Advanced Mass Spectrometry-Based Biomarker Identification for Metabolomics of Diabetes Mellitus and Its Complications
Abstract
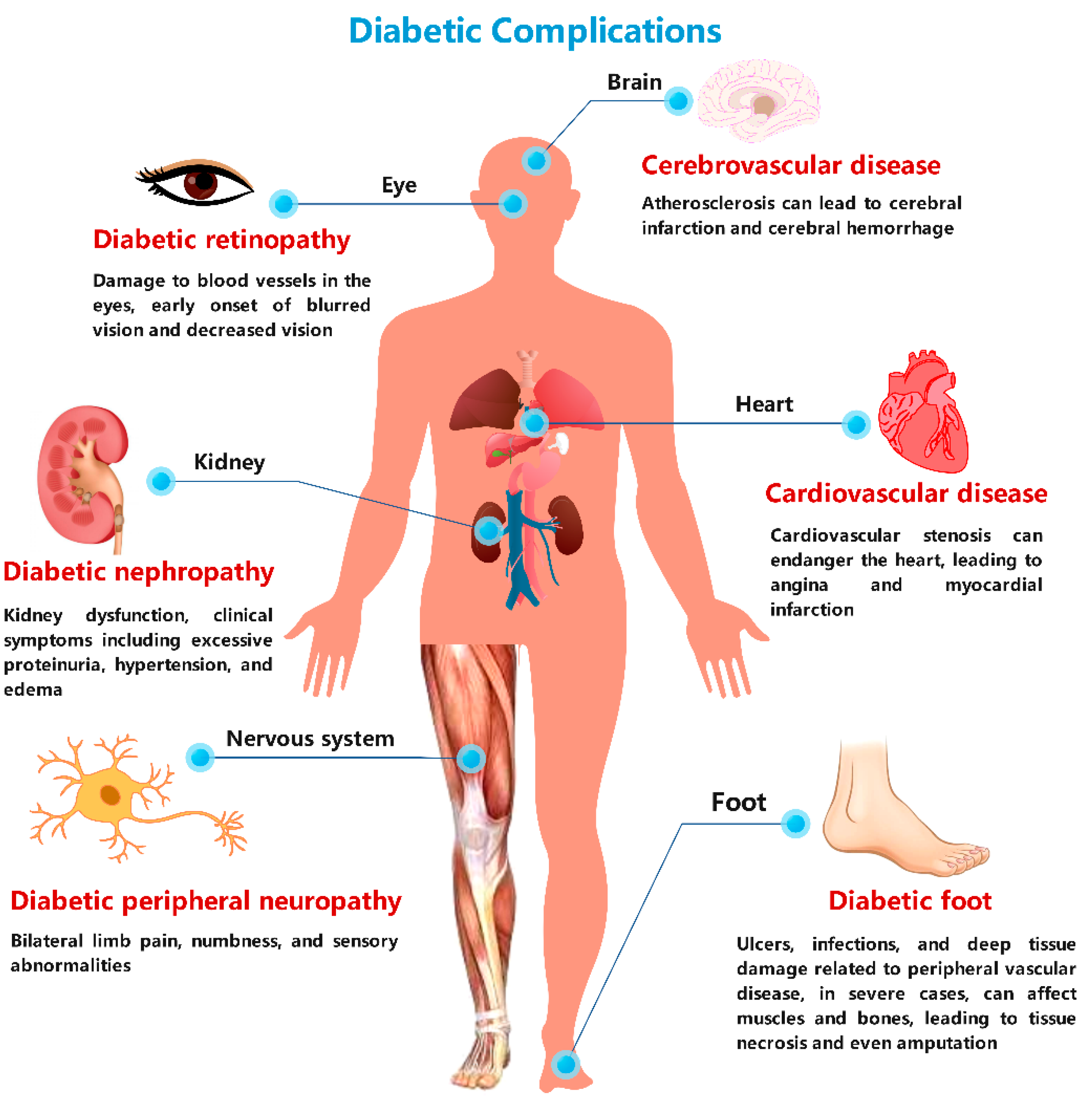
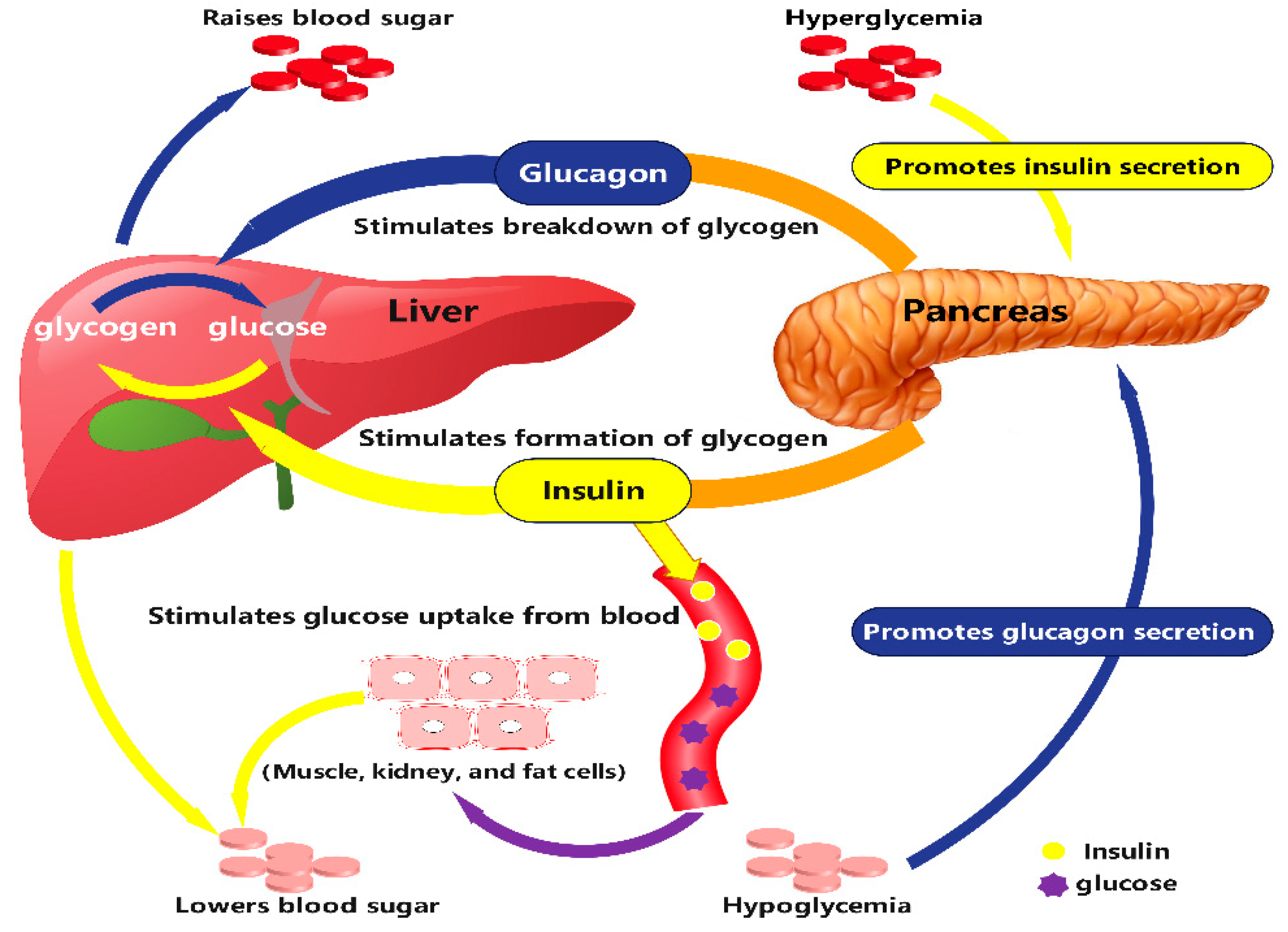
:1. Introduction
2. MS Technology for Biomarker Identification in Metabolomics of Diabetes Mellitus and Its Complications
3. MS-Based Metabolomics for Diabetes Clinic Research
3.1. MS-Based Research in Diabetes
3.2. MS-Based Research in Gestational Diabetes Mellitus
| Disease | Objective | Methods | Sample Source | Major Findings | Levels of Metabolites | Reference |
|---|---|---|---|---|---|---|
| Diabetes mellitus (DM) | Untargeted metabolomics analysis of anti-diabetic effects | UHPLC-MS/MS | Rat serum samples | Red ginseng extract intervention regulated 50 biomarkers, involving multiple metabolic pathways, including amino acid metabolism, glycerol–phospholipid metabolism, and fatty acid metabolism 5-HTP should be used as a potential lead compound | MOD: Phe↓, TUDCA↓, LysoPCs↓, LysoPEs↓, 2-methylbutyroylcarnitine↓, Gamma-tocotrienol↓, equo l4′-O-glucuronide↓ | [73] |
| Plasma metabolomics applied to type 2 diabetes research | FIE -FTICR MS | Mouse plasma samples | Successfully detected over 300 statistically significant metabolic features, identifying novel T2DM biomarker candidates | Among the non-polar metabolites: lipids with shorter acyl chains↑, myristic↑, palmitic↑, arachidonic acids↑, PCs of fewer total carbons↓, PCs with more total carbons↑, hexoses↑ | [74] | |
| Searching early marker for dysglycemia | UHPLC-MS/MS, GC-MS | Human plasma samples | Identified α-hydroxybutyric acid as a potential biomarker for both insulin resistance and impaired glucose regulation | Insulin resistant subjects: a-HB↑, 1-linoleoyl-GPC↓, glycine↓, 3-methyl-2-oxobutyrate↑, 1-oleoyl-GPC↓, creatine↑, decanoylcarnitine↓, octanylcarnitine↓, 1-stearoyl-GPC↓, adrenate (22:4n6)↑, stearate↑, 1-palmitoyl-GPC↓, palmitate (16:0)↑, margarate↑ | [75] | |
| Metabolite profiles during oral glucose challenge | LC-MS/MS | Human plasma samples | 91 out of 110 metabolites significantly changed with OGTT Downregulation of metabolites like β-hydroxybutyrate and upregulation of metabolites like hippurate observed | Metabolite changes following OGTT in insulin-resistant patients: b-Hydroxybutyrate↑, Isoleucine↑, Lactate↓, Orotate↓, Pyridoxate↑ | [76] | |
| Metabolomics insights into early type 2 diabetes pathogenesis | LC-MS/MS | Human plasma samples | Discovered changes in 19 metabolites, including lipids, amino acids, and small organic acids, associated with the onset of type 2 diabetes | Type 2 diabetes: glycine↑, taurine↑, phenylalanine↑ | [77] | |
| To examine the relationships between amino acid levels and the hyperinsulinemic–euglycemic clamp | LC-MS | Human serum samples | Found positive correlation of glycine with insulin resistance and negative correlation of leucine and isoleucine with insulin resistance and type 2 diabetes | T2DM: Gly↓, Leu/Ile↑, Val↑, His↓, Asx↑, Glx↑ | [78] | |
| Searching for novel molecular markers that arise before and after hyperglycemia | LC-MS, GC-MS | Women’s plasma and urine samples | Detected significant differences in 42 metabolites, including amino acids and sugars, between normal and type 2 diabetes groups, as well as in 14 metabolites between normal and impaired fasting glucose groups | IFG and subjects with T2DM: Valine↑, Isoleucine↑, Leucine↑, 3-methyl-2-oxovalerate↑, 4-methyl-2-oxopentanoate↑, 3-methyl-2-oxobutyrate↑ | [79] | |
| Plasma acylcarnitine profiling in T2D | LC-MS | Women’s plasma samples | Discovered decreased levels of acylcarnitines (fatty acid derivatives) and increased levels of amino acids such as glycine and lysine in type 2 diabetes patients | T2DM: Calculated total acylcarnitines (total free)↑, Acyl/free ratio↑, C2↑, C3↓, C6↑, C8↑, C10↑, C14↑, C18:1↑, C8-dicarb↑, summed C10-C14 acylcarnitines↑, total acylcarnitines↑ | [80] | |
| UPLC-oaTOF-MS for serum profiling in diabetic patients | UPLC-oaTOF-MS | Human serum samples | The metabolomics based on UPLC–oaTOF-MS could reflect the balance of homeostasis and metabolism of nourishment | T2DM: Phytosphingosine↓, Dihydrosphingosine↓, Leucine↓ | [81] | |
| Determine the differences in metabolite concentrations between T2D patients and healthy volunteers | UPLC-ESI-Q-TOF-MS | Human urine samples | Identified 12 metabolites, including acylcarnitines, citric acid, canine urea, and taurine, distinguishing between normal and type 2 diabetes groups | T2DM: Adiponectin↑, Acylcarnitines↑, Citric acid↓, Kynurenic acid↓, 3-Indoxyl sulfate↑, bile acids↑, Urate, glucose↑, Glycine↑, Glucuronolactone↓, Lysine↓, Phosphate↓ | [82] | |
| Identify biomarker signatures to differentiate pancreatic cancer from type 2 diabetes mellitus in early diagnosis | UPLC-MS/MS | Human plasma samples | Successful screening of differential metabolite ions between pancreatic cancer and DM patients and healthy individuals | DM:LysoPC (20: 4)↑, Deoxyadenosine↑, Asparaginyl-histidine↑, Vaccenyl carnitine↑, Phytal↓, 2 (R)-hydroxydocosanoic acid↓, Behenic acid↓, Catelaidic acid↓, 2-hydroxyphytanic acid↓, Phytosphingosine↓, Cerebronic acid↓, Docosanamide↓, Eicosenoic acid↓ | [83] | |
| Gestational diabetes mellitus (GDM) | Identify early risk indicators for GDM | LC-MS/MS | Women’s peripheral blood | Arginine assists in distinguishing GDM from NGT, early detection of GDM, and predicting increased risk of T2DM in women | GDM group: Arg↓, Gln↓, His↓, Met↓, Phe↓, Ser | [91] |
| Investigate estrogen metabolism imbalance in GDM | UPLC-MS/MS | GDM women’s urine samples | Successfully detected and quantified thirteen estrogens in different samples | GDM group: E1↑, E2↑, E3↓, 16epiE3↑, 17epiE3↑, 16α-OHE1↑, 2-OHE2↑, 2MeOE1↑, 4MeOE1↓, 2MeOE2↑, 4MeOE2↓, 2-OHE1↓, 4-OHE1↑ | [92] | |
| Unique biomarker characteristics in gestational diabetes mellitus | LC-MS | Human urine samples | 184 metabolites were increased and 86 metabolites were decreased in the positive ion mode, and 65 metabolites were increased and 71 were decreased in the negative ion mode | GDM group: Eicosapentaenoic Acid↓; Docosahexaenoic Acid↑; Docosapentaenoic Acid; Arachidonic Acid↑; α-Ketoglutaric Acid↑; Phosphoric Acid↑; Citric Acid↑; Genistein↓; Daidzein↓; 2-Furoic Acid↑ | [93] | |
