Enhancement of Biodegradability of Chicken Manure via the Addition of Zeolite in a Two-Stage Dry Anaerobic Digestion Configuration
Abstract
:1. Introduction
2. Results
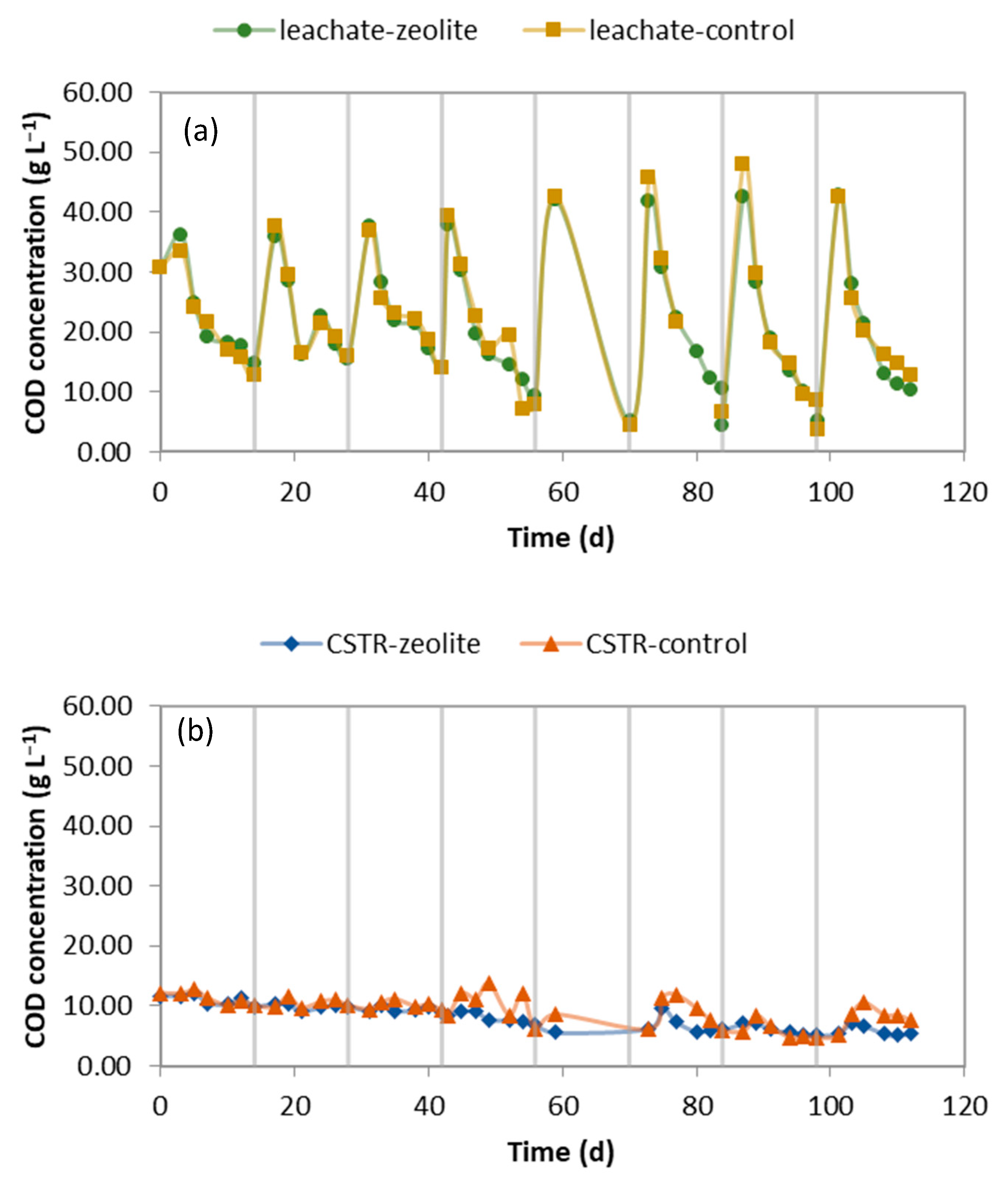
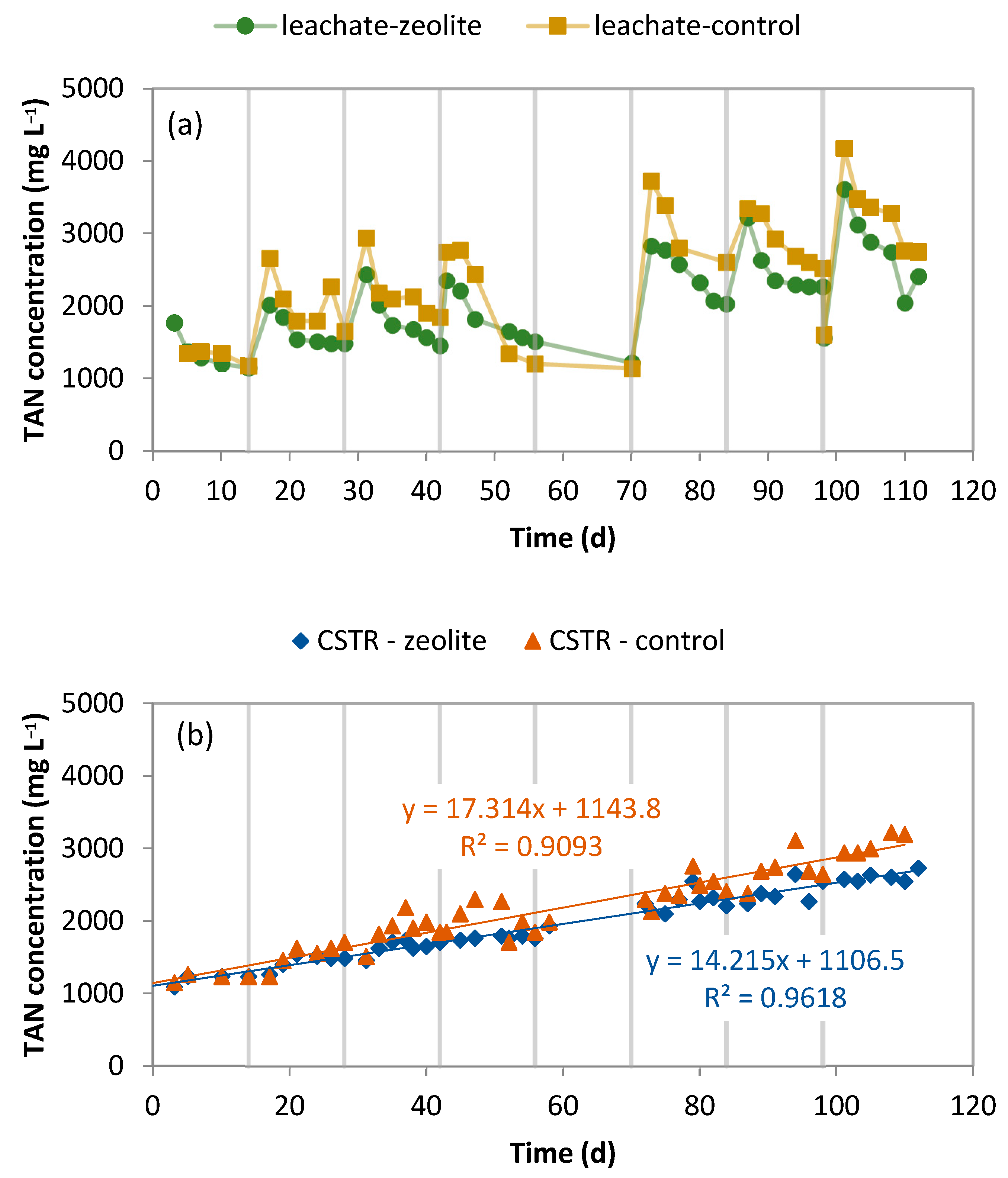
