural orders of wheat starch do not determine the in vitro enzymatic digestibility. J. Agric. Food Chem. 2017, 65, 1697–1706. [Google Scholar] [CrossRef]
- Wang, Y.; Chao, C.; Huang, H.; Wang, S.; Wang, S.; Wang, S.; Copeland, L. Revisiting mechanisms underlying digestion of starches. J. Agric. Food Chem. 2019, 67, 8212–8226. [Google Scholar] [CrossRef]
- Dahlqvist, A.; Borgström, B. Digestion and absorption of disaccharides in man. Biochem. J. 1961, 81, 411. [Google Scholar] [CrossRef]
- Srere, P.A. Enzyme concentrations in tissues. Science 1967, 158, 936–937. [Google Scholar] [CrossRef]
- Mukhopadhya, I.; Moraïs, S.; Laverde-Gomez, J.; Sheridan, P.O.; Walker, A.W.; Kelly, W.; Klieve, A.V.; Ouwerkerk, D.; Duncan, S.H.; Louis, P. Sporulation capability and amylosome conservation among diverse human colonic and rumen isolates of the keystone starch-degrader Ruminococcus bromii. Environ. Microbiol. 2018, 20, 324–336. [Google Scholar] [CrossRef] [Green Version]
- Jung, D.-H.; Kim, G.-Y.; Kim, I.-Y.; Seo, D.-H.; Nam, Y.-D.; Kang, H.; Song, Y.; Park, C.-S. Bifidobacterium adolescentis P2P3, a human gut bacterium having strong non-gelatinized resistant starch-degrading activity. J. Microbiol. Biotechnol. 2019, 29, 1904–1915. [Google Scholar] [CrossRef] [PubMed]
- Bui, A.T.; Williams, B.A.; Hoedt, E.C.; Morrison, M.; Mikkelsen, D.; Gidley, M.J. High amylose wheat starch structures display unique fermentability characteristics, microbial community shifts and enzyme degradation profiles. Food Funct. 2020, 11, 5635–5646. [Google Scholar] [CrossRef]
- Hardy, K.; Brand-Miller, J.; Brown, K.D.; Thomas, M.G.; Copeland, L. The Importance of Dietary Carbohydrate in Human Evolution. Q. Rev. Biol. 2015, 90, 251–268. [Google Scholar] [CrossRef] [Green Version]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Y.; Wang, Y.; Zhong, Y.; Møller, M.S.; Westh, P.; Svensson, B.; Blennow, A. Interfacial Catalysis during Amylolytic Degradation of Starch Granules: Current Understanding and Kinetic Approaches. Molecules 2023, 28, 3799. https://doi.org/10.3390/molecules28093799
Tian Y, Wang Y, Zhong Y, Møller MS, Westh P, Svensson B, Blennow A. Interfacial Catalysis during Amylolytic Degradation of Starch Granules: Current Understanding and Kinetic Approaches. Molecules. 2023; 28(9):3799. https://doi.org/10.3390/molecules28093799
Chicago/Turabian StyleTian, Yu, Yu Wang, Yuyue Zhong, Marie Sofie Møller, Peter Westh, Birte Svensson, and Andreas Blennow. 2023. "Interfacial Catalysis during Amylolytic Degradation of Starch Granules: Current Understanding and Kinetic Approaches" Molecules 28, no. 9: 3799. https://doi.org/10.3390/molecules28093799