| Metabolic alteration of circulating steroid hormones in women with gestational diabetes mellitus | UPLC-MS/MS | Human urine samples | 16OHE1 may be a strong marker associated with the risk for GDM | GDM group: 16OHE1↑; E1-G/S↑ | [94] | |
| Effects of pregnancy on plasma sphingolipids using metabolomics | LC/MS/MS | Women’s blood samples | A wide range of sphingolipids have altered plasma concentrations during pregnancy compared to postpartum, including ceramides, sphingomyelins, and sphingosines | During pregnancy, the most altered metabolite of interest was sphingomyelin (d18:1/20:2, d18:2/20:1, d16:1/22:2)↑ | [95] | |
| Diabetic cardiomyopathy (DCM) | Multi-omics of a preclinical model of diabetic cardiomyopathy | LC-MS/MS | Rat blood and ventricle samples | Metabolomics detected 19 amino acids, of which 12 were significantly altered | High-fat diet with STZ group: Arg↓, Asn↓, Asp↓, Gln↓, Gly↓, His↓, Met↓, Ser↓, Glu↑, Val↑ | [100] |
| To distinguish T2DM patients with or without damp-heat syndrome (DHS) and discover biomarkers | UPLC-TOFMS/MS | Human plasma samples | Vitamin and amino acid metabolism were changed in T2DM patients with the syndrome of DHS | DHS: imidazole↓, L-pipecolic acid↓, L-citrulline↓, L-carnitine↓, 3′-O-methylguanosine↓, pantothenate↑, sphingomyelin↑, thioetheramide-PC↑ | [101] | |
| Metabolic markers in patients with chronic heart failure before and after LVAD implantation | LC-MS/MS | Human plasma samples | Some acylcarnitines were more prominently altered in ICM or DCM when compared to the control, which could be interesting for the specific metabolic characterization of ICM and DCM | LVAD implantation postoperatively: SM (OH) C14:1↑, SM (OH) C22:1↑, SM C16:0↑, and SM C24:0↑ Potential prognostic markers: lysoPCs, proline (Pro) | [102] | |
| To investigate the differences in circulating LCAC levels in HF patients with and without DM | MS | Human plasma samples | LCAC biomarkers were associated with exercise status and clinical outcomes differentially in HF patients | DCM group: LCACs (C16, C16:1, C18, C18:1, C18:2)↑ | [103] | |
| Metabolic profiling, mitochondrial dysfunction, ob/ob mouse heart | LC-MS | Mouse heart tissue | There was an age-dependent decrease in myocardial acylcarnitine concentrations in ob/ob mice fed either RCD or HFD compared with those in the corresponding WT mice | HFD ob/ob mouse: myocardial acylcarnitine↓ | [104] | |
| Diabetic encephalopathy (DE) | Identify hippocampal metabolic alterations in a rat model of diabetic encephalopathy induced by STZ | GC-MS | Rat hippocampus Samples | Lower levels of NAA and DHAP and higher levels of homocysteine and glutamate in DE group rats, indicating a potential correlation between cognitive impairment and these metabolic changes | DM group: NAA↓, DHAP↓; homocysteine↑, glutamate↑ | [105] |
| To test the hypothesis that brain glycogen metabolism is impaired in type 2 diabetes | GC-MS | Rat cortex, hippocampus, striatum, and hypothalamus samples | Impaired brain glycogen metabolism related to T2D, suggesting a connection between brain glycogen metabolism and type 2 diabetes | The phosphorylation rate of glycogen synthase was increased | [106] | |
| Diabetic nephropathy (DN) | Plasma esterified and non-esterified fatty acid metabolic profiling in diabetic nephropathy | GC-MS | Human plasma samples | Developed a new method for simultaneous identification of 25 NEFAs and EFAs | Control–DM: EFAs↓, NEFAs↑; DM-DNШ: EFAs↑ | [107] |
| UPLC-oaTOF-MS for serum profiling in diabetic patients | UPLC-oaTOF-MS | Human serum samples | The metabolomics based on UPLC–oaTOF-MS could reflect the balance of homeostasis and metabolism of nourishment | DN group: Phytosphingosine↓, Dihydrosphingosine↓, Leucine↓ | [81] | |
| Identification of potential serum metabolic biomarkers of diabetic kidney disease | UPLC-ESI-MS/MS | Human serum samples | Identified 11 new metabolites closely related to DKD through comprehensive targeted metabolomic profiling. Provides insights into various early metabolic signs of DKD, aiding prediction and prevention in populations | Hexadecanoic Acid (C16:0)↑, Linolelaidic Acid (C18:2N6T)↑, Linoleic Acid (C18:2N6C)↑, Trans-4-Hydroxy-L-Proline↓, Aminocaproic Acid↓, L-Dihydroorotic Acid↓, Methylmercaptopurine↓, Piperidine↓, Azoxystrobin Acid↑, Lysopc 20:4↑, Cuminaldehyde↓ | [108] | |
| Diabetic peripheral neuropathy (DPN) | To examine the serum lipidomic profile associated with neuropathy in type 2 diabetes | MS | Human serum samples | Circulating acylcarnitines, free fatty acids, phosphatidylcholines, and lysophosphatidylcholines are associated with neuropathy status in type 2 diabetes | Pima participants with T2D: medium-chain acylcarnitines↓, total free fatty acids↑, phosphatidylcholines↓, lysophosphatidylcholines↑ | [109] |
| To investigate the neuroprotective effect of Jin-Mai-Tong (JMT) decoction on diabetic rats with peripheral neuropathy and to elucidate the potential mechanism | UPLC/QTOF-MS | Rat serum samples | 21 metabolites were identified; JMT decoction has an obvious protective effect against DPN; lipid metabolism, TCA cycle, amino acid metabolism | STZ group: 2-Ketobutyric acid↑, Paraxanthine↑, Leucyl-Cysteine↑, Artonin K↑, Deoxycytidine↑, Oxalacetic acid↑, LysoPC (18:3)↑, LysoPE (0:0/18:2)↑, Lysophosphatidic acid (0:0/18:2)↑, Delcorine↑, LysoPE (0:0/22:6)↑, Hexadec-2-enoyl carnitine↑, LysoPE (0:0/16:0)↑Lithocholic acid glycine conjugate↑, N-(1-Deoxy-1fructosyl) leucine↑, Stearoylcarnitine↑, Glycerol tripropanoate↑, Retinyl beta-glucuronide↑, C46H74NO10P↑, Hexyl dodecanoate↑, Tyr-Pro-Phe↓ | [110] | |