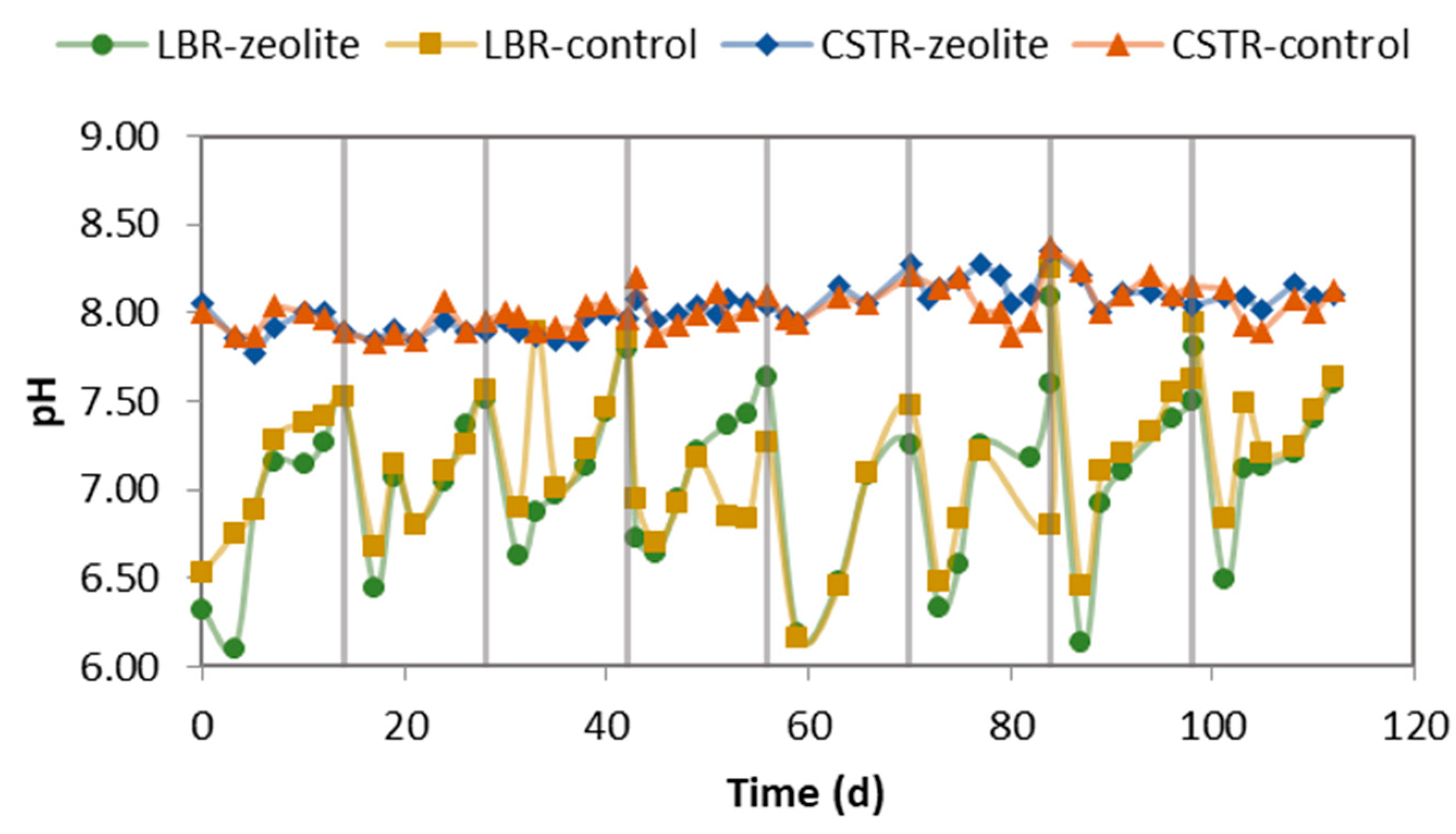
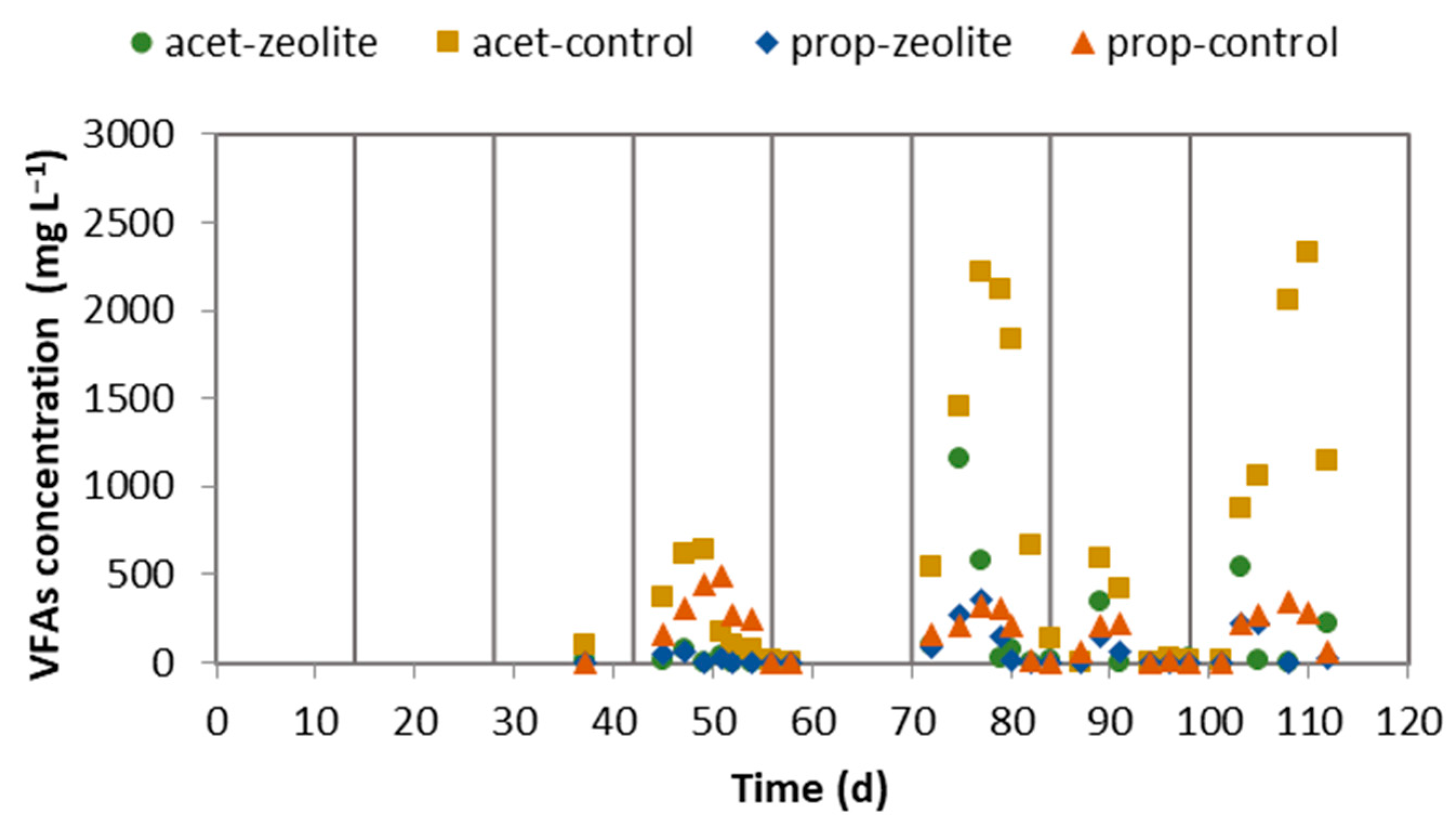
2.1. Performance of the LBR-CSTR Systems
2.2. Microbial Composition and Dynamics
3. Discussion
4. Materials and Methods
4.1. CM and Zeolite Origin and Characteristics
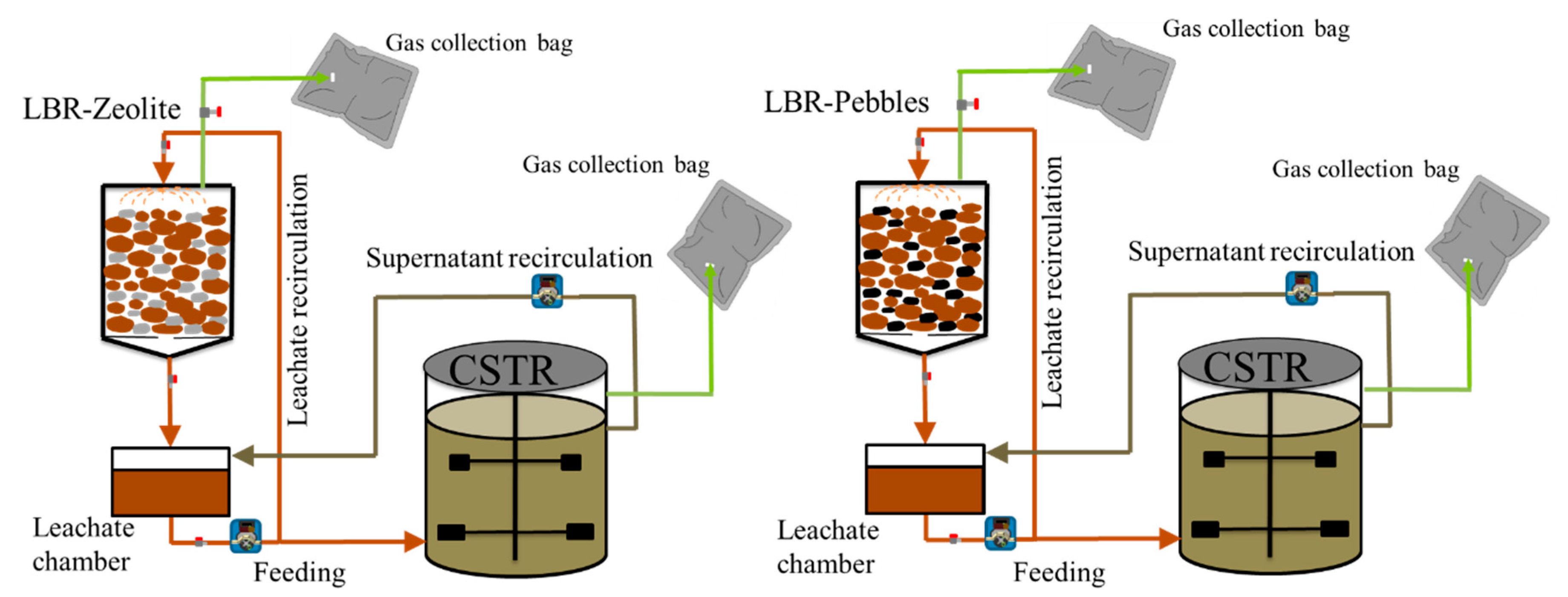
4.2. Experimental Set-Up and Operation
4.3. Analytical Methods
4.4. Microbial Analysis
4.4.1. DNA Extraction and 16S rDNA Amplification
4.4.2. Library Preparation and Illumina MiSeq Sequencing
4.4.3. Bioinformatics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- OECD/FAO. OECD-FAO Agricultural Outlook 2022–2031; OECD Publishing: Paris, France, 2022. [Google Scholar]
- AVEC Annual Report. 2023. Available online: https://avec-poultry.eu/wp-content/uploads/2023/09/AVEC_AnnualReport_2023.pdf (accessed on 8 January 2024).