| Investigate the effects of Tang Luo Ning (TLN) on DPN in rats using an LC-MS metabolomics approach | HPLC-IT-TOF/MS | Rat serum sample | 14 potential biomarkers; TLN could improve the peripheral nerve function and reduce the demyelination of the sciatic nerve in DPN rats; TCA cycle; glycine, serine, and threonine metabolism; glyoxylate and dicarboxylate metabolism | MOD: 3-Butenoic acid↓, Acetylcarnitine↓, Citrate↑, Creatine↑, Creatinine↓, Fumarate↑, Glyceric acid↓, Glycine↑, Lactate↓, LysoPC (22:5)↑, Palmitoyl glucuronide↑, Riboflavin↓, Succinate↓, Tryptophan↓ | [111] | |
| Diabetic foot ulcers (DFUs) | To evaluate and identify specific amino acids associated with the healing outcomes of patients with DFUs | LC-MS/MS | Human blood samples | Higher levels of serum arginine, isoleucine, leucine, and serine were observed in the healed ulcer group compared to the non-healing group, indicating potential biomarkers for wound healing in DFUs | DFUs: Arginine↑, Leucine↑, Isoleucine↑, Threonine↑ | [112] |
| Diabetic retinopathy (DR) | To identify tear fluid biomarkers for differentiating between PDR and NPDR in T2D patients | GC-MS | Human tear samples | D-Glutamine and D-glutamate metabolism was significantly highlighted in the PDR group as compared to the non-diabetic group. The metabolites in tears could be potential biomarkers in DR analysis | PDR group: Guanosine↑, Uric acid↑, D-(+)-malic acid↑, Pimelic acid↑, Azelaic acid↑, 2-Hydroxybenzothiazole↑, N, N-Diethyl-4-methoxybenzamide↓, Homovanillic acid↓, Phenol↓, Pipecolic acid↓, Guvacoline↓, Spironolactone↓, N,N-Diethyl-3-methoxybenzamide↓, Diazepam↓, Prostaglandin F2α 1-11-lactone↓, 2-Methoxyestrone↓, Tretinoin↓ | [113] |
| Identify a specific plasma metabolic profile associated with DR as distinct from diabetes alone | GC-MS | Human plasma samples | Elevated levels of 2-deoxyribonucleic acid, 3,4-dihydroxybutyric acid, erythritol, gluconic acid, and ribose were found in patients. These were validated in an independent sample set and considered as potential biomarkers for diabetic eye disease | DR group: 2-deoxyribonucleic acid↑, 3,4-dihydroxybutyric acid, erythritol↑, gluconic acid↑, ribose↑ | [114] | |
| Identify serum metabolite biomarkers for DR using various metabolomics platforms | LC-MS, GC-MS | Human serum samples | Identified 348 metabolites with significant differences between groups. 12-Hydroxyeicosatetraenoic acid and 2-pyrrolidinone were significantly elevated in patients with diabetic eye disease | DR group: 12-Hydroxyeicosatetraenoic acid↑, 2-pyrrolidino↑ne | [115] |
4. MS-Based Metabolomics in Clinical Cases of Diabetes-Induced Complications
4.1. MS-Based Research in Diabetic Cardiomyopathy
4.2. MS-Based Research in Diabetic Encephalopathy
4.3. MS-Based Research in Diabetic Nephropathy
4.4. MS-Based Research in Diabetic Peripheral Neuropathy
4.5. MS-Based Research in Diabetic Foot Ulcers
4.6. MS-Based Research in Diabetic Eye Disease
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Zhou, B.; Lu, Y.; Hajifathalian, K.; Bentham, J.; Di Cesare, M.; Danaei, G.; Bixby, H.; Cowan, M.J.; Ali, M.K.; Taddei, C.; et al. Worldwide trends in diabetes since 1980: A pooled analysis of 751 population-based studies with 4.4 million participants. Lancet 2016, 387, 1513–1530. [Google Scholar] [CrossRef]
- Ong, K.L.; Stafford, L.K.; McLaughlin, S.A.; Boyko, E.J.; Vollset, S.E.; Smith, A.E.; Dalton, B.E.; Duprey, J.; Cruz, J.A.; Hagins, H.; et al. Global, regional, and national burden of diabetes from 1990 to 2021, with projections of prevalence to 2050: A systematic analysis for the Global Burden of Disease Study 2021. Lancet 2023, 402, 203–234. [Google Scholar] [CrossRef]
- Soriguer, F.; Goday, A.; Bosch-Comas, A.; Bordiu, E.; Calle-Pascual, A.; Carmena, R.; Casamitjana, R.; Castano, L.; Castell, C.; Catala, M.; et al. Prevalence of diabetes mellitus and impaired glucose regulation in Spain: The Di@bet.es Study. Diabetologia 2012, 55, 88–93. [Google Scholar] [CrossRef]
- Rossi, T.; Panozzo, G.; Della Mura, G.; Giannarelli, D.; Ferrari, D.; Alessio, G.; Palmisano, C.; Telani, S.; Ripandelli, G. Diabetes and diabetic retinopathy in patients undergoing cataract surgery: A prevalence study-DiCat study report #2. Acta Diabetol. 2020, 57, 645–650. [Google Scholar]
- Glovaci, D.; Fan, W.; Wong, N.D. Epidemiology of Diabetes Mellitus and Cardiovascular Disease. Curr. Cardiol. Rep. 2019, 21, 21. [Google Scholar] [CrossRef]
- Boyko, E.J.; Zelnick, L.R.; Braffett, B.H.; Pop-Busui, R.; Cowie, C.C.; Lorenzi, G.M.; Gubitosi-Klug, R.; Zinman, B.; de Boer, I.H. Risk of Foot Ulcer and Lower-Extremity Amputation Among Participants in the Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Study. Diabetes Care 2022, 45, 357–364. [Google Scholar] [CrossRef]
- Koye, D.N.; Magliano, D.J.; Nelson, R.G.; Pavkov, M.E. The Global Epidemiology of Diabetes and Kidney Disease. Adv. Chronic Kidney Dis. 2018, 25, 121–132. [Google Scholar] [CrossRef]
- Ostergaard, L.; Mogensen, U.M.; Bundgaard, J.S.; Dahl, A.; Wang, A.; Torp-Pedersen, C.; Gislason, G.; Kober, L.; Kober, N.; Dejgaard, T.F.; et al. Duration and complications of diabetes mellitus and the associated risk of infective endocarditis. Int. J. Cardiol. 2019, 278, 280–284. [Google Scholar] [CrossRef]
- Moxey, P.W.; Gogalniceanu, P.; Hinchliffe, R.J.; Loftus, I.M.; Jones, K.J.; Thompson, M.M.; Holt, P.J. Lower extremity amputations--a review of global variability in incidence. Diabet. Med. 2011, 28, 1144–1153. [Google Scholar] [CrossRef] [PubMed]
- Grarup, N.; Sandholt, C.H.; Hansen, T.; Pedersen, O. Genetic susceptibility to type 2 diabetes and obesity: From genome-wide association studies to rare variants and beyond. Diabetologia 2014, 57, 1528–1541. [Google Scholar] [CrossRef] [PubMed]
- Jenkinson, C.P.; Goring, H.H.; Arya, R.; Blangero, J.; Duggirala, R.; DeFronzo, R.A. Transcriptomics in type 2 diabetes: Bridging the gap between genotype and phenotype. Genom. Data 2016, 8, 25–36. [Google Scholar] [CrossRef]
- Sohail, W.; Majeed, F.; Afroz, A. Differential proteome analysis of diabetes mellitus type 2 and its pathophysiological complications. Diabetes Metab. Syndr. 2018, 12, 1125–1131. [Google Scholar] [CrossRef]
- Guasch-Ferre, M.; Hruby, A.; Toledo, E.; Clish, C.B.; Martinez-Gonzalez, M.A.; Salas-Salvado, J.; Hu, F.B. Metabolomics in Prediabetes and Diabetes: A Systematic Review and Meta-analysis. Diabetes Care 2016, 39, 833–846. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. M‘etabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef]
- Bjerrum, J.T.; Nielsen, O.H. Metabonomics in Gastroenterology and Hepatology. Int. J. Mol. Sci. 2019, 20, 3638. [Google Scholar] [CrossRef]
- Wang-Sattler, R.; Yu, Z.; Herder, C.; Messias, A.C.; Floegel, A.; He, Y.; Heim, K.; Campillos, M.; Holzapfel, C.; Thorand, B.; et al. Novel biomarkers for pre-diabetes identified by metabolomics. Mol. Syst. Biol. 2012, 8, 615. [Google Scholar] [CrossRef]
- Lee, H.S.; Park, T.J.; Kim, J.M.; Yun, J.H.; Yu, H.Y.; Kim, Y.J.; Kim, B.J. Identification of metabolic markers predictive of prediabetes in a Korean population. Syst. Biol. 2020, 10, 22009. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.C.; Watkins, S.M.; Lorenzo, C.; Wagenknecht, L.E.; Il’yasova, D.; Chen, Y.D.; Haffner, S.M.; Hanley, A.J. Branched-Chain Amino Acids and Insulin Metabolism: The Insulin Resistance Atherosclerosis Study (IRAS). Diabetes Care 2016, 39, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Roberts, L.D.; Koulman, A.; Griffin, J.L. Towards metabolic biomarkers of insulin resistance and type 2 diabetes: Progress from the metabolome. Lancet Diabetes Endocrinol. 2014, 2, 65–75. [Google Scholar] [CrossRef]
- Wang, T.J.; Larson, M.G.; Vasan, R.S.; Cheng, S.; Rhee, E.P.; McCabe, E.; Lewis, G.D.; Fox, C.S.; Jacques, P.F.; Fernandez, C.; et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 2011, 17, 448–453. [Google Scholar] [CrossRef]
- Ahola-Olli, A.V.; Mustelin, L.; Kalimeri, M.; Kettunen, J.; Jokelainen, J.; Auvinen, J.; Puukka, K.; Havulinna, A.S.; Lehtimäki, T.; Kähönen, M.; et al. Circulating metabolites and the risk of type 2 diabetes: A prospective study of 11,896 young adults from four Finnish cohorts. Diabetologia 2019, 62, 2298–2309. [Google Scholar] [CrossRef]
- Al-Sulaiti, H.; Diboun, I.; Agha, M.V.; Mohamed, F.F.S.; Atkin, S.; Dömling, A.S.; Elrayess, M.A.; Mazloum, N.A. Metabolic signature of obesity-associated insulin resistance and type 2 diabetes. J. Transl. Med. 2019, 17, 348. [Google Scholar] [CrossRef]
- Würtz, P.; Tiainen, M.; Mäkinen, V.P.; Kangas, A.J.; Soininen, P.; Saltevo, J.; Keinänen-Kiukaanniemi, S.; Mäntyselkä, P.; Lehtimäki, T.; Laakso, M.; et al. Circulating metabolite predictors of glycemia in middle-aged men and women. Diabetes Care 2012, 35, 1749–1756. [Google Scholar] [CrossRef]
- Floegel, A.; Stefan, N.; Yu, Z.; Mühlenbruch, K.; Drogan, D.; Joost, H.G.; Fritsche, A.; Häring, H.U.; Hrabě de Angelis, M.; Peters, A.; et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes 2013, 62, 639–648. [Google Scholar] [CrossRef]
- Padberg, I.; Peter, E.; González-Maldonado, S.; Witt, H.; Mueller, M.; Weis, T.; Bethan, B.; Liebenberg, V.; Wiemer, J.; Katus, H.A.; et al. A new metabolomic signature in type-2 diabetes mellitus and its pathophysiology. PLoS ONE 2014, 9, e85082. [Google Scholar] [CrossRef]
- Tokarz, J.; Haid, M.; Cecil, A.; Prehn, C.; Artati, A.; Moller, G.; Adamski, J. Endocrinology Meets Metabolomics: Achievements, Pitfalls, and Challenges. Trends Endocrinol. Metab. 2017, 28, 705–721. [Google Scholar] [CrossRef]
- Liu, X.; Locasale, J.W. Metabolomics: A Primer. Trends Biochem. Sci. 2017, 42, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Chaleckis, R.; Meister, I.; Zhang, P.; Wheelock, C.E. Challenges, progress and promises of metabolite annotation for LC-MS-based metabolomics. Curr. Opin. Biotechnol. 2019, 55, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C. Metabolomics takes its place as latest up-and-coming “omic” science. J. Natl. Cancer Inst. 2004, 96, 732–734. [Google Scholar] [CrossRef]
- Dumas, M.E.; Kinross, J.; Nicholson, J.K. Metabolic phenotyping and systems biology approaches to understanding metabolic syndrome and fatty liver disease. Gastroenterology 2014, 146, 46–62. [Google Scholar] [CrossRef]
- Del Coco, L.; Vergara, D.; De Matteis, S.; Mensà, E.; Sabbatinelli, J.; Prattichizzo, F.; Bonfigli, A.R.; Storci, G.; Bravaccini, S.; Pirini, F.; et al. NMR-Based Metabolomic Approach Tracks Potential Serum Biomarkers of Disease Progression in Patients with Type 2 Diabetes Mellitus. J. Clin. Med. 2019, 8, 720. [Google Scholar] [CrossRef]
- Palomino-Schätzlein, M.; Lamas-Domingo, R.; Ciudin, A.; Gutiérrez-Carcedo, P.; Marés, R.; Aparicio-Gómez, C.; Hernández, C.; Simó, R.; Herance, J.R. A Translational In Vivo and In Vitro Metabolomic Study Reveals Altered Metabolic Pathways in Red Blood Cells of Type 2 Diabetes. J. Clin. Med. 2020, 9, 1619. [Google Scholar] [CrossRef]
- Jin, H.; Zhu, B.; Liu, X.; Jin, J.; Zou, H. Metabolic characterization of diabetic retinopathy: An (1)H-NMR-based metabolomic approach using human aqueous humor. J. Pharm. Biomed. Anal. 2019, 174, 414–421. [Google Scholar] [CrossRef]
- Tian, J.S.; Zhao, L.; Shen, X.L.; Liu, H.; Qin, X.M. (1)H NMR-based metabolomics approach to investigating the renal protective effects of Genipin in diabetic rats. Chin. J. Nat. Med. 2018, 16, 261–270. [Google Scholar] [CrossRef]
- Zheng, H.; Lin, Q.; Wang, D.; Xu, P.; Zhao, L.; Hu, W.; Bai, G.; Yan, Z.; Gao, H. NMR-based metabolomics reveals brain region-specific metabolic alterations in streptozotocin-induced diabetic rats with cognitive dysfunction. Metab. Brain Dis. 2017, 32, 585–593. [Google Scholar] [CrossRef]
- Nagana Gowda, G.A.; Raftery, D. Can NMR solve some significant challenges in metabolomics? J. Magn. Reson. 2015, 260, 144–160. [Google Scholar] [CrossRef]
- Kim, H.K.; Choi, Y.H.; Verpoorte, R. NMR-based plant metabolomics: Where do we stand, where do we go? Trends Biotechnol. 2011, 29, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Roca, M.; Alcoriza, M.I.; Garcia-Cañaveras, J.C.; Lahoz, A. Reviewing the metabolome coverage provided by LC-MS: Focus on sample preparation and chromatography-A tutorial. Anal. Chim. Acta 2021, 1147, 38–55. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.M.; Gao, Y.; Teo, G.S.; Koh, H.W.L.; Tai, E.S.; Khoo, C.M.; Choi, K.P.; Zhou, L.; Choi, H. Plasma Metabolome and Lipidome Associations with Type 2 Diabetes and Diabetic Nephropathy. Metabolites 2021, 11, 228. [Google Scholar] [CrossRef] [PubMed]