- Gržinić, G.; Piotrowicz-Cieślak, A.; Klimkowicz-Pawlas, A.; Górny, R.L.; Ławniczek-Wałczyk, A.; Piechowicz, L.; Olkowska, E.; Potrykus, M.; Tankiewicz, M.; Krupka, M.; et al. Intensive Poultry Farming: A Review of the Impact on the Environment and Human Health. Sci. Total Environ. 2023, 858, 160014. [Google Scholar] [CrossRef] [PubMed]
- Tańczuk, M.; Junga, R.; Kolasa-Więcek, A.; Niemiec, P. Assessment of the Energy Potential of Chicken Manure in Poland. Energies 2019, 12, 1244. [Google Scholar] [CrossRef]
- Kreidenweis, U.; Breier, J.; Herrmann, C.; Libra, J.; Prochnow, A. Greenhouse Gas Emissions from Broiler Manure Treatment Options Are Lowest in Well-Managed Biogas Production. J. Clean. Prod. 2021, 280, 124969. [Google Scholar] [CrossRef]
- Manogaran, M.D.; Shamsuddin, R.; Mohd Yusoff, M.H.; Lay, M.; Siyal, A.A. A Review on Treatment Processes of Chicken Manure. Clean. Circ. Bioecon. 2022, 2, 100013. [Google Scholar] [CrossRef]
- Kalogiannis, A.; Vasiliadou, I.A.; Spyridonidis, A.; Diamantis, V.; Stamatelatou, K. Biogas Production from Chicken Manure Wastes Using an LBR-CSTR Two-Stage System: Process Efficiency, Economic Feasibility, and Carbon Dioxide Footprint. J. Chem. Technol. Biotechnol. 2022, 97, 2952–2961. [Google Scholar] [CrossRef]
- Rajagopal, R.; Massé, D.I.; Singh, G. A Critical Review on Inhibition of Anaerobic Digestion Process by Excess Ammonia. Bioresour. Technol. 2013, 143, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, W.; He, Y.; Zhang, R.; Liu, G. Effect of Ammonia on Methane Production, Methanogenesis Pathway, Microbial Community and Reactor Performance under Mesophilic and Thermophilic Conditions. Renew Energy 2018, 125, 915–925. [Google Scholar] [CrossRef]
- Bayrakdar, A.; Sürmeli, R.Ö.; Çalli, B. Anaerobic Digestion of Chicken Manure by a Leach-Bed Process Coupled with Side-Stream Membrane Ammonia Separation. Bioresour. Technol. 2018, 258, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Palakodeti, A.; Rupani, P.F.; Azman, S.; Dewil, R.; Appels, L. Novel Approach to Ammonia Recovery from Anaerobic Digestion via Side-Stream Stripping at Multiple PH Levels. Bioresour. Technol. 2022, 361, 127685. [Google Scholar] [CrossRef]
- Molaey, R.; Bayrakdar, A.; Çalli, B. Long-Term Influence of Trace Element Deficiency on Anaerobic Mono-Digestion of Chicken Manure. J. Environ. Manag. 2018, 223, 743–748. [Google Scholar] [CrossRef]
- Molaey, R.; Bayrakdar, A.; Sürmeli, R.Ö.; Çalli, B. Influence of Trace Element Supplementation on Anaerobic Digestion of Chicken Manure: Linking Process Stability to Methanogenic Population Dynamics. J. Clean. Prod. 2018, 181, 794–800. [Google Scholar] [CrossRef]
- Cai, Y.; Zhu, M.; Meng, X.; Zhou, J.L.; Zhang, H.; Shen, X. The Role of Biochar on Alleviating Ammonia Toxicity in Anaerobic Digestion of Nitrogen-Rich Wastes: A Review. Bioresour. Technol. 2022, 351, 126924. [Google Scholar] [CrossRef] [PubMed]
- Fotidis, I.A.; Kougias, P.G.; Zaganas, I.D.; Kotsopoulos, T.A.; Martzopoulos, G.G. Inoculum and Zeolite Synergistic Effect on Anaerobic Digestion of Poultry Manure. Environ. Technol. 2014, 35, 1219–1225. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Wan, C.; Liu, X.; Lei, Z.; Lee, D.J.; Zhang, Y.; Tay, J.H.; Zhang, Z. Anaerobic Digestion of Swine Manure under Natural Zeolite Addition: VFA Evolution, Cation Variation, and Related Microbial Diversity. Appl. Microbiol. Biotechnol. 2013, 97, 10575–10583. [Google Scholar] [CrossRef] [PubMed]
- Tzenos, C.A.; Kalamaras, S.D.; Economou, E.A.; Romanos, G.E.; Veziri, C.M.; Mitsopoulos, A.; Menexes, G.C.; Sfetsas, T.; Kotsopoulos, T.A. The Multifunctional Effect of Porous Additives on the Alleviation of Ammonia and Sulfate Co-Inhibition in Anaerobic Digestion. Sustainability 2023, 15, 9994. [Google Scholar] [CrossRef]
- Palominos, N.; Castillo, A.; Guerrero, L.; Borja, R.; Huiliñir, C. Coupling of Anaerobic Digestion and Struvite Precipitation in the Same Reactor: Effect of Zeolite and Bischofite as Mg2+ Source. Front. Environ. Sci. 2021, 9, 706730. [Google Scholar] [CrossRef]
- Montalvo, S.; Huiliñir, C.; Borja, R.; Sánchez, E.; Herrmann, C. Application of Zeolites for Biological Treatment Processes of Solid Wastes and Wastewaters—A Review. Bioresour. Technol. 2020, 301, 122808. [Google Scholar] [CrossRef]