- Cajka, T.; Fiehn, O. Toward Merging Untargeted and Targeted Methods in Mass Spectrometry-Based Metabolomics and Lipidomics. Anal. Chem. 2016, 88, 524–545. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.B.; Lin, L.; Xu, X.S.; Cheng, X.X.; Zhang, Y.Y.; Hall, R.; Xu, P. A Critical Review of Analytical Methods for Comprehensive Characterization of Produced Water. Water 2021, 13, 183. [Google Scholar] [CrossRef]
- Beccaria, M.; Cabooter, D. Current developments in LC-MS for pharmaceutical analysis. Analyst 2020, 145, 1129–1157. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Shao, C.; Li, C.; Zhou, H.; Yu, L.; Yang, J.; Wan, H.; He, Y. Metabolomics: A useful tool for ischemic stroke research. J. Pharm. Anal. 2023, 13, 968–983. [Google Scholar] [CrossRef] [PubMed]
- Chan, E.C.Y.; Pasikanti, K.K.; Nicholson, J.K. Global urinary metabolic profiling procedures using gas chromatography-mass spectrometry. Nat. Protoc. 2011, 6, 1483–1499. [Google Scholar] [CrossRef]
- Chen, X.; Lin, L.J.; Cai, H.T.; Gao, X.Y. Identification and Analysis of Metabolites That Contribute to the Formation of Distinctive Flavour Components of Laoxianghuang. Foods 2023, 12, 425. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.H. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Methods Mol. Biol. 2015, 1277, 161–193. [Google Scholar] [PubMed]
- Liu, X.Y.; Zhou, L.N.; Shi, X.Z.; Xu, G.W. New advances in analytical methods for mass spectrometry-based large-scale metabolomics study. Trac-Trends Anal. Chem. 2019, 121, 115665. [Google Scholar] [CrossRef]
- Zhu, X.C.; Chen, Y.P.; Subramanian, R. Comparison of Information-Dependent Acquisition, SWATH, and MS (All) Techniques in Metabolite Identification Study Employing Ultrahigh-Performance Liquid Chromatography-Quadrupole Time-of-Flight Mass Spectrometry. Anal. Chem. 2014, 86, 1202–1209. [Google Scholar] [CrossRef]
- Rost, H.L.; Rosenberger, G.; Navarro, P.; Gillet, L.; Miladinovic, S.M.; Schubert, O.T.; Wolski, W.; Collins, B.C.; Malmstrom, J.; Malmstrom, L.; et al. Publisher Correction: OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat. Biotechnol. 2020, 38, 374. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Li, H.; Cai, Y.; Guo, Y.; Chen, F.; Zhu, Z.J. MetDIA: Targeted Metabolite Extraction of Multiplexed MS/MS Spectra Generated by Data-Independent Acquisition. Anal. Chem. 2016, 88, 8757–8764. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, R.; Cai, Y.; Wang, Z.; Zhu, Z.J. DecoMetDIA: Deconvolution of Multiplexed MS/MS Spectra for Metabolite Identification in SWATH-MS-Based Untargeted Metabolomics. Anal. Chem. 2019, 91, 11897–11904. [Google Scholar] [CrossRef]
- Huang, Q.; Tan, Y.; Yin, P.; Ye, G.; Gao, P.; Lu, X.; Wang, H.; Xu, G. Metabolic characterization of hepatocellular carcinoma using nontargeted tissue metabolomics. Cancer Res. 2013, 73, 4992–5002. [Google Scholar] [CrossRef]
- Schillemans, T.; Shi, L.; Donat-Vargas, C.; Hanhineva, K.; Tornevi, A.; Johansson, I.; Koponen, J.; Kiviranta, H.; Rolandsson, O.; Bergdahl, I.A.; et al. Plasma metabolites associated with exposure to perfluoroalkyl substances and risk of type 2 diabetes—A nested case-control study. Environ. Int. 2021, 146, 106180. [Google Scholar] [CrossRef]
- Fu, Y.; Zhou, Z.; Kong, H.; Lu, X.; Zhao, X.; Chen, Y.; Chen, J.; Wu, Z.; Xu, Z.; Zhao, C.; et al. Nontargeted Screening Method for Illegal Additives Based on Ultrahigh-Performance Liquid Chromatography-High-Resolution Mass Spectrometry. Anal. Chem. 2016, 88, 8870–8877. [Google Scholar] [CrossRef]
- Jacob, M.; Malkawi, A.; Albast, N.; Al Bougha, S.; Lopata, A.; Dasouki, M.; Abdel Rahman, A.M. A targeted metabolomics approach for clinical diagnosis of inborn errors of metabolism. Anal. Chim. Acta 2018, 1025, 141–153. [Google Scholar] [CrossRef]
- Kitagawa, H.; Ohbuchi, K.; Munekage, M.; Fujisawa, K.; Kawanishi, Y.; Namikawa, T.; Kushida, H.; Matsumoto, T.; Shimobori, C.; Nishi, A.; et al. Phenotyping analysis of the Japanese Kampo medicine maoto in healthy human subjects using wide-targeted plasma metabolomics. J. Pharm. Biomed. Anal. 2019, 164, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, W.J.; Koal, T.; Wang, Y.; Kohl, M.; Enot, D.P.; Deigner, H.P. Targeted metabolomics for biomarker discovery. Angew. Chem. 2010, 49, 5426–5445. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Luo, X.; Guo, R.; Jing, W.; Lu, H. Cell Metabolomics Reveals Berberine-Inhibited Pancreatic Cancer Cell Viability and Metastasis by Regulating Citrate Metabolism. J. Proteome Res. 2020, 19, 3825–3836. [Google Scholar] [CrossRef] [PubMed]
- Scholz, K.; Dekant, W.; Volkel, W.; Pahler, A. Rapid detection and identification of N-acetyl-L-cysteine thioethers using constant neutral loss and theoretical multiple reaction monitoring combined with enhanced product-ion scans on a linear ion trap mass spectrometer. J. Am. Soc. Mass Spectrom. 2005, 16, 1976–1984. [Google Scholar] [CrossRef]
- Ikeda, K.; Taguchi, R. Highly sensitive localization analysis of gangliosides and sulfatides including structural isomers in mouse cerebellum sections by combination of laser microdissection and hydrophilic interaction liquid chromatography/electrospray ionization mass spectrometry with theoretically expanded multiple reaction monitoring. Rapid Commun. Mass Spectrom. 2010, 24, 2957–2965. [Google Scholar]
- Zhou, J.; Liu, H.; Liu, Y.; Liu, J.; Zhao, X.; Yin, Y. Development and Evaluation of a Parallel Reaction Monitoring Strategy for Large-Scale Targeted Metabolomics Quantification. Anal. Chem. 2016, 88, 4478–4486. [Google Scholar] [CrossRef]
- Ying, L.W.; He, X.X.; Ma, X.J.; Shen, Y.; Su, H.; Peng, J.H.; Wang, Y.F.; Bao, Y.Q.; Zhou, J.; Jia, W.P. Serum 1,5-anhydroglucitol when used with fasting plasma glucose improves the efficiency of diabetes screening in a Chinese population. Sci. Rep. 2017, 7, 11968. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Vijayakumar, A.; Kahn, B.B. Metabolites as regulators of insulin sensitivity and metabolism. Nat. Rev. Mol. Cell Biol. 2018, 19, 654–672. [Google Scholar] [CrossRef]
- Kane, J.P.; Pullinger, C.R.; Goldfine, I.D.; Malloy, M.J. Dyslipidemia and diabetes mellitus: Role of lipoprotein species and interrelated pathways of lipid metabolism in diabetes mellitus. Curr. Opin. Pharmacol. 2021, 61, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Sokooti, S.; Flores-Guerrero, J.L.; Kieneker, L.M.; Heerspink, H.J.L.; Connelly, M.A.; Bakker, S.J.L.; Dullaart, R.P.F. HDL Particle Subspecies and Their Association With Incident Type 2 Diabetes: The PREVEND Study. J. Clin. Endocrinol. Metab. 2021, 106, 1761–1772. [Google Scholar] [CrossRef] [PubMed]
- Giesbertz, P.; Daniel, H. Branched-chain amino acids as biomarkers in diabetes. Curr. Opin. Clin. Nutr. Metab. Care 2016, 19, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Luc, K.; Schramm-Luc, A.; Guzik, T.J.; Mikolajczyk, T.P. Oxidative stress and inflammatory markers in prediabetes and diabetes. J. Physiol. Pharmacol. 2019, 70, 809–824. [Google Scholar]
- Stanimirovic, J.; Radovanovic, J.; Banjac, K.; Obradovic, M.; Essack, M.; Zafirovic, S.; Gluvic, Z.; Gojobori, T.; Isenovic, E.R. Role of C-Reactive Protein in Diabetic Inflammation. Mediat. Inflamm. 2022, 2022, 3706508. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Vanhoutte, P.M. Macro- and microvascular endothelial dysfunction in diabetes. J. Diabetes 2017, 9, 434–449. [Google Scholar] [CrossRef]
- Yang, Z.; Dan, W.; Li, Y.; Zhou, X.; Liu, T.; Shi, C.; Li, R.; Zhang, Y.; Zhang, J.; Yan, J.; et al. Untargeted metabolomics analysis of the anti-diabetic effect of Red ginseng extract in Type 2 diabetes Mellitus rats based on UHPLC-MS/MS. Biomed. Pharmacother. 2022, 146, 112495. [Google Scholar] [CrossRef]
- Zhu, Y.; Wancewicz, B.; Schaid, M.; Tiambeng, T.N.; Wenger, K.; Jin, Y.; Heyman, H.; Thompson, C.J.; Barsch, A.; Cox, E.D.; et al. Ultrahigh-Resolution Mass Spectrometry-Based Platform for Plasma Metabolomics Applied to Type 2 Diabetes Research. J. Proteome Res. 2021, 20, 463–473. [Google Scholar] [CrossRef]
- Gall, W.E.; Beebe, K.; Lawton, K.A.; Adam, K.P.; Mitchell, M.W.; Nakhle, P.J.; Ryals, J.A.; Milburn, M.V.; Nannipieri, M.; Camastra, S.; et al. α-Hydroxybutyrate Is an Early Biomarker of Insulin Resistance and Glucose Intolerance in a Nondiabetic Population. PLoS ONE 2010, 5, e10883. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.E.; Larson, M.G.; Vasan, R.S.; Ghorbani, A.; Cheng, S.; Rhee, E.P.; Florez, J.C.; Clish, C.B.; Gerszten, R.E.; Wang, T.J. Metabolite Profiles During Oral Glucose Challenge. Diabetes 2013, 62, 2689–2698. [Google Scholar] [CrossRef]
- Merino, J.; Leong, A.; Liu, C.T.; Porneala, B.; Walford, G.; von Grotthuss, M.; Wang, T.; Flannick, J.; Dupuis, J.; Levy, D.; et al. Metabolomics insights into early type 2 diabetes pathogenesis and detection in individuals with normal fasting glucose. Diabetologia 2018, 61, 1315–1324. [Google Scholar] [CrossRef]
- Thalacker-Mercer, A.E.; Ingram, K.H.; Guo, F.J.; Ilkayeva, O.; Newgard, C.B.; Garvey, W.T. BMI, RQ, Diabetes, and Sex Affect the Relationships Between Amino Acids and Clamp Measures of Insulin Action in Humans. Diabetes 2014, 63, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Menni, C.; Fauman, E.; Erte, I.; Perry, J.R.B.; Kastenmüller, G.; Shin, S.Y.; Petersen, A.K.; Hyde, C.; Psatha, M.; Ward, K.J.; et al. Biomarkers for Type 2 Diabetes and Impaired Fasting Glucose Using a Nontargeted Metabolomics Approach. Diabetes 2013, 62, 4270–4276. [Google Scholar] [CrossRef]
- Adams, S.H.; Hoppel, C.L.; Lok, K.H.; Zhao, L.; Wong, S.W.; Minkler, P.E.; Hwang, D.H.; Newman, J.W.; Garvey, W.T. Plasma Acylcarnitine Profiles Suggest Incomplete Long-Chain Fatty Acid β-Oxidation and Altered Tricarboxylic Acid Cycle Activity in Type 2 Diabetic African-American Women. J. Nutr. 2009, 139, 1073–1081. [Google Scholar] [CrossRef]
- Zhang, J.; Yan, L.J.; Chen, W.G.; Lin, L.; Song, X.Y.; Yan, X.M.; Hang, W.; Huang, B.L. Metabonomics research of diabetic nephropathy and type 2 diabetes mellitus based on UPLC-oaTOF-MS system. Anal. Chim. Acta 2009, 650, 16–22. [Google Scholar] [CrossRef]
- Zhang, A.H.; Sun, H.; Yan, G.L.; Yuan, Y.; Han, Y.; Wang, X.J. Metabolomics study of type 2 diabetes using ultra-performance LC-ESI/quadrupole-TOF high-definition MS coupled with pattern recognition methods. J. Physiol. Biochem. 2014, 70, 117–128. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, L.; Kang, H.; Liu, J.; Zhang, J.; Zhao, J.; Liu, S. Metabolomics Identifies Biomarker Signatures to Differentiate Pancreatic Cancer from Type 2 Diabetes Mellitus in Early Diagnosis. Int. J. Endocrinol. 2021, 2021, 9990768. [Google Scholar] [CrossRef]
- Wang, W.; Zhou, W.; Wang, S.; Huang, J.; Le, Y.; Nie, S.; Wang, W.; Guo, Q. Accuracy of breath test for diabetes mellitus diagnosis: A systematic review and meta-analysis. BMJ Open Diabetes Res. Care 2021, 9, e002174. [Google Scholar] [CrossRef] [PubMed]
- Trefz, P.; Schmidt, S.C.; Sukul, P.; Schubert, J.K.; Miekisch, W.; Fischer, D.C. Non-Invasive Assessment of Metabolic Adaptation in Paediatric Patients Suffering from Type 1 Diabetes Mellitus. J. Clin. Med. 2019, 8, 1797. [Google Scholar] [CrossRef]
- Jiang, C.; Sun, M.; Wang, Z.; Chen, Z.; Zhao, X.; Yuan, Y.; Li, Y.; Wang, C. A Portable Real-Time Ringdown Breath Acetone Analyzer: Toward Potential Diabetic Screening and Management. Sensors 2016, 16, 1199. [Google Scholar] [CrossRef]
- Weinert, L.S. International Association of Diabetes and Pregnancy Study Groups recommendations on the diagnosis and classification of hyperglycemia in pregnancy: Comment to the International Association of Diabetes and Pregnancy Study Groups Consensus Panel. Diabetes Care 2010, 33, e97. [Google Scholar] [CrossRef]
- Li, J.; Wang, P.; Zhang, C.; Leng, J.; Li, N.; Wang, L.; Li, W.; Liu, H.; Yu, Z.; Hu, G.; et al. Short Body Height and Pre-pregnancy Overweight for Increased Risk of Gestational Diabetes Mellitus: A Population-Based Cohort Study. Front. Endocrinol. 2018, 9, 349. [Google Scholar] [CrossRef]