- Nizami, A.S.; Murphy, J.D. Optimizing the Operation of a Two-Phase Anaerobic Digestion System Digesting Grass Silage. Environ. Sci. Technol. 2011, 45, 7561–7569. [Google Scholar] [CrossRef]
- Collins, B.A.; Birzer, C.H.; Harris, P.W.; Kidd, S.P.; McCabe, B.K.; Medwell, P.R. Two-Phase Anaerobic Digestion in Leach Bed Reactors Coupled to Anaerobic Filters: A Review and the Potential of Biochar Filters. Renew. Sustain. Energy Rev. 2023, 175, 113187. [Google Scholar] [CrossRef]
- Spyridonidis, A.; Vasiliadou, I.A.; Stamatelatou, K. Effect of Zeolite on the Methane Production from Chicken Manure Leachate. Sustainability 2022, 14, 2207. [Google Scholar] [CrossRef]
- Pelletier, E.; Kreimeyer, A.; Bocs, S.; Rouy, Z.; Gyapay, G.; Chouari, R.; Rivière, D.; Ganesan, A.; Daegelen, P.; Sghir, A.; et al. “Candidatus Cloacamonas Acidaminovorans”: Genome Sequence Reconstruction Provides a First Glimpse of a New Bacterial Division. J. Bacteriol. 2008, 190, 2572–2579. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Lang, Q.; Pan, Z.; Jiang, Y.; Liebetrau, J.; Nelles, M.; Dong, H.; Dong, R. Performance Evaluation of a Novel Anaerobic Digestion Operation Process for Treating High-Solids Content Chicken Manure: Effect of Reduction of the Hydraulic Retention Time at a Constant Organic Loading Rate. Waste Manag. 2017, 64, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Alba Reyes, Y.; Barrera, E.L.; Cheng, K.K. A Review on the Prospective Use of Chicken Manure Leachate in High-Rate Anaerobic Reactors. J. Environ. Chem. Eng. 2021, 9, 104695. [Google Scholar] [CrossRef]
- Yang, J.; Wang, D.; Luo, Z.; Zeng, W. Anaerobic Mono-Digestion of Pig Manure in a Leach Bed Coupled with a Methanogenic Reactor: Effects of the Filter Media. J. Clean. Prod. 2019, 234, 1094–1101. [Google Scholar] [CrossRef]
- Spyridonidis, A.; Vasiliadou, I.A.; Akratos, C.S.; Stamatelatou, K. Performance of a Full-Scale Biogas Plant Operation in Greece and Its Impact on the Circular Economy. Water 2020, 12, 3074. [Google Scholar] [CrossRef]
- Blackburn, S.D.; Wherry, E.J. IL-10, T Cell Exhaustion and Viral Persistence. Trends Microbiol. 2007, 15, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Li, X.; Yekta, S.S.; Björn, A.; Mu, B.-Z.; Masuda, L.S.M.; Schnürer, A.; Enrich-Prast, A. Absence of Oxygen Effect on Microbial Structure and Methane Production during Drying and Rewetting Events. Sci. Rep. 2022, 12, 16570. [Google Scholar] [CrossRef] [PubMed]
- Rotaru, A.E.; Shrestha, P.M.; Liu, F.; Shrestha, M.; Shrestha, D.; Embree, M.; Zengler, K.; Wardman, C.; Nevin, K.P.; Lovley, D.R. A New Model for Electron Flow during Anaerobic Digestion: Direct Interspecies Electron Transfer to Methanosaeta for the Reduction of Carbon Dioxide to Methane. Energy Environ. Sci. 2014, 7, 408–415. [Google Scholar] [CrossRef]
- Wasserfallen, A.; Nolling, J.; Pfister, P.; Reeve, J.; Conway de Macario, E. Phylogenetic analysis of 18 thermophilic Methanobacterium isolates supports the proposals to create a new genus, Methanothermobacter gen. nov., and to reclassify several isolates in three species, Methanothermobacter thermautotrophicus comb. nov., Methanothermobacter wolfeii comb. nov., and Methanothermobacter marburgensis sp. nov. Int. J. Syst. Evol. Microbiol. 2000, 50, 43–53. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sowers, K.R. Methanogenesis. In Encyclopedia of Microbiology; Elsevier: Amsterdam, The Netherlands, 2009; pp. 265–286. [Google Scholar] [CrossRef]
- Yi, X.; Brandt, K.K.; Xue, S.; Peng, J.; Wang, Y.; Li, M.; Deng, Y.; Duan, G. Niche Differentiation and Biogeography of Bathyarchaeia in Paddy Soil Ecosystems: A Case Study in Eastern China. Environ. Microbiome 2024, 19, 13. [Google Scholar] [CrossRef]
- Duan, C.; Liu, Y.; Liu, Y.; Liu, L.; Cai, M.; Zhang, R.; Zeng, Q.; Koonin, E.V.; Krupovic, M.; Li, M. Diversity of Bathyarchaeia Viruses in Metagenomes and Virus-Encoded CRISPR System Components. ISME Commun. 2024, 4, ycad011. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Wang, Y.; Zhu, P.; Yang, N.; Liang, L.; Yu, T.; Niu, M.; Konhauser, K.; Woodcroft, B.J.; Wang, F. Taxonomic and Carbon Metabolic Diversification of Bathyarchaeia during Its Coevolution History with Early Earth Surface Environment. Sci. Adv. 2023, 9, eadf5069. [Google Scholar] [CrossRef] [PubMed]