- Wen, L.; Wu, Y.; Yang, Y.; Han, T.L.; Wang, W.; Fu, H.; Zheng, Y.; Shan, T.; Chen, J.; Xu, P.; et al. Gestational Diabetes Mellitus Changes the Metabolomes of Human Colostrum, Transition Milk and Mature Milk. Med. Sci. Monit. 2019, 25, 6128–6152. [Google Scholar] [CrossRef]
- Shaikh, A.S.; Li, W.; Yuan, G.; Gao, M.; Geng, C.; Guo, N.; Guo, R. Simple, rapid and highly sensitive HPLC method for measurement of Lamotrigine in human plasma and its clinical applications. Pak. J. Pharm. Sci. 2016, 29, 2245–2250. [Google Scholar]
- Burzynska-Pedziwiatr, I.; Jankowski, A.; Kowalski, K.; Sendys, P.; Zieleniak, A.; Cypryk, K.; Zurawska-Klis, M.; Wozniak, L.A.; Bukowiecka-Matusiak, M. Associations of Arginine with Gestational Diabetes Mellitus in a Follow-Up Study. Int. J. Mol. Sci. 2020, 21, 7811. [Google Scholar] [CrossRef]
- Zhang, X.L.; Yang, N.; Wang, M.; Ge, J.J.; Zhu, H.J.; He, J.; Ge, W.H. Establishment and validation of a sensitive LC-MS/MS method for quantification of urinary estrogens in women with gestational diabetes mellitus. J. Pharm. Biomed. Anal. 2022, 221, 115044. [Google Scholar] [CrossRef]
- Meng, X.J.; Zhu, B.; Liu, Y.; Fang, L.; Yin, B.B.; Sun, Y.N.; Ma, M.N.; Huang, Y.L.; Zhu, Y.N.; Zhang, Y.L. Unique Biomarker Characteristics in Gestational Diabetes Mellitus Identified by LC-MS-Based Metabolic Profiling. J. Diabetes Res. 2021, 2021, 6689414. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Zhang, W.; Ji, C.; Ge, J.J.; Zhang, X.L.; Li, M.J.; Wang, M.; Zhang, T.Q.; He, J.; Zhu, H.J. Metabolic alteration of circulating steroid hormones in women with gestational diabetes mellitus and the related risk factors. Front. Endocrinol. 2023, 14, 1196935. [Google Scholar] [CrossRef]
- Enthoven, L.F.; Shi, Y.; Fay, E.; Kim, A.; Moreni, S.; Mao, J.; Isoherranen, N.; Totah, R.A.; Hebert, M.F. Effects of Pregnancy on Plasma Sphingolipids Using a Metabolomic and Quantitative Analysis Approach. Metabolites 2023, 13, 1026. [Google Scholar] [CrossRef]
- Sharma, A.; Kandlakunta, H.; Nagpal, S.J.S.; Feng, Z.; Hoos, W.; Petersen, G.M.; Chari, S.T. Model to Determine Risk of Pancreatic Cancer in Patients With New-Onset Diabetes. Gastroenterology 2018, 155, 730–739.e3. [Google Scholar] [CrossRef]
- Crowley, G.; Kim, J.; Kwon, S.; Lam, R.; Prezant, D.J.; Liu, M.; Nolan, A. PEDF, a pleiotropic WTC-LI biomarker: Machine learning biomarker identification and validation. PLoS Comput. Biol. 2021, 17, e1009144. [Google Scholar] [CrossRef]
- Zhu, Y.; Barupal, D.K.; Ngo, A.L.; Quesenberry, C.P.; Feng, J.; Fiehn, O.; Ferrara, A. Predictive Metabolomic Markers in Early to Mid-pregnancy for Gestational Diabetes Mellitus: A Prospective Test and Validation Study. Diabetes 2022, 71, 1807–1817. [Google Scholar] [CrossRef]
- Taylor, J.M.; Ankerst, D.P.; Andridge, R.R. Validation of biomarker-based risk prediction models. Clin. Cancer Res. 2008, 14, 5977–5983. [Google Scholar] [CrossRef]
- Li, D.K.; Smith, L.E.; Rookyard, A.W.; Lingam, S.J.; Koay, Y.C.; McEwen, H.P.; Twigg, S.M.; Don, A.S.; O’Sullivan, J.F.; Cordwell, S.J.; et al. Multi-omics of a pre-clinical model of diabetic cardiomyopathy reveals increased fatty acid supply impacts mitochondrial metabolic selectivity. J. Mol. Cell. Cardiol. 2022, 164, 92–109. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Hu, G.; Lu, Y.; Li, M.; Shen, B.; Kong, W.; Guan, Y.; Yang, X.; Fang, J.; Liu, J.; et al. Discrimination of Traditional Chinese Medicine Syndromes in Type 2 Diabetic Patients Based on Metabolomics-Proteomics Profiles. Int. J. Anal. Chem. 2023, 2023, 5722131. [Google Scholar] [CrossRef] [PubMed]
- Hilse, M.S.; Kretzschmar, T.; Pistulli, R.; Franz, M.; Bekfani, T.; Haase, D.; Neugebauer, S.; Kiehntopf, M.; Gummert, J.F.; Milting, H.; et al. Analysis of Metabolic Markers in Patients with Chronic Heart Failure before and after LVAD Implantation. Metabolites 2021, 11, 615. [Google Scholar] [CrossRef] [PubMed]
- Truby, L.K.; Regan, J.A.; Giamberardino, S.N.; Ilkayeva, O.; Bain, J.; Newgard, C.B.; O’Connor, C.M.; Felker, G.M.; Kraus, W.E.; McGarrah, R.W.; et al. Circulating long chain acylcarnitines and outcomes in diabetic heart failure: An HF-ACTION clinical trial substudy. Cardiovasc. Diabetol. 2021, 20, 161. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Z.; West, J.A.; Murray, A.J.; Griffin, J.L. Comprehensive Metabolic Profiling of Age-Related Mitochondrial Dysfunction in the High-Fat-Fed/Mouse Heart. J. Proteome Res. 2015, 14, 2849–2862. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wang, Y.; Li, Y.; Zhang, L.; Dong, M. A novel hippocampus metabolite signature in diabetes mellitus rat model of diabetic encephalopathy. Metab. Brain Dis. 2020, 35, 895–904. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.F.; Nissen, J.D.; Garcia-Serrano, A.M.; Nussbaum, S.S.; Waagepetersen, H.S.; Duarte, J.M.N. Glycogen metabolism is impaired in the brain of male type 2 diabetic Goto-Kakizaki rats. J. Neurosci. Res. 2019, 97, 1004–1017. [Google Scholar] [CrossRef] [PubMed]
- Han, L.D.; Xia, J.F.; Liang, Q.L.; Wang, Y.; Wang, Y.M.; Hu, P.; Li, P.; Luo, G.A. Plasma esterified and non-esterified fatty acids metabolic profiling using gas chromatography-mass spectrometry and its application in the study of diabetic mellitus and diabetic nephropathy. Anal. Chim. Acta 2011, 689, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zuo, J.J.; Dong, S.S.; Lan, Y.; Wu, C.W.; Mao, G.Y.; Zheng, C. Identification of Potential Serum Metabolic Biomarkers of Diabetic Kidney Disease: A Widely Targeted Metabolomics Study. J. Diabetes Res. 2020, 2020, 3049098. [Google Scholar] [CrossRef]
- Afshinnia, F.; Reynolds, E.L.; Rajendiran, T.M.; Soni, T.; Byun, J.; Savelieff, M.G.; Looker, H.C.; Nelson, R.G.; Michailidis, G.; Callaghan, B.C.; et al. Serum lipidomic determinants of human diabetic neuropathy in type 2 diabetes. Ann. Clin. Transl. Neurol. 2022, 9, 1392–1404. [Google Scholar] [CrossRef]
- Zhang, Q.; Song, W.; Liang, X.C.; Xie, J.; Shi, Y.; Shi, X.H.; Qiu, B.T.; Chen, X.T. A Metabolic Insight Into the Neuroprotective Effect of Jin-Mai-Tong (JMT) Decoction on Diabetic Rats With Peripheral Neuropathy Using Untargeted Metabolomics Strategy. Front. Pharmacol. 2020, 11, 221. [Google Scholar] [CrossRef]
- Li, Y.F.; Yao, W.J.; Gao, Y.B. Effects of Tang Luo Ning on diabetic peripheral neuropathy in rats revealed by LC-MS metabolomics approach. Biomed. Chromatogr. 2022, 36, e5374. [Google Scholar] [CrossRef] [PubMed]
- Hung, S.Y.; Tsai, J.R.S.O.; Yeh, J.T.; Chen, K.H.; Lin, C.N.; Yang, H.M.; Lin, C.W.; Chen, H.Y.; Huang, C.H.E.; Yu, Y.H. Amino acids and wound healing in people with limb-threatening diabetic foot ulcers. J. Diabetes Its Complicat. 2019, 33, 107403. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Ng, T.K.; Liu, Q.P.; Wu, Z.G.; Zhang, G.H.; Zhang, M.Z. Azelaic acid and guanosine in tears improve discrimination of proliferative from non-proliferative diabetic retinopathy in type-2 diabetes patients: A tear metabolomics study. Heliyon 2023, 9, e16109. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Cheng, C.Y.; Choi, H.; Ikram, M.K.; Sabanayagam, C.; Tan, G.S.; Tian, D.; Zhang, L.; Venkatesan, G.; Tai, E.S.; et al. Plasma Metabonomic Profiling of Diabetic Retinopathy. Diabetes 2016, 65, 1099–1108. [Google Scholar] [CrossRef] [PubMed]
- Xuan, Q.H.; Ouyang, Y.; Wang, Y.F.; Wu, L.; Li, H.T.; Luo, Y.Y.; Zhao, X.J.; Feng, D.S.; Qin, W.S.; Hu, C.X.; et al. Multiplatform Metabolomics Reveals Novel Serum Metabolite Biomarkers in Diabetic Retinopathy Subjects. Adv. Sci. 2020, 7, 2001714. [Google Scholar] [CrossRef] [PubMed]
- Pappachan, J.M.; Varughese, G.I.; Sriraman, R.; Arunagirinathan, G. Diabetic cardiomyopathy: Pathophysiology, diagnostic evaluation and management. World J. Diabetes 2013, 4, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Dillmann, W.H. Diabetic Cardiomyopathy. Circ. Res. 2019, 124, 1160–1162. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Han, Q.; Gang, X.K.; Wang, G.X. Altered brain metabolites in patients with diabetes mellitus and related complications—Evidence from H MRS study. Biosci. Rep. 2018, 38, BSR20180660. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Han, Q.; Lv, Y.; Sun, L.; Gang, X.; Wang, G. Biomarkers for cognitive decline in patients with diabetes mellitus: Evidence from clinical studies. Oncotarget 2018, 9, 7710–7726. [Google Scholar] [CrossRef]
- Chen, R.; Shi, J.; Yin, Q.; Li, X.; Sheng, Y.; Han, J.; Zhuang, P.; Zhang, Y. Morphological and Pathological Characteristics of Brain in Diabetic Encephalopathy. J. Alzheimer’s Dis. 2018, 65, 15–28. [Google Scholar] [CrossRef]
- Volmer-Thole, M.; Lobmann, R. Neuropathy and Diabetic Foot Syndrome. Int. J. Mol. Sci. 2016, 17, 917. [Google Scholar] [CrossRef] [PubMed]
- Feldman, E.L.; Callaghan, B.C.; Pop-Busui, R.; Zochodne, D.W.; Wright, D.E.; Bennett, D.L.; Bril, V.; Russell, J.W.; Viswanathan, V. Diabetic neuropathy. Nat. Rev. Dis. Primers 2019, 5, 42. [Google Scholar] [CrossRef]
- Rumora, A.E.; Alakwaa, F.; Andersen, S.T.; Jorgensen, M.E.; Charles, M.; Callaghan, B.C.; Jensen, T.S.; Feldman, E.L. Metabolomics Identifies Novel Plasma Metabolomic Signatures Associated with Diabetic Neuropathy in a Cohort with Screen-Tested Type 2 Diabetes: ADDITION-Denmark. Diabetes 2020, 69. [Google Scholar] [CrossRef]
- Walsh, J.W.; Hoffstad, O.J.; Sullivan, M.O.; Margolis, D.J. Association of diabetic foot ulcer and death in a population-based cohort from the United Kingdom. Diabet. Med. 2016, 33, 1493–1498. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Shao, T.T.; Wang, J.L.; Huang, X.T.; Deng, X.F.; Cao, Y.M.; Zhou, M.M.; Zhao, C. An update on potential biomarkers for diagnosing diabetic foot ulcer at early stage. Biomed. Pharmacother. 2021, 133, 110991. [Google Scholar] [CrossRef]
- Cheung, N.; Wong, T.Y. Diabetic retinopathy and systemic vascular complications. Prog. Retin. Eye Res. 2008, 27, 161–176. [Google Scholar] [CrossRef]
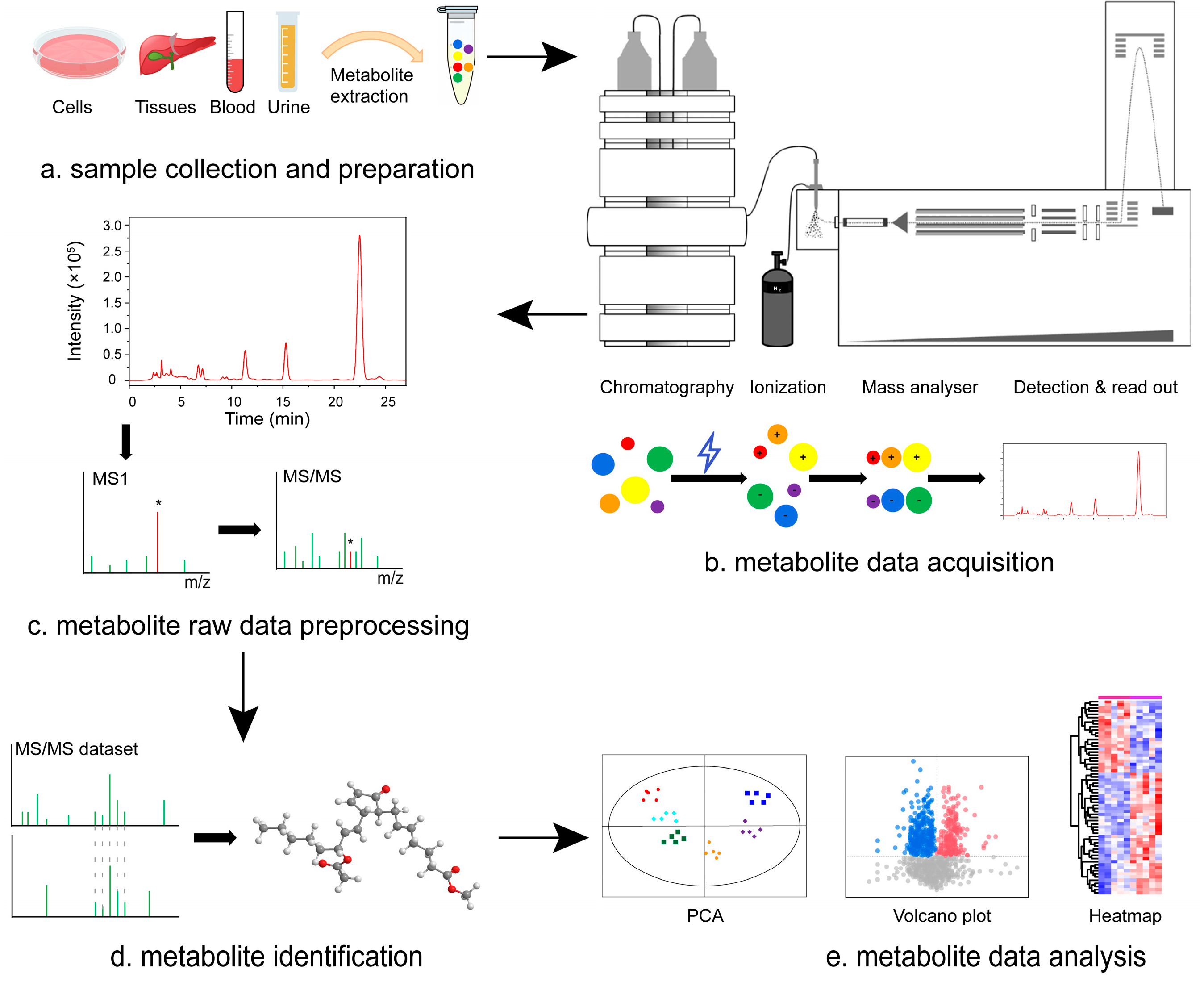
| Ionization Source Type | Characteristics | Applicable Compounds | Instrument Type |
|---|---|---|---|
| Electron Ionization (EI) | Hard ionization, generates molecular ions | Suitable for thermally stable, volatile substances | Gas chromatography mass spectrometer (GC-MS) |
| Chemical Ionization (CI) | Soft ionization | Suitable for volatile, thermally stable substances | Gas chromatography mass spectrometer (GC-MS) |
| Electrospray Ionization (ESI) | Soft ionization source at atmospheric pressure | Suitable for less volatile, thermally unstable compounds | Liquid chromatography mass spectrometer (LC-MS) or capillary electrophoresis mass spectrometer (CE-MS) |
| Atmospheric Pressure Chemical Ionization (APCI) | Soft ionization; ionization of oxygen or nitrogen with a corona needle | Suitable for volatile, thermally stable substances | Liquid chromatography mass spectrometer (LC-MS) |
| Matrix-Assisted Laser Desorption Ionization (MALDI) | Soft ionization; the matrix cocrystallizes with the compound, and the compound ion is produced by laser hitting the matrix | Suitable for large molecules | Mass spectrometer (MS) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, F.; Shan, S.; Fu, C.; Guo, S.; Liu, C.; Wang, S. Advanced Mass Spectrometry-Based Biomarker Identification for Metabolomics of Diabetes Mellitus and Its Complications. Molecules 2024, 29, 2530. https://doi.org/10.3390/molecules29112530
Zhang F, Shan S, Fu C, Guo S, Liu C, Wang S. Advanced Mass Spectrometry-Based Biomarker Identification for Metabolomics of Diabetes Mellitus and Its Complications. Molecules. 2024; 29(11):2530. https://doi.org/10.3390/molecules29112530
Chicago/Turabian StyleZhang, Feixue, Shan Shan, Chenlu Fu, Shuang Guo, Chao Liu, and Shuanglong Wang. 2024. "Advanced Mass Spectrometry-Based Biomarker Identification for Metabolomics of Diabetes Mellitus and Its Complications" Molecules 29, no. 11: 2530. https://doi.org/10.3390/molecules29112530
APA StyleZhang, F., Shan, S., Fu, C., Guo, S., Liu, C., & Wang, S. (2024). Advanced Mass Spectrometry-Based Biomarker Identification for Metabolomics of Diabetes Mellitus and Its Complications. Molecules, 29(11), 2530. https://doi.org/10.3390/molecules29112530