- Weiland, P. Biogas Production: Current State and Perspectives. Appl. Microbiol. Biotechnol. 2010, 85, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Liu, L.; Wang, S.; Wang, Q.; Jiang, Y.; Hou, X.; Tan, T. Semi-Continuous Anaerobic Digestion for Biogas Production: Influence of Ammonium Acetate Supplement and Structure of the Microbial Community. Biotechnol. Biofuels 2015, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Detman, A.; Bucha, M.; Treu, L.; Chojnacka, A.; Pleśniak, Ł.; Salamon, A.; Łupikasza, E.; Gromadka, R.; Gawor, J.; Gromadka, A.; et al. Evaluation of Acidogenesis Products’ Effect on Biogas Production Performed with Metagenomics and Isotopic Approaches. Biotechnol. Biofuels 2021, 14, 125. [Google Scholar] [CrossRef] [PubMed]
- Bouanane-Darenfed, A.; Fardeau, M.L.; Olivier, B. The Family Caldicoprobacteraceae. In The Prokaryotes; Springer: Berlin/Heidelberg, Germany, 2014; pp. 13–14. [Google Scholar] [CrossRef]
- Dyksma, S.; Jansen, L.; Gallert, C. Syntrophic Acetate Oxidation Replaces Acetoclastic Methanogenesis during Thermophilic Digestion of Biowaste. Microbiome 2020, 8, 105. [Google Scholar] [CrossRef] [PubMed]
- Ziganshina, E.E.; Ziganshin, A.M. Anaerobic Digestion of Chicken Manure in the Presence of Magnetite, Granular Activated Carbon, and Biochar: Operation of Anaerobic Reactors and Microbial Community Structure. Microorganisms 2022, 10, 1422. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Yu, Z.; Zhao, J.; Zhang, H. Do Microbial Communities in an Anaerobic Bioreactor Change with Continuous Feeding Sludge into a Full-Scale Anaerobic Digestion System? Bioresour. Technol. 2018, 249, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.A.; Hug, L.A. Cloacimonadota metabolisms include adaptations in engineered environments that are reflected in the evolutionary history of the phylum. Environ. Microbiol. Rep. 2022, 14, 520–529. [Google Scholar] [CrossRef]
- Williams, T.J.; Allen, M.A.; Berengut, J.F.; Cavicchioli, R. Shedding Light on Microbial “Dark Matter”: Insights Into Novel Cloacimonadota and Omnitrophota from an Antarctic Lake. Front. Microbiol. 2021, 12, 741077. [Google Scholar] [CrossRef]
- Saha, S.; Jeon, B.H.; Kurade, M.B.; Chatterjee, P.K.; Chang, S.W.; Markkandan, K.; Salama, E.S.; Govindwar, S.P.; Roh, H.S. Microbial Acclimatization to Lipidic-Waste Facilitates the Efficacy of Acidogenic Fermentation. Chem. Eng. J. 2019, 358, 188–196. [Google Scholar] [CrossRef]
- Li, C.; He, P.; Hao, L.; Lü, F.; Shao, L.; Zhang, H. Diverse Acetate-Oxidizing Syntrophs Contributing to Biogas Production from Food Waste in Full-Scale Anaerobic Digesters in China. Renew Energy 2022, 193, 240–250. [Google Scholar] [CrossRef]
- Cardona, L.; Mazéas, L.; Chapleur, O. Zeolite Favours Propionate Syntrophic Degradation during Anaerobic Digestion of Food Waste under Low Ammonia Stress. Chemosphere 2021, 262, 127932. [Google Scholar] [CrossRef] [PubMed]
- Owen, W.F.; Stuckey, D.C.; Healy, J.B., Jr.; Young, L.Y.; McCarty, P.L. Bioassay for Monitoring Biochemical Methane Potential and Anaerobic Toxicity. Water Res. 1979, 13, 485–492. [Google Scholar] [CrossRef]
- Angelidaki, I.; Petersen, S.P.; Ahring, B.K. Effects of Lipids on Thermophilic Anaerobic Digestion and Reduction of Lipid Inhibition upon Addition of Bentonite. Appl. Microbiol. Biotechnol. 1990, 33, 469–472. [Google Scholar] [CrossRef] [PubMed]
- APHA. Standard Methods for the Examination of Water and Waste Water, 20th ed.; Standard Methods for the Examination of Water and Wastewater; American Public Health Association: Washington, DC, USA, 1999. [Google Scholar]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Thijs, S.; Op De Beeck, M.; Beckers, B.; Truyens, S.; Stevens, V.; Van Hamme, J.D.; Weyens, N.; Vangronsveld, J. Comparative Evaluation of Four Bacteria-Specific Primer Pairs for 16S RRNA Gene Surveys. Front. Microbiol. 2017, 8, 494. [Google Scholar] [CrossRef] [PubMed]
- Pinto, A.J.; Raskin, L. PCR Biases Distort Bacterial and Archaeal Community Structure in Pyrosequencing Datasets. PLoS ONE 2012, 7, e43093. [Google Scholar] [CrossRef] [PubMed]
- Yaman, C.; Rehman, S.; Ahmad, T.; Kucukaga, Y.; Pala, B.; AlRushaid, N.; Riyaz Ul Hassan, S.; Yaman, A.B. Community Structure of Bacteria and Archaea Associated with Geotextile Filters in Anaerobic Bioreactor Landfills. Processes 2021, 9, 1377. [Google Scholar] [CrossRef]
- Li, L.; Zhang, W.; Zhang, S.; Song, L.; Sun, Q.; Zhang, H.; Xiang, H.; Dong, X. Bacteria and Archaea Synergistically Convert Glycine Betaine to Biogenic Methane in the Formosa Cold Seep of the South China Sea. mSystems 2021, 6, e0070321. [Google Scholar] [CrossRef]
- Ntougia, S.; Polkowska, Ż.; Nikolaki, S.; Dionyssopoulou, E.; Stathopoulou, P.; Doudoumis, V.; Ruman, M.; Kozak, K.; Namieśnik, J.; Tsiamis, G. Bacterial Community Structures in Freshwater Polar Environments of Svalbard. Microbes Environ. 2016, 31, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and Clustering Orders of Magnitude Faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly Accurate OTU Sequences from Microbial Amplicon Reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UNCROSS2: Identification of Cross-Talk in 16S RRNA OTU Tables. bioRxiv 2018. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2.2-0. 2014. Available online: https://CRAN.Rproject.org/package=vegan (accessed on 1 April 2024).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009; Volume VIII, p. 213. [Google Scholar] [CrossRef]
- Basualdo, C. Choosing the Best Non-Parametric Richness Estimator for Benthic Macroinvertebrates Databases. Rev. Soc. Entomol. Argent. 2011, 70, 27–38. [Google Scholar]
- Anderson, M.J.; Willis, T.J. Canonical Analysis of Principal Coordinates: A Useful Method of Constrained Ordination for Ecology. Ecology 2003, 84, 511–525. [Google Scholar] [CrossRef]
- Anderson, M.J. A New Method for Non-Parametric Multivariate Analysis of Variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Anderson, M.J.; Gorley, R.N.; Clarke, K.R. PERMANOVA+ for PRIMER: Guide to Software and Statistical Methods; Primer-E Limited: Devon, UK, 2008; p. 841889821. [Google Scholar]
- Clarke, K.R.; Gorley, R.N. PRIMER v6: User Manual/Tutorial; PRIMER-E Limited: Devon, UK, 2006; Volume 29, pp. 1060–1065. [Google Scholar]





| Cycle No | Methane Yield (NL CH4 gVSin−1) | |
|---|---|---|
| Zeolite | Control | |
| 4 | 0.178 | 0.113 |
| 5 | 0.150 | 0.146 |
| 6 | 0.204 | 0.174 |
| 7 | 0.159 | 0.157 |
| 8 | 0.165 | 0.120 |
| CM | Parameter | |||
|---|---|---|---|---|
| TS (g kg−1FM) | VS (g kg−1FM) | COD (g kg−1VS) | TKN (g kg−1VS) | |
| 1st batch | 828 ± 0.8 | 677 ± 2.76 | 1250 ± 78 | 59 ± 4.4 |
| 2nd batch | 619 ± 9.2 | 502 ± 9.3 | 1307 ± 70 | 63 ± 2.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalogiannis, A.; Vasiliadou, I.A.; Tsiamis, A.; Galiatsatos, I.; Stathopoulou, P.; Tsiamis, G.; Stamatelatou, K. Enhancement of Biodegradability of Chicken Manure via the Addition of Zeolite in a Two-Stage Dry Anaerobic Digestion Configuration. Molecules 2024, 29, 2568. https://doi.org/10.3390/molecules29112568
Kalogiannis A, Vasiliadou IA, Tsiamis A, Galiatsatos I, Stathopoulou P, Tsiamis G, Stamatelatou K. Enhancement of Biodegradability of Chicken Manure via the Addition of Zeolite in a Two-Stage Dry Anaerobic Digestion Configuration. Molecules. 2024; 29(11):2568. https://doi.org/10.3390/molecules29112568
Chicago/Turabian StyleKalogiannis, Achilleas, Ioanna A. Vasiliadou, Athanasios Tsiamis, Ioannis Galiatsatos, Panagiota Stathopoulou, George Tsiamis, and Katerina Stamatelatou. 2024. "Enhancement of Biodegradability of Chicken Manure via the Addition of Zeolite in a Two-Stage Dry Anaerobic Digestion Configuration" Molecules 29, no. 11: 2568. https://doi.org/10.3390/molecules29112568
APA StyleKalogiannis, A., Vasiliadou, I. A., Tsiamis, A., Galiatsatos, I., Stathopoulou, P., Tsiamis, G., & Stamatelatou, K. (2024). Enhancement of Biodegradability of Chicken Manure via the Addition of Zeolite in a Two-Stage Dry Anaerobic Digestion Configuration. Molecules, 29(11), 2568. https://doi.org/10.3390/molecules29112568











